Abstract
In oral cavity chronic inflammation has been observed at various stages of oral squamous cell carcinomas (OSCC). This inflammation could result from persistent mucosal or epithelial cell colonization by microorganisms. There is an increasing evidence of the involvement of oral bacteria in inflammation and warrant further studies on the association of bacteria in the progression of OSCC. The objective of this study was to evaluate the diversity and relative abundance of bacteria in the saliva of subjects with OSCC. Using 454 parallel DNA sequencing, ~58,000 PCR amplicons that span the V4-V5 hypervariable region of ribosomal RNAs from 5 subjects were sequenced. Members of 8 phyla (divisions) of bacteria were detected. The majority of classified sequences belonged to phyla, Firmicutes (45%) and Bacteroidetes (25%). Further, a total of 52 different genera containing approximately 860 (16.51%) known species were identified, 1077 (67%) sequences belonged to various uncultured bacteria or unclassified group. The species diversity estimates obtained with abundance-based coverage estimators (ACE) and Chao1 were greater than published analyses of other microbial profiles from the oral cavity. Fifteen unique phylotypes were present in all three OSCC subjects.
Keywords: oral squamous cell carcinoma, microbial diversity, denaturing gradient gel electrophoresis, 454 pyrosequencing
Introduction
Microbes induce an estimated 20% of all the fatal cancers in human beings (Blaser, 2008) and numerous bacterial species are associated with different cancers (Lax & Thomas, 2002, Vogelmann & Amieva, 2007, Kurago, et al., 2008). The best documented relationship between a bacterial infection and cancer is that of Helicobacter pylori and two different forms of gastric cancer: MALT lymphoma and the more common gastric adenocarcinoma (Marshall & Windsor, 2005). It is estimated that H. pylori is causally related to 60 to 90% of all gastric cancers (Malfertheiner, et al., 2005). Other known associations between bacterial infections and human cancer include Salmonella typhi infection and gall bladder cancer in people that develop chronic carriage after typhoid fever (Shukla, et al., 2000); Streptococcus bovis and colon cancer (Ellmerich, et al., 2000); Chlamydia pneumonia and lung cancer (Littman, et al., 2005); Bartonella species and vascular tumor formation (Dehio, 2005); and infections linked to prostatitis and carcinoma (Wagenlehner, et al., 2007a, Wagenlehner, et al., 2007b).
Microorganisms and their products, including endotoxins (lipopolysaccharides), enzymes (e.g. proteases, collagenases, fibrinolysin, phospholipase), and metabolic byproducts (e.g. hydrogen sulfide, ammonia, fatty acids), are toxic to host cells and may directly induce mutations or alter signaling pathways that affect cell proliferation and/or survival of epithelial cells (Kuper, et al., 2000, Lax & Thomas, 2002, Lax, 2005). Moreover, microorganisms can also have indirect effects through inflammation by activating host cells, such as neutrophils, macrophages, monocytes, lymphocytes, fibroblasts, and epithelial cells, to generate reactive oxygen species (e.g. hydrogen peroxide, and oxygen radicals), reactive nitrogen species (nitric oxides), reactive lipids and metabolites (e.g. malondialdehyde, 4-hydroxy-2-nonenal), and matrix metalloproteases, which can induce damage of DNA in epithelial cells (Wogan, et al., 2004, Peek, et al., 2005, Chen, et al., 2006).
The hypothesis that certain bacteria are capable of causing cancer is further supported by studies of animal-specific pathogens that promote tumor formation. For example, Helicobacter hepaticus was discovered in 1992 as a cause of chronic active hepatitis that progressed to hepatocellular carcinoma in A/JCr mice (Ward, et al., 1994). Formation of colon cancer in genetically altered mice is promoted by infection with H. hepaticus, either by itself (Engle, et al., 2002, Erdman, et al., 2003) or in conjunction with Helicobacter bilis (Maggio-Price, et al., 2006). Chronic infection with Citrobacter rodentium, a mouse pathogen that is genetically similar to enteropathogenic Escherichia coli, can result in colon cancer (Newman, et al., 2001). H. hepaticus was shown to promote cancer formation in the mammary gland of mice (Rao, et al., 2006, Rao, et al., 2007).
Most oral/pharyngeal cancers (OPC) are oral squamous cell carcinomas (OSCC) (Scully & Bagan, 2009, Bagan, et al., 2010) and in the United States, the primary risk factors are tobacco, smoking, and elevated levels of alcohol consumption (Jemal, et al., 2008). Other potential risk factors include diet, the human papillomavirus (particularly HPV16), and various oral factors, including oral hygiene (Jemal, et al., 2008). High levels of colonization of OSCC by facultative oral streptococci were observed in the saliva of OSCC subjects (Sasaki, et al., 1998, Sakamoto, et al., 1999, Tateda, et al., 2000, Shiga, et al., 2001). More recently, viable bacteria have been isolated from both superficial and deep portions of OSCC (Hooper, et al., 2006, Hooper, et al., 2007), revealing that the tumor microenvironment is well suited for bacterial survival. The role of bacteria in the development of oral cancer has not been delineated, but the persistent presence of bacteria at tumor sites in the oral cavity raises intriguing questions about the role of bacteria in the progression of OSCC. Unfortunately, most of such studies to date have included only cultured oral bacterial species, using classical cloning and sequencing approaches (Nagy, et al., 1998, Mager, et al., 2005, Hong, et al., 2006). To establish the association of oral bacteria in the progression of OSCC, the complete bacterial profile (cultured and uncultured) in the oral cavity of OSCC subjects needs to be first determined. Here we report a preliminary study on the use of 454 parallel DNA sequencing to assess the bacterial diversity and their relative abundance in the saliva of OSCC subjects.
Materials and methods
Saliva samples and total bacterial DNA extraction
Stimulated saliva samples were collected from three cases of OSCC and two matched controls (non-malignant). The study protocol was approved by the Institutional Review Board of New York University for human subjects. All three OSCC cases had a floor of mouth cancer. Each case including control was a male over 50 years of age and had a history of smoking at least one pack of cigarettes per day and consuming more than 5 drinks per day of beer or hard liquor. The total bacterial genomic DNA from saliva samples was isolated using DNA purification kit (MasterPure; Epicenter, Madison, WI). In brief, 300 μL of tissue and cell lysis solution (Epicenter) was added to the suspended pellet obtained from 1 ml of saliva. The suspension was sonicated for 30 seconds. Ten μL of a proteinase K (QIAGEN) stock solution of 10 mg mL−1 in TES buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 100 mM NaCl), and 2 μL of mutanolysin (Sigma-Aldrich, St. Louis, MO)(5,000 U mL−1 in phosphate-buffered saline) and 10 μL of lysozyme stock solution (100 mg mL−1 in TES buffer) was added. The mixture was incubated at 37°C for 1 h, with gentle mixing at 15-minintervals. Proteinase K was inactivated via incubation at 65°Cfor 1 h. One μl of 5-μg μL−1 RNase A was added and the mixture incubated at 37°C for 30 min. The samples were then placed on ice for 3 to 5 min, 400μL of MPC protein precipitation reagent (Epicenter) was added, vortexed vigorously for 10 seconds, and subsequently extracted extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with isopropanol. The quality and quantity of DNA samples were measured at 260 nm and 280 nm (DU 640 spectrophotometer; Beckman, Hayward, CA). The final concentration of each DNA sample was adjusted to 20 ng μL−1 for all PCR applications.
Denaturing gradient gel electrophoresis (DGGE)
Aliquots of bacterial DNA (~20 ng) extracted from OSCC subjects were amplified using a set of universal 16S rDNA (V4-V5 region) sequence primers (Bac1 and Bac2) to generate an amplicon of ~300 bp as reported elsewhere (Li, et al., 2005, Li, et al., 2006, Li, et al., 2007). DGGE was performed with the Bio-Rad DCode System (Hercules, CA) using conditions previously reported (Li, et al., 2005, Li, et al., 2006). The DGGE profile images were digitally captured (Alpha Innotech, San Leandro, CA) and analyzed.
Primer design for pyrosequencing
PCR primers used in this study were designed based on the V4-V5 hypervariable region (~300 bp) of the 16S rRNA gene locus (Li, et al., 2007). The oligonucleotide design included the 454 Life Science’s A or B sequencing adapter (shown in lower case in the following) fused to the 5′ end of primerA-BAC1, 5′-gcctccctcgcgccatcag -ACTACGTGCCAGCAGCC 3′, and B-BAC2, 5′-gccttgccagcccgctcag-GGACTACCAGGGTATCTAATCC 3′ and synthesized by IDT technologies.
PCR amplicon library construction and sequencing
The amplified PCR products were generated by amplifying 20 ng of genomic DNA with primers designed based on the V4-V5 hypervariable region (~300 bp) of the 16S rRNA gene locus. The PCR Master mix consisted of Qiagen’s Taq PCR master mix kit, 0.2 μM concentration of each primer in a volume of 100μl. Cycling conditions were: an initial denaturation at 94°C for 1 min; 25 cycles of 94°Cfor 30 s, 55°C for 45 s, and 72°C for 45 s; and a final7-min extension at 72°C. The products were pooled after cycling and cleaned by using the MinElute PCR purification kit (Qiagen, Valencia, CA). The quality of the product was assessed on a Bioanalyzer 2100 (Agilent, Palo Alto, CA) using a DNA1000LabChip. Only sharp, distinct amplification products with a total yield of >200 ng were used for sequencing. The amplicons were amplified using the 454 Life Science’s A or B sequencing adapter primersA-BAC1 and B-BAC2, under the same amplification conditions, purified and re-bioanalyzed for QC purposes.
The amplicon libraries were bound to beads (one DNA molecule per bead) and subjected to the emPCR process using, the Amplicom Primer A kit (Roche). After emulsion PCR, the DNA strands were enriched and deposited onto PicoTiterPlates (454 Life Sciences) for pyrosequencing on a Genome Sequencer GS FLX system (LR 70 100 cycles) (Roche, Basel, Switzerland).
Preprocessing of sequence reads
The reads obtained from GS-FLX were preprocessed to identify sequencing errors. A read was eliminated if: 1) it contained no forward primer (exact match) at the proximal end; 2) it was less than 50 bases before reaching the reverse primer sequences on the distal end (a read was retained if it was shorter than 50 bp but had the reverse primer sequence); and 3) it did not contain at least 12 bases (with at most 2 errors (gap or mismatch)) of the reverse primer sequence before the read ended. To identify distal primers, Vmatch was used (Kurtz, et al., 2001, Herold, et al., 2008). Both proximal and distal primer sequences were trimmed from the reads before further analysis. Reads were grouped into a cluster if they were 99% similar over 99% of the length. A representative unique read sequence was selected randomly from each cluster for further analyses.
Construction of V4-V5 database, assignment of phylotype operational taxonomic units (OTUs) and species richness estimators
To assign phylotypes to the 454 sequence reads, a reference database of 77,085 non identical V4-V5 sequences extracted from 511,847 bacterial rRNAs derived from Ribosomal Database Project II (Cole, et al., 2007) was constructed. Each unique 454 sequence tag was BLASTed against the V4-V5 reference database. Reads greater than 200 bp were used for multiple sequence alignment using MUSCLE (Edgar, 2004). Top 50 hits with an e-value of ≤ 1e-05 (Turnbaugh, 2009) were aligned to each other using MUSCLE (with parameters-diags and -maxiters 3) (Sogin, et al., 2006). Phylotype assignments were made according to the V4-V5 reference sequence(s) that displayed the minimum distance to the query sequence. The frequency of observed best matches to V4-V5 database (OTUs) for each site was used to calculate rarefaction curves with the program Analytic Rarefaction 1.3 (http://www.uga.edu/~strata/software/).
Pairwise distance between sequences was calculated using ClustalW (Larkin, 2007). These pairwise distances were used with DOTUR (Distance-Based OTU and Richness) (Schloss & Handelsman, 2005) to cluster OTUs, generate rarefaction curves, and calculate the species richness estimator (abundance-based coverage estimators; ACE and Chao1) values by using samples without replacement, a parameter of 100 for precision of distance, jumbled input order, and 50,000 iterations. Bacterial phylotypes were identified after removing the duplicated sequences and compared among the groups and between the groups.
The sequences of unknowns and unique phylotypes would be submitted to genebank database. The V4-V5 database would also be available at this web site.
Results and Discussion
To assess the role of bacteria in the development of OSCC, the first step is to identify both cultured and unculturerd bacteria in the saliva samples of OSCC subjects. These bacteria may have the potential to cause inflammation, which may possibly support the progression of OSCC. Initially, the bacterial profiles of OSCC and control subjects were identified by amplifying 300-bp fragments of 16S rDNA gene fragments using DGGE. Subsequently, same 300-bp products were subjected to 454 pyrosequencing, to determine the total bacterial diversity and relative abundance of bacterial species in the samples.
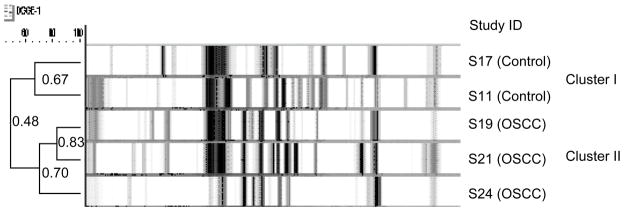
We used DGGE (Li, et al., 2007) to examine the total bacterial profile of OSCC. DGGE profiles were obtained from the saliva samples of the 5 subjects (Fig. 1). A total of 86 distinct amplicons were detected from the overall DGGE profiles after gel normalization. The number of distinct bands (amplicons) ranged from 23 to 37, with a mean ± SD of 32.2 ± 4.0 for each individual sample. On an average, the total number of detectable bands was significantly higher in the control group (36.9 ± 2.7) than in the OSCC group (29.4 ± 1.5). The results of our study also demonstrated that the DGGE profiles of OSCC and control formed significant group-specific clusters (Fig 1). The categorization of two distinct clusters was reflected by the number of bands detected (richness), the intensity, and the migration distribution of the PCR 16S amplicons.
Figure 1.
Cluster analysis. The difference in microbial diversity was clearly distinguished by cluster analysis with Ward’s algorithm based on the Dice coefficient. A distinct cluster from the OSCC group was observed, and 3 OSCC profiles were grouped into one dendrogram branch. The DGGE profiles of the control subjects were differentiated from those of the OSCC subjects in a separate cluster. The difference in the mean similarity values (52.3% ± 2.6%; P = 0.003 by ANOVA) between the two groups was statistically significant.
This finding is noteworthy, considering that OSCC and control profiles were more similar within each group than between the groups, which suggests the presence of common phylotypes associated with OSCC or controls. These results demonstrated that OSCC group can be predicted with reasonable accuracy based on DGGE banding patterns.
Using 454 pyrosequencing approximately 58,000 PCR amplicons that spanned the V4-V5 hypervariable region of ribosomal RNAs were sequenced. The number of reads per sample ranged from 7,977 to 15,170 sequences (Table 1). A systematic trimming procedure was used to eliminate sequences with multiple undetermined residues or mismatches to the PCR primers at the beginning of a read. On an average, this trimming procedure reduced the size of the data set by approximately 20%.
Table 1.
Data summary and OTUsa
| Sample | Total reads | Trimmed tags | Unique tags | OTUs (unique) |
|---|---|---|---|---|
| S11 | 11,644 | 11,351 | 3,613 | 596 |
| S17 | 7,977 | 7,799 | 2,338 | 562 |
| S19 | 11,481 | 11,141 | 5,837 | 443 |
| S21 | 15,170 | 14,831 | 7,351 | 587 |
| S24 | 12,293 | 11,996 | 5,743 | 445 |
Operational taxonomic units
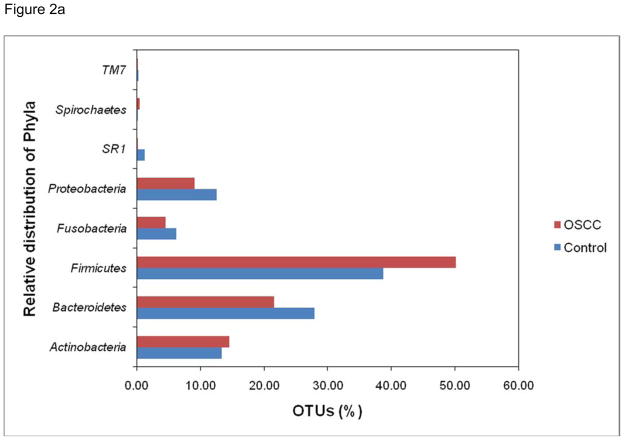
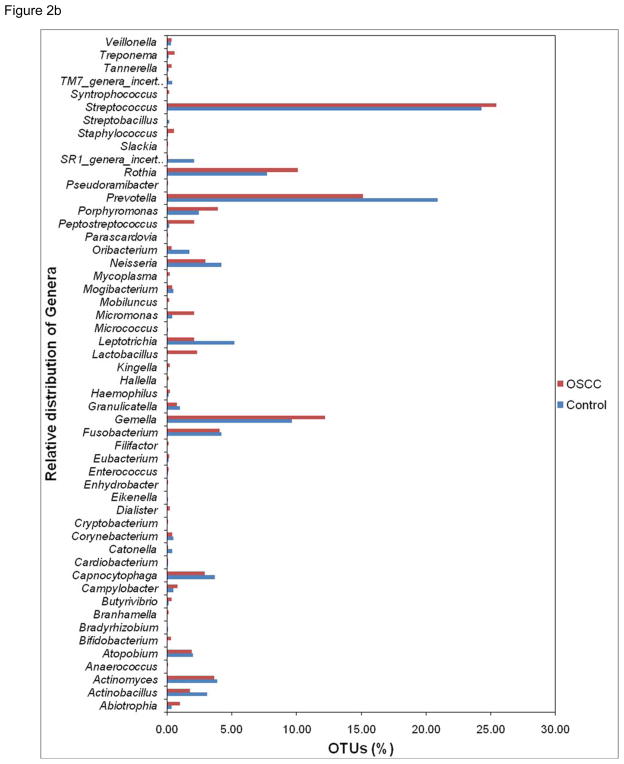
To assess taxonomic diversity, each trimmed 454 read (tag sequence) served as a query to identify its closest match in a reference database (V4-V5 database) of ~50,000 unique V4-V5 hypervariable sequences extracted from nearly 550,000 published rRNA genes for the bacteria domain. The ensemble of sequences in this study provides a far-reaching view of the salivary microbiota. Members of 8 phyla (divisions) of bacteria were detected (Fig 2a). The majority of sequences in combined libraries belonged to Firmicutes (45% of classified sequences) and to Bacteroidetes (25%). The phylum Firmicutes showed most abundance populations in OSCC library as compared to control library. The other phyla represented in both the libraries were Actinobacteria (14%); Proteobacteria (10%); Fusobacteria (5%); SR1 (0.6%); Spirochaetes (0.2%); and uncultured TM7 (0.2%). More than 810 16S rRNA gene sequences that passed a chimera-checking algorithm could not be assigned to known phyla, based on BLAST searches against the RDP taxonomy annotations. Further, out of 2633 unique sequences (no duplicated) used, to determine the species level identification at 80% or higher sequence similarity, a total of 52 different genera (Fig 2b) containing approximately 860 (33%) known species and 1773 (67%) sequences belonging to various uncultured or unclassified group were observed. The most prevalent genera in the OSCC library were Streptococcus, Gemella, Rothia, Peptostreptococcus, Lactobacillus, Porphyromonas and Lactobacillus. In the control group, genera Prevotella, Neisseria, Leptotrichia, Capnocytophaga, Actinobacillus, Oribacterium, SR1 and TM7 were predominantly observed. This shift may be due to the alterations in oral microenvironment disturbing the normal microbiota conducive for opportunistic pathogens to repopulate. The information content ofV4-V5 sequences is assumed to be sufficient to identify phylogenetic affinity with full-length sequences in a reference database. Rarefaction analysis was based on best matches for each tag to sequences in the V4-V5 database and their frequency of recovery. These rarefaction curves described unparalleled levels of bacterial complexity for all saliva samples; the curve clearly showed no plateau and gave an indication of the extensive species richness.
Figure 2.
Figure 2a. Percentage of phlya represented in the saliva samples of OSCC and control.
Figure 2b. Percentage of genera represented in the saliva samples of OSCC and control.
As an alternative to defining OTUs by their best matches against theV4-V5 database, sequence tags were clustered into groups of defined sequence variation ranging from unique sequences (no variation) to10% differences using DOTUR. These clusters served as OTUs for generating rarefaction curves and for making calculations with the ACE andChao1 estimator of species diversity. This approach is frequently used in microbial diversity research especially when there is enough representation of all of the species (Sogin, et al., 2006). As a result, these coverage-based nonparametric estimates of microbial richness probably represent the true bacterial diversity. The data in Table 2 show that the estimates species diversity of bacteria obtained with ACE and Chao1 were significantly greater than published analyses of other microbial profiles from the oral cavity (Paster, et al., 1994, Paster, et al., 2001, Paster, et al., 2002).
Table 2.
Estimatesa of similarity-based OTUs and species richness.
| Cluster distance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.05 | 0.1 | ||||||||
| Sample ID | Reads | OTU | ACE | Chao 1 | OTU | ACE | Chao 1 | OTU | ACE | Chao 1 |
| S11 | 7229 | 784 | 1327 | 1301 | 514 | 747 | 723 | 200 | 235 | 234 |
| S17 | 6309 | 782 | 1386 | 1392 | 476 | 707 | 701 | 207 | 243 | 234 |
| S19 | 3986 | 389 | 680 | 707 | 232 | 327 | 318 | 85 | 96 | 111 |
| S21 | 6677 | 905 | 1548 | 1492 | 564 | 774 | 770 | 198 | 222 | 219 |
| S24 | 4326 | 520 | 900 | 886 | 321 | 496 | 521 | 131 | 160 | 171 |
ACE and Chao1 were calculated using DOTUR.
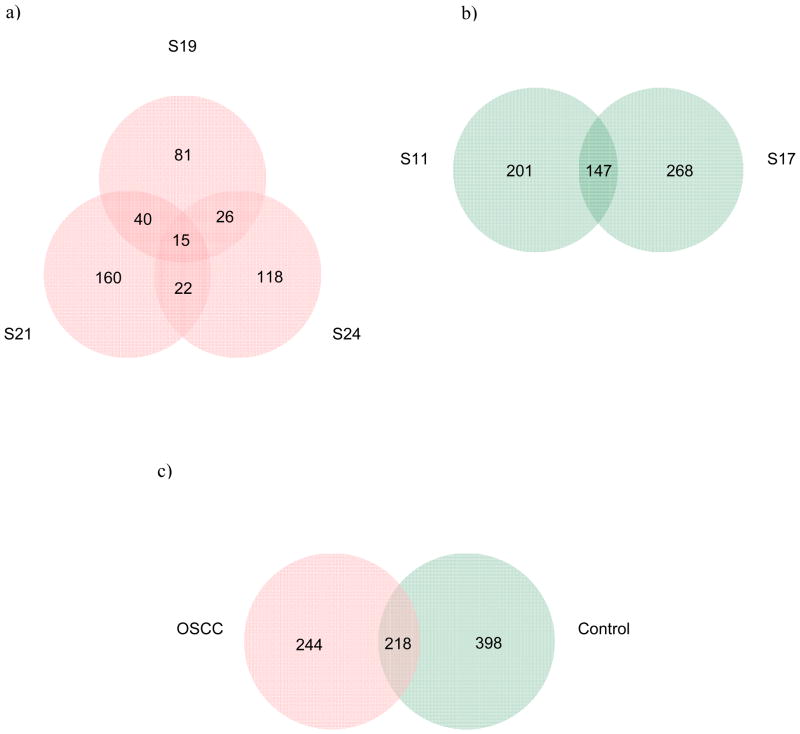
The alteration of the oral microflora in OSCC is of particular interest because of its potential application as a diagnostic tool to predict oral cancer (Mager, et al., 2005, Mager, 2006). Unfortunately, most of such studies to date have included only cultured oral bacterial species (Nagy, et al., 1998, Mager, et al., 2005, Hooper, et al., 2006), using classical cloning and sequencing approaches. To understand the role of bacteria in the development of oral cancer, the first step is to identify both cultured and uncultured organisms in the saliva as these organisms have the potential to cause inflammation that may support OSCC progression. After removing the duplicate sequences we identified over 860 bacterial phylotypes, and each subject harbored more than 162 known bacterial species. The number of species shared by OSCC samples S19/S21/S24, S19/S21, S19/S24 and S21/S24 were 15, 40, 26 and 22; whereas that uniquely observed in S19, S21 and S24 were 81, 160 and 118, respectively (Fig 3). In total, 462 bacterial species were present in the three samples which constituted the OSCC group. The control samples S11 and S17 comprised of 616 total species with 201 and 268 species exclusive to each samples and 147 common species. The combined group, OSCC and control, showed 860 species with 218 shared species, and 244 and 398 species unique to respective group. Comparison within OSCC samples demonstrated fifteen unique bacterial phylotypes were present in all three OSCC subjects (Table 3). There are evidence that these unique phylotypes and uncultured/unidentified forms may lead to continued inflammation and other pathological conditions (Table 3). Further bacterial products, such as the cell wall product of gram-negative bacteria (lipopolysaccharide, LPS), have a significant role in a variety of pathologic conditions due to bacterial association with vast mucosal and skin surfaces, and LPS stimulates many cell types (Kurago, et al., 2008).
Figure 3.
Venn diagram of the number of species common/unique within: a) OSCC group; b) Control group. c) Combined, OSCC and control. The interior of each circle symbolically represents the number of observed bacterial species in the certain sample/group. The overlapping area or intersection would represent the set of bacteria commonly present in the counterpart samples/groups. Likewise, the single-layer zone represents the number of bacteria uniquely found in the certain sample/group.
Table 3.
Selected bacterial phylotypes identified in either of OSCC subjects and their putative virulence properties.
| Bacterial Phylotypes | Characteristics | Reference/s |
|---|---|---|
| Actinomyces georgiae | oral biofilm | (Henssge, et al., 2009) |
| Actinomyces naeslundii | oral biofilm | (Henssge, et al., 2009) |
| Bifidobacterium breve | intestinal inflammation | (Heuvelin, et al., 2009) |
| Capnocytophaga spp | periodontal diseases, septicaemia | (Jolivet-Gougeon, et al., 2007) |
| Clostridium butyricum | intestinal toxemia botulism | (Fenicia, et al., 2002) |
| Prevotella melaninogenica | oral cancer | (Mager, et al., 2005) |
| Tissierella praeacuta | (Papaparaskevas, 2005) | |
| Fusobacterium necrophorum | septicemia, tonsillitis | (Riordan, 2007) |
| Gemella haemolysans | Meningitis, renal failure | (Eisenhut, et al., 2004, Anil, et al., 2007) |
| Parvimonas | apical abscesses | (Siqueira & Rocas, 2009) |
| Peptostreptococcus micros | chronic inflammatory | (Tanabe, et al., 2007) |
| Porphyromonas gingivalis | periodontitis, rheumatoid arthritis, OSCC | (Mager, et al., 2005, Rosenstein, et al., 2009) |
| Prevotella intermedia/nigrescens | extraoral and some odontogenic Infections | (Mättö, 1997) |
| Rothia mucilaginosa | bacteremia | (Vaccher, et al., 2007) |
| Staphylococcus saccharolyticus | infective endocarditis | (Westblom, et al., 1990) |
In recent study viable bacteria have been isolated from both superficial and deep portions of OSCC (Hooper, et al., 2006, Hooper, et al., 2007), revealing that the tumor microenvironment is well suited for bacterial survival. Hooper et al. (Hooper, et al., 2006, Hooper, et al., 2007) using PCR and sequencing partial sequencing of the 16S rRNA gene fragments identified a total of 70 distinct taxa; 52different phylotypes isolated from the tumorous tissues, and37 taxa from within the non-tumorous specimens. Differences between the composition of the microbiotas within the tumorous and non-tumorous mucosae were apparent in saccharolytic and aciduric species. Several unusual bacteria, such as Clavibacter michiganensis, Plantibacter flavus, Tepidimonasaquatica and Thermus scotoductus, were present in deep tumors. These rare species were previously been isolated from non-clinical sources (Hooper, et al., 2006, Hooper, et al., 2007), whereas in our study using saliva from OSCC subjects, we identified different bacterial phylotypes (Table 3). These differences are due to the fact that we used saliva samples and Hooper et al. (Hooper, et al., 2006, Hooper, et al., 2007) used tumor samples. In our study, we preferred saliva samples as to develop further as a salivary diagnostic tool for OSCC. Similarly, in other study using saliva samples high levels of colonization of OSCC by facultative oral streptococci (Sasaki, et al., 1998, Sakamoto, et al., 1999, Tateda, et al., 2000, Shiga, et al., 2001) and species of anaerobic bacteria, such as Prevotella, Veillonella, Porphyromonas, Streptococcus anginosus, and Capnocytophaga, have been demonstrated relative to uninvolved mucosa (Sakamoto, et al., 1999, Tateda, et al., 2000, Crean, et al., 2002, Sasaki, et al., 2005). Our results indicated that the bacterial diversity in oral samples is more than an order of magnitude higher than obtained previously (Socransky, et al., 1998, Paster, et al., 2001, Paster, et al., 2002, Hooper, et al., 2006, Hooper, et al., 2007). Of these 2633 OTUs from 5 subjects, more than 67% can either not be grown in the laboratory or were unknowns. These uncultured and sometimes dormant bacteria occupy different ecological microniches, and they are involved in latent infections (Mackowiak, 1978a, Mackowiak, 1978b, Domingue & Woody, 1997, Nystrom, 2003, Anderson, et al., 2004). Further, these cultured and uncultured organisms colonize the tumor and have the potential to cause chronic inflammation that may support OSCC progression. Although the role of bacteria in the development of OSCC has not been determined, chronic inflammation has been observed in the oral cavity at various stages of cancer (Shafer, et al., 1983, Coussens & Werb, 2002, Peek, et al., 2005, Kurago, et al., 2008). This inflammation could also result from persistent colonization of mucosal or epithelial cells by bacteria as reported for other cancers (Lee, et al., 2003, Wogan, et al., 2004, Peek, et al., 2005, Chen, et al., 2006). Moreover, there is frequent loss of surface integrity in OSCC, with gram-positive and gram-negative oral and enteric bacteria found in 75% of cervical lymph nodes containing metastatic OSCC (Kurago, et al., 2008). We recognize the fact that our data is based on limited samples size however using DGGE in combination with high throughput sequencing allowed us to determine the total bacterial diversity by minimizing the effect of individual variation of OSCC subjects and matched controls.
It is estimated that in 2008 approximately 35,000 new cases of and 7,600 deaths attributable to cancer of the oral cavity and pharynx in the United States (Jemal, et al., 2008) and early detection of OSCC is critical for patient survival. In addition, early detection of OSCC requires less radical treatment of patients and results in a higher quality of life (Morse & Kerr, 2006). The results of our study on saliva microbiome are of particular interest as it reflects the repopulation or shift in microbial communities. These changes may have potential application as a diagnostic tool to predict oral cancer before its develops into malignant tumor (Mager, et al., 2005, Mager, 2006). However, longitudinal studies are required on bacterial diversity in saliva and tissue samples using cohorts from different stages of OSCC progression. This may provide the critical foundation for the identification of bacterial indicators of carcinogenesis in OSCC and, in turn, suggest strategies for more effective diagnosis and treatment.
Acknowledgments
This work was supported by NIDCR Grants U54-DE14257, U19-DE018385, RO3-DE019178, and RO1-DE020891.
References
- Anderson M, Bollinger D, Hagler A, Hartwell H, Rivers B, Ward K, Steck TR. Viable but nonculturable bacteria are present in mouse and human urine specimens. J Clin Microbiol. 2004;42:753–758. doi: 10.1128/JCM.42.2.753-758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anil M, Ozkalay N, Helvaci M, Agus N, Guler O, Dikerler A, Kanar B. Meningitis due to Gemella haemolysans in a pediatric case. J Clin Microbiol. 2007;45:2337–2339. doi: 10.1128/JCM.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncol. 2010;46:414–417. doi: 10.1016/j.oraloncology.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. Understanding microbe-induced cancers. Cancer Prev Res. 2008:1940–6207. doi: 10.1158/1940-6207.CAPR-08-0024. CAPR-1908–0024. [DOI] [PubMed] [Google Scholar]
- Chen PC, Pan CC, Kuo C, Lin CP. Risk of oral nonmalignant lesions associated with human papillomavirus infection, betel quid chewing, and cigarette smoking in Taiwan: an integrated molecular and epidemiologic study. Arch Pathol Lab Med. 2006;130:57–61. doi: 10.5858/2006-130-57-ROONLA. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crean, Nair, Fardy, Wilson, Spratt Identification of bacterial DNA using PCR cloning within oral squamous cell carcinomas (abstract) J Dent Res. 2002;81:A364. [Google Scholar]
- Dehio C. Bartonella-host-cell interactions and vascular tumour formation. Nat Rev Microbiol. 2005;3:621–631. doi: 10.1038/nrmicro1209. [DOI] [PubMed] [Google Scholar]
- Domingue GJ, Sr, Woody HB. Bacterial persistence and expression of disease. Clin Microbiol Rev. 1997;10:320–344. doi: 10.1128/cmr.10.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;19:113–131. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Jones C, Hughes D, Herrington S, Kokai G. Acute renal failure associated with Gemella haemolysans pneumonia. Pediatric Nephrology. 2004;19:448–450. doi: 10.1007/s00467-003-1344-5. [DOI] [PubMed] [Google Scholar]
- Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, Raul F. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21:753–756. doi: 10.1093/carcin/21.4.753. [DOI] [PubMed] [Google Scholar]
- Engle SJ, Ormsby I, Pawlowski S, Boivin GP, Croft J, Balish E, Doetschman T. Elimination of colon cancer in germ-free transforming growth factor beta 1-deficient mice. Cancer Res. 2002;62:6362–6366. [PubMed] [Google Scholar]
- Erdman SE, Poutahidis T, Tomczak M, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162:691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenicia L, Da D, et al. A case of infant botulism due to neurotoxigenic Clostridium butyricum Type E associated with Clostridium difficile colitis. Eur J Clin Microbiol Infect Dis. 2002;21:736–738. doi: 10.1007/s10096-002-0816-z. [DOI] [PubMed] [Google Scholar]
- Henssge U, Do T, Radford DR, Gilbert SC, Clark D, Beighton D. Emended description of Actinomyces naeslundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov., previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int J Syst Evol Microbiol. 2009;59:509–516. doi: 10.1099/ijs.0.000950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J, Kurtz S, Giegerich R. Efficient computation of absent words in genomic sequences. BMC Bioinformatics. 2008;9:167. doi: 10.1186/1471-2105-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvelin E, Lebreton C, Grangette C, Pot B, Cerf-Bensussan N, Heyman M. Mechanisms involved in alleviation of intestinal inflammation by Bifidobacterium breve soluble factors. PLoS ONE. 2009;4:e5184. doi: 10.1371/journal.pone.0005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Bunge J, Jeon SO, Epstein SS. Predicting microbial species richness. Proc Natl Acad Sci U S A. 2006;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SJ, Crean SJ, Lewis MAO, Spratt DA, Wade WG, Wilson MJ. Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol. 2006;44:1719–1725. doi: 10.1128/JCM.44.5.1719-1725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SJ, Crean SJ, Fardy MJ, Lewis MA, Spratt DA, Wade WG, Wilson MJ. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. 2007;56:1651–1659. doi: 10.1099/jmm.0.46918-0. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Jolivet-Gougeon A, Sixou J-L, Tamanai-Shacoori Z, Bonnaure-Mallet M. Antimicrobial treatment of Capnocytophaga infections. Int J Antimicrob Ag. 2007;29:367–373. doi: 10.1016/j.ijantimicag.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248:171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- Kurago Z, Lam-ubol A, Stetsenko A, De La Mater C, Chen Y, Dawson D. Lipopolysaccharide-squamous cell carcinoma-monocyte interactions induce cancer-supporting factors leading to rapid STAT3 activation. Head Neck Pathol. 2008;2:1–12. doi: 10.1007/s12105-007-0038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lax AJ. Bacterial toxins and cancer -a case to answer? Nat Rev Micro. 2005;3:343–349. doi: 10.1038/nrmicro1130. [DOI] [PubMed] [Google Scholar]
- Lax AJ, Thomas W. How bacteria could cause cancer: one step at a time. Trends Microbiol. 2002;10:293–299. doi: 10.1016/s0966-842x(02)02360-0. [DOI] [PubMed] [Google Scholar]
- Lee CH, Ko YC, Huang HL, Chao YY, Tsai CC, Shieh TY, Lin LM. The precancer risk of betel quid chewing, tobacco use and alcohol consumption in oral leukoplakia and oral submucous fibrosis in southern Taiwan. Br J Cancer. 2003;88:366–372. doi: 10.1038/sj.bjc.6600727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbiol. 2007;45:81–87. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ku CY, Xu J, Saxena D, Caufield PW. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res. 2005;84:559–564. doi: 10.1177/154405910508400614. [DOI] [PubMed] [Google Scholar]
- Li Y, Saxena D, Barnes VM, Trivedi HM, Ge Y, Xu T. Polymerase chain reaction-based denaturing gradient gel electrophoresis in the evaluation of oral microbiota. Oral Microbiol Immunol. 2006;21:333–339. doi: 10.1111/j.1399-302X.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- Littman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomarkers Prev. 2005;14:773–778. doi: 10.1158/1055-9965.EPI-04-0599. [DOI] [PubMed] [Google Scholar]
- Mackowiak PA. Microbial synergism in human infections. N Engl J Med. 1978a;298:21–26. doi: 10.1056/NEJM197801052980105. [DOI] [PubMed] [Google Scholar]
- Mackowiak PA. Microbial synergism in human infections. N Engl J Med. 1978b;298:83–87. doi: 10.1056/NEJM197801122980206. [DOI] [PubMed] [Google Scholar]
- Mager DL. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. doi: 10.1186/1479-5876-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio-Price L, Treuting P, Zeng W, Tsang M, Bielefeldt-Ohmann H, Iritani BM. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 2006;66:828–838. doi: 10.1158/0008-5472.CAN-05-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfertheiner P, Sipponen P, Naumann M, et al. Helicobacter pylori eradication has the potential to prevent gastric cancer: a state-of-the-art critique. Am J Gastroenterol. 2005;100:2100–2115. doi: 10.1111/j.1572-0241.2005.41688.x. [DOI] [PubMed] [Google Scholar]
- Marshall BJ, Windsor HM. The relation of Helicobacter pylori to gastric adenocarcinoma and lymphoma: pathophysiology, epidemiology, screening, clinical presentation, treatment, and prevention. Med Clin North Am. 2005;89:313–344. viii. doi: 10.1016/j.mcna.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Mättö J, Asikainen S, Väisänen ML, Rautio M, Saarela M, Summanen P, Finegold S, Jousimies-Somer H. Role of Porphyromonas gingivalis, Prevotella intermedia, and Prevotella nigrescens in extraoral and some odontogenic infections. Clin Infect Dis. 1997;25:S194–S198. doi: 10.1086/516205. [DOI] [PubMed] [Google Scholar]
- Morse DE, Kerr AR. Disparities in oral and pharyngeal cancer incidence, mortality and survival among black and white Americans. J Am Dent Assoc. 2006;137:203–212. doi: 10.14219/jada.archive.2006.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy KN, Sonkodi I, Szoke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34:304–308. [PubMed] [Google Scholar]
- Newman JV, Kosaka T, Sheppard BJ, Fox JG, Schauer DB. Bacterial infection promotes colon tumorigenesis in Apc(−/+) mice. J Infect Dis. 2001;184:227–230. doi: 10.1086/321998. [DOI] [PubMed] [Google Scholar]
- Nystrom T. Nonculturable bacteria: programmed survival forms or cells at death’s door? Bioessays. 2003;25:204–211. doi: 10.1002/bies.10233. [DOI] [PubMed] [Google Scholar]
- Papaparaskevas J, Pantazatou A, Katsandri A, Legakis NJ the Hellenic Study Group for Gram-Negative Anaerobic Bacteria and Avlamis, A. Multicentre survey of the in-vitro activity of seven antimicrobial agents, including ertapenem, against recently isolated Gram-negative anaerobic bacteria in Greece. Clin Microbiol Infect. 2005;11:820–824. doi: 10.1111/j.1469-0691.2005.01233.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE, Olsen I, Fraser GJ. Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J Bacteriol. 1994;176:725–732. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Falkler WA, Jr, Enwonwu CO, et al. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J Clin Microbiol. 2002;40:2187–2191. doi: 10.1128/JCM.40.6.2187-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek RM, Jr, Mohla S, DuBois RN. Inflammation in the genesis and perpetuation of cancer: summary and recommendations from a national cancer institute-sponsored meeting. Cancer Res. 2005;65:8583–8586. doi: 10.1158/0008-5472.CAN-05-1777. [DOI] [PubMed] [Google Scholar]
- Rao VP, Poutahidis T, Fox JG, Erdman SE. Breast cancer: should gastrointestinal bacteria be on our radar screen? Cancer Res. 2007;67:847–850. doi: 10.1158/0008-5472.CAN-06-3468. [DOI] [PubMed] [Google Scholar]
- Rao VP, Poutahidis T, Ge Z, et al. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 2006;66:7395–7400. doi: 10.1158/0008-5472.CAN-06-0558. [DOI] [PubMed] [Google Scholar]
- Riordan T. Human Infection with Fusobacterium necrophorum (Necrobacillosis), with a focus on Lemierre’s syndrome. Clin Microbiol Rev. 2007;20:622–659. doi: 10.1128/CMR.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein ED, Weissmann G, Greenwald RA. Porphyromonas gingivalis, periodontitis and rheumatoid arthritis. Eur J Cardiovasc Prev Rehabil. 2009;73:457–458. doi: 10.1016/j.mehy.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Naito H, Ohta Y, Tanakna R, Maeda N, Sasaki J, Nord CE. Isolation of bacteria from cervical lymph nodes in patients with oral cancer. Arch Oral Biol. 1999;44:789–793. doi: 10.1016/s0003-9969(99)00079-5. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Ishizuka T, Muto M, et al. Presence of Streptococcus anginosus DNA in esophageal cancer, dysplasia of esophagus, and gastric cancer. Cancer Res. 1998;58:2991–2995. [PubMed] [Google Scholar]
- Sasaki M, Yamaura C, Ohara-Nemoto Y, et al. Streptococcus anginosus infection in oral cancer and its infection route. Oral Diseases. 2005;11:151–156. doi: 10.1111/j.1601-0825.2005.01051.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45:301–308. doi: 10.1016/j.oraloncology.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Shafer W, Hine M, Levy B. A Textbook of Oral Pathology. 4. Philadelphia, Pa: WB Saunders; 1983. Benign and malignant tumors of oral cavity; pp. 86–93. [Google Scholar]
- Shiga K, Tateda M, Saijo S, et al. Presence of Streptococcus infection in extra-oropharyngeal head and neck squamous cell carcinoma and its implication in carcinogenesis. Oncol Rep. 2001;8:245–248. [PubMed] [Google Scholar]
- Shukla VK, Singh H, Pandey M, Upadhyay SK, Nath G. Carcinoma of the Gallbladder - Is it a sequel of typhoid? Dig Dis Sci. 2000;45:900–903. doi: 10.1023/a:1005564822630. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN. The Microbiota of Acute Apical Abscesses. J Dent Res. 2009;88:61–65. doi: 10.1177/0022034508328124. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S-i, Bodet C, Grenier D. Peptostreptococcus micros cell wall elicits a pro-inflammatory response in human macrophages. J Endotoxin Res. 2007;13:219–226. doi: 10.1177/0968051907081869. [DOI] [PubMed] [Google Scholar]
- Tateda M, Shiga K, Saijo S, et al. Streptococcus anginosus in head and neck squamous cell carcinoma: implication in carcinogenesis. Int J Mol Med. 2000;6:699–703. doi: 10.3892/ijmm.6.6.699. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccher S, Cordiali R, Osimani P, Manso E, de Benedictis F. Bacteremia caused by Rothia mucilaginosa in a patient with shwachman-diamond syndrome. Infection. 2007;35:209–210. doi: 10.1007/s15010-007-6284-8. [DOI] [PubMed] [Google Scholar]
- Vogelmann R, Amieva MR. The role of bacterial pathogens in cancer. Curr Opin Microbiol. 2007;10:76–81. doi: 10.1016/j.mib.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Wagenlehner FM, Elkahwaji JE, Algaba F, Bjerklund-Johansen T, Naber KG, Hartung R, Weidner W. The role of inflammation and infection in the pathogenesis of prostate carcinoma. BJU Int. 2007a;100:733–737. doi: 10.1111/j.1464-410X.2007.07091.x. [DOI] [PubMed] [Google Scholar]
- Wagenlehner FME, Weidner W, Naber KG. Therapy for prostatitis, with emphasis on bacterial prostatitis. Expert Opin Pharmacother. 2007b;8:1667–1674. doi: 10.1517/14656566.8.11.1667. [DOI] [PubMed] [Google Scholar]
- Ward JM, Fox JG, Anver MR, et al. Chronic Active Hepatitis and Associated Liver Tumors in Mice Caused by a Presistent Bacterial Infection With a Novel Helicobacter Species. J Natl Cancer Inst. 1994;86:1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- Westblom TU, Gorse GJ, Milligan TW, Schindzielorz AH. Anaerobic endocarditis caused by Staphylococcus saccharolyticus. J Clin Microbiol. 1990;28:2818–2819. doi: 10.1128/jcm.28.12.2818-2819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Sem Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]