Abstract
Increasing evidence has shown that a fraction of the wild-type (wt) form of the tumor suppressor p53, can translocate to mitochondria due to genotoxic stress. The mitochondrial targets of wt p53 have also been studied. However, whether mutant p53, which exists in 50% of human cancers, translocates to mitochondria and affects mitochondrial functions is unclear. In this study, we used doxorubicin, a chemotherapeutic drug, to treat five human lymphoma cell lines with wt, mutant or deficient in p53, to induce p53 activation and mitochondrial translocation. Our results demonstrated that mutant p53, like wt p53, was induced upon doxorubicin treatment. Similarly, a fraction of mutant p53 also translocated to mitochondria. However, Complex I and II activities in the mitochondria were compromised only in wt p53-bearing cells after doxorubicin treatment, but not in mutant p53-bearing cells. Similarly, doxorubicin treatment caused greater cell death only in wt p53-bearing cells, but not in mutant p53-bearing cells. When p53 deficient Ramos cells were transfected with mutant p53 (249S), the cells showed resistance to doxorubicin-induced cell death and decreases in complex activities. To reactivate mutant p53 and reverse chemoresistance, ellipticine (5,11-dimethyl-6H-pyrido[4,3-b]carbazole) was used to treat mutant p53 cells. Ellipticine enhanced p53 mitochondrial translocation, decreased Complex I activity, and sensitized p53 mutant cells to doxorubicin-induced apoptosis. In summary, our studies suggest that mutations in p53 may not hinder p53’s mitochondrial translocation, but impair its effects on mitochondrial functions. Therefore, restoring mutant p53 by ellipticine may sensitize these cells to chemotherapy.
Keywords: Mitochondria, Apoptosis, Lymphoma, p53
Introduction
p53, considered as a pivotal tumor suppressor, can initiate apoptosis in response to expression of viral or cellular oncogenes [1] It is also the most well-known and commonly mutated gene in human cancers [2]. Increasing the activity of p53 has been an attractive strategy for cancer prevention and therapy. Indeed, activated p53 has been confirmed to exhibit enhanced tumor resistance [3] or a reduction of simultaneous tumor growth [4] in mouse models. It is not surprising that p53 has become the first approved gene-therapy treatment for head-and-neck squamous cell carcinoma [5].
Activation of apoptosis is an important mechanism in p53-induced tumor suppression. As a transcription factor, p53 induces the expression of pro-apoptotic genes [6, 7]. Recently, evidence linking p53 transcription-independent or mitochondria-targeted apoptosis has received considerable attention. A fraction of p53 translocates to mitochondria prior to changes in mitochondrial membrane potential, cytochrome c release, and caspase activation [8–10]. Our previous study further suggests that p53’s mitochondrial translocation precedes its nuclear translocation; subsequently blocking mitochondrial reactive oxygen species which prevents p53’s nuclear translocation [11]. These results suggest that cellular cross-talk may exist between mitochondrial and nuclear p53.
Nevertheless, the above mentioned properties correspond to wt p53. Whether mutant p53, existing in 50% of human cancers, translocates to mitochondria, and thereafter modulates mitochondrial functions has not been thoroughly studied. Numerous studies have suggested mitochondrial localization of mutant p53. Mihara et al. [12] demonstrated the mitochondrial localization of mutant p53 in cancer cells which lose the ability to interact with Bcl-XL; Heyne et al. [13] suggested that mutant p53 exists as a monomeric protein in mitochondria; Tang et al. [14] demonstrated that mutant p53 translocates to mitochondria in UVB-irradiated murine skin carcinoma cells; Mahyar-Roemer et al. [15] suggested that mutant p53 is present in mitochondria independent of apoptotic signals.
We have focused on studying whether mutant p53 affects mitochondrial functions using human lymphoma cells as a model. As the sixth most common cancer among males and the fifth most common cancer among females in the United States, non-Hodgkin’s lymphoma is increasing at an annual percentage of 2.6% with about 67,000 Americans being diagnosed with lymphoma in 2006. Numerous lymphoma cell lines have been established and the p53 status (wt or mutant) has been revealed in a number of the cell lines. Lymphoma cells are also chosen for this study, because apoptosis is a profound mechanism during lymphogenesis. Lymphoma cells have well adapted this mechanism. The lymphoma cell lines we chose include wt, mutant, and deficient in p53, and we aim to demonstrate whether mutant p53 translocates to mitochondria during cancer therapy; whether mutant p53 affects mitochondrial functions including complex activities and apoptosis; and whether reactivating mutant p53 by ellipticine mediates mutant p53’s effects on mitochondrial functions.
Materials and methods
Cell lines and treatments
Human B-cell lymphoma-derived B-cell lines (DoHH2 [kindly provided by Dr. Mitchell Smith at Fox Chase Cancer Center, Philadelphia, PA], Raji, Su-DHL-4, and Ramos) and B-cell chronic lymphocytic leukemia JOK-1 cells were used for the studies. These four cell lines were kindly provided by Dr. Heinz Kohler at the University of Kentucky. Only lower-passage cells were used; and experiments were conducted within three constant passages. p53 is wt in DoHH2 cells [16] and JOK-1; mutated in Su-DHL-4 (named DHL-4 thereafter, [16]) and Raji cells [17]. Ramos cells have been reported to either express mutant p53 [18, 19] or to be deficient in p53 [20]. We did not detect the p53 protein in the Ramos cell line and therefore referred to it as p53 deficient.
DoHH2 (wt p53) and DHL-4 (mutant p53) were grown in RPMI 1640 medium supplemented with 10% FBS and sodium pyruvate. JOK-1 (wt p53), Raji (mutant p53), and Ramos (p53 deficient) were grown in RPMI 1640 supplemented with 10% FBS, sodium pyruvate, and β-mercaptoethanol.
Doxorubicin in Saline (2 mg/ml) was purchased from Bedford Laboratories (Bedford, OH). For treatment of lymphoma cells, the final concentration was 200 ng/ml. Ellipticine (purchased from Sigma, St. Louis, MO) dissolved in dimethyl sulfoxide (DMSO) (stocking solution: 10 mM) was used to reactivate mutant p53.
Transfection of Ramos cells with mutant p53
For mitochondrial p53 translocation and mitochondrial complex activity assays, Ramos cells were seeded into p60 plates (2 × 106/ml) in growth medium. The next day, cells were collected and resuspended in the transfection mix (Opti-MEM medium and lipofectamine 2000, both purchased from Invitrogen [Carlsbad, CA]) containing 15 μg pcDNA3.1 vectors (containing the green fluorescent protein [GFP] or mutant p53 in Codon 249 [G → T transition, arginine to serine amino acid substitution, which is one of many mutations with high frequency in human cancers], which were kindly provided by Dr. Xinbin Chen at University of California-Davis). Six hours later, cells were collected and resuspended in fresh growth medium and incubated for 48 h (at 48 h, the transfection efficiency was approximately 20–25%). Cells were then treated with doxorubicin or the vehicle control for 24 h. For each treatment group, cells from ten plates were combined for isolation of mitochondria.
For preparation of total cell lysate and nucleosome fragmentation assays, Ramos cells were seeded into 24-well plates (5 × 105/ml) in 0.5 ml growth medium. Cells were transfected with either the GFP or the mutant p53 (249S) vector as described previously, except only 2 μg DNA were used. Forty-eight hours after transfection, cells were treated with doxorubicin for 24 h. Total cell lysate was prepared for detection of transfected mutant p53 and nucleosome fragmentation assay.
Preparation of total cell lysate from lymphoma cells
Collected lymphoma cells were resuspended in 500 μl RIPA buffer (50 mM Tris base, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% NP-40, adjusted to pH 7.4 with a few drops of 12 M HCl, protease inhibitor cocktail [100×] added immediately before use), sonicated on ice for three strokes (10 s/stroke), and stored on ice for 30 min. Samples were then centrifuged at 18,000×g for 20 min, and the supernatant was collected as total cell lysate.
Isolation of mitochondrial fractions from lymphoma cells
After treatment with vehicle (Saline) or doxorubicin, cells were collected and suspended in 2 ml of mitochondrial isolation buffer (0.225 M mannitol, 0.075 M sucrose, 1 mM EGTA, pH 7.4) in a 10-ml Wheaton homogenizer tube, and carefully homogenized for 30 strokes on ice. The cell debris was removed by centrifugation at 800×g for 10 min. The supernatant was filtered through a nylon screen cloth (Small Parts Inc., Miami Lakes, FL), then centrifuged at 10,000×g for 10 min. The supernatant was designated as cytosolic fraction and stored at −80°C. The pellet was washed by adding 0.5 ml of mitochondrial isolation buffer and centrifuged again at 10,000×g for 5 min. This washing step was performed twice. The mitochondrial pellet was resuspended in 200 μl of various buffers, depending on the intended use of the mitochondria [11]. The purity of the mitochondrial fraction was monitored by detecting the following markers using Western blot analysis: SDHB (succinate dehydrogenase complex subunit B, a mitochondrial marker), lamin B or PCNA (proliferating cell nuclear antigen, a nuclear marker), and GAPDH (glyceraldehyde 3-phosphate dehydrogenase, a cytoplasmic marker).
Western blot analysis
The total cell lysate was used to detect the protein levels of p53 and Bax (Bcl-2-associated X protein); the mitochondrial fraction was used to detect the protein levels of p53 and cytochrome c in mitochondria; and the cytosolic fraction was used to detect cytochrome c release from mitochondria. Anti-p53 antibody (FL393, to detected both wt and mutant p53) was purchased from Santa Cruz Bio-technology (Santa Cruz, CA). Anti-mutant p53 (Ab-3) and anti-wt p53 (Ab-5) antibodies were purchased from Cal-biochem (Boston, MA). Anti-Bax, anti-GAPDH, anti-lamin B, and anti-PCNA antibodies were purchased from Santa Cruz Biotechnology. Anti-cytochrome c antibody was purchased from Cell Signaling Technology (Danvers, MA). The corresponding bands were scanned, and the densities were quantitatively assessed using the Image-Quant 5.1 software (Bio-Rad, Hercules, CA).
Cell growth assay
1 × 104 lymphoma cells were seeded in 100 μl growth medium in 96-well plates. Cells were then treated with 200 ng/ml doxorubicin. At the indicated time points, cells were collected and stained with Trypan Blue. Viable cells were counted and the numbers were plotted as a function of doxorubicin treatment time. N = 3 for each sample.
Mitochondrial complex I activity assay
The assay was performed at 30°C with a Bio-Rad spectrophotometer, as described by Birch-Machin et al. with slight modifications [21, 22]. Mitochondrial samples were subjected to three fast freeze–thaw cycles in hypotonic buffer before the assay. The assay mixtures, containing 25 mM potassium phosphate buffer (pH 7.2), 5 mM MgCl2, 2 mM KCN, 2.5 mg/ml bovine serum albumin (fraction V), 0.13 mM NADH (Nicotinamide adenine dinucleotide), 65 mM coenzyme Q1, and 2 mg/ml anti-mycin A, were incubated at 30°C for 1 min. Mitochondria were then added to initiate the reaction, and the initial rate of NADH oxidation was monitored at 340 nm (ε = 6.81 mM−1 cm−1) for 1 min (ΔA). The reaction was inhibited by 2 μl of 2 mg/ml rotenone and the rate of NADH oxidation was monitored for 1 min (ΔAr). The relative complex activity was calculated and plotted.
Mitochondrial complex II activity assay
Mitochondria isolated from lymphoma cells were added to a buffer containing 5 mM MgCl2, 25 mM KPO3, and 20 mM succinate. After 10 min, 2 μg antimycin A, 2 μM rotenone, 2 mM KCN, and 50 μM 2,6-dichlorophenolindophenol were added and the basal rate of succinate oxidation was measured at 600 nm (ε = 19.1 mM−1 cm−1) for 1 min (ΔAb). Then co-enzyme Q was added and the activity was measured again (ΔA). The relative complex activity was calculated and plotted.
Nucleosome fragmentation assay
Cells were first lysed following the manufacturer’s instructions. DNA fragmentation was detected using cell death detection ELISA (Roche, Indianapolis, IN) [11]. This method applies an anti-histone antibody to capture the histone-DNA fragments, then applies a HRP-labeled anti-DNA antibody to detect fragmented DNA.
Statistical analysis
Statistical analysis was performed using both student t-test (for two-group comparison) and one-way ANOVA (for multiple group comparison) followed by Newman–Keuls post-test. Data were reported as means ± standard error (SEM). P < 0.05 was considered as being significant.
Results
The expression levels of both wt and mutant p53 were increased in lymphoma cells upon doxorubicin treatment
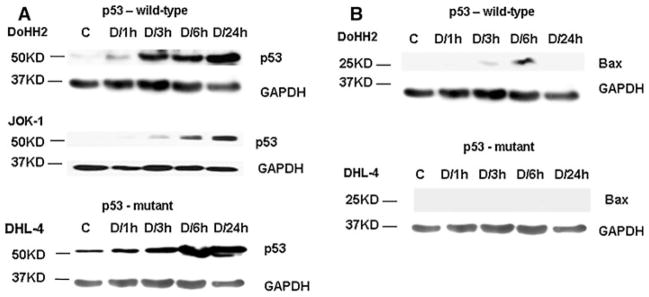
To detect whether p53 is increased upon doxorubicin treatment, lymphoma cells were treated with 200 ng/ml doxorubicin (the concentration is in line with the doses reported in the literature) at four time points within a 24-h period. Figure 1a demonstrates that in the total cell lysate of virtually all the lymphoma cell lines with either wild-type (wt) or mutant p53, doxorubicin treatment increased p53 protein levels starting at 3h, and the increases were sustained at 6 and 24 h (the results from DoHH2, JOK-1, and DHL-4 cells are shown; anti-p53 antibody: FL393). Next, the expression levels of Bax, a p53 target gene, were detected as an indicator of wt p53’s activity. As shown in Fig. 1b, the protein levels of Bax were increased at 3 and 6 h in wt p53-bearing DoHH2 cells, but remained undetectable from 1 to 24 h in mutant p53-containing DHL-4 cells. The results from JOK-1 (wt p53) and Raji (mutant p53) cells show similar trends (data not shown).
Fig. 1.
Western blot analysis of p53 (a) and Bax (b) in B-cell lymphoma cells after doxorubicin (doxorubicin [D], 200 ng/ml) treatment. A representative result was shown. Total cell lysate was used. GAPDH served as the loading control. Control (c), Saline treatment for 24 h
Both wt and mutant p53 translocated to mitochondria upon doxorubicin treatment
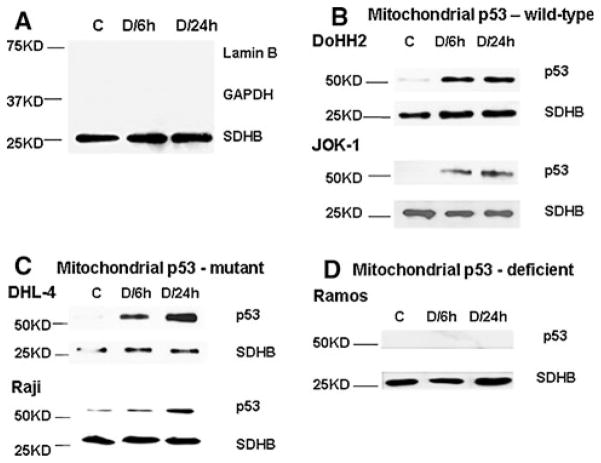
To detect mitochondrial p53, mitochondrial fractions were prepared following the method described previously in the Materials and methods section. The 6 and 24 h post doxorubicin treatments were selected based on the more apparent increases in total p53 expression levels (Fig. 1a).
Figure 2a demonstrates the purity of the isolated mitochondrial fractions (samples from DHL-4 cells were used as an example): as a mitochondrial Complex II component, succinate dehydrogenase subunit B (SDHB) was clearly detectable; whereas, glyceraldehyde-3-phosphate dehydrogenase (GAPDH, a cytoplasmic enzyme) and lamin B (a nuclear protein) were almost undetectable.
Fig. 2.
Western blot analysis of mitochondrial p53. A representative result was shown. (a) Detection of the purity of isolated mitochondrial fractions (samples from DHL-4 cells were used as examples). Lamin B served as a nuclear marker; GAPDH served as a cytoplasmic marker; and SDHB served as a mitochondrial marker. Mitochondrial p53: wild-type in DoHH2 and JOK-1 cells (b); mutant in DHL-4 and Raji cells (c); and deficient in Ramos cells (d) after doxorubicin (doxorubicin [D], 200 ng/ml) treatment. Mitochondrial fractions were used. SDHB served as the loading control. C control, Saline treatment for 24 h
Next, the p53 levels in the mitochondrial fractions from both wt and mutant p53 cells were determined. Figure 2b demonstrates increased mitochondrial levels of wt p53 at both 6 and 24 h post doxorubicin treatment (detected by an antibody for wt p53: Ab5). Similarly, Fig. 2c demonstrates that mutant p53 also translocated to mitochondria upon doxorubicin treatment, as detected using an antibody against mutant p53 (Ab3). As the control, the isolated mitochondria from p53-deficient Ramos cells were used. Not surprisingly, neither wt nor mutant p53 was detectable in Ramos cells (Fig. 2d, only the detection of mutant p53 is shown).
Mutant p53 cells were more resistant to doxorubicin treatment than wt p53 cells
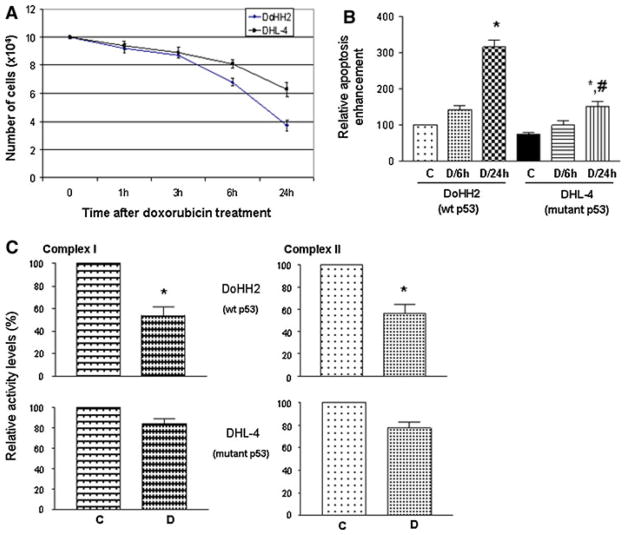
Figure 3a shows that wt p53-bearing DoHH2 cells were more sensitive to doxorubicin-induced cell death than mutant p53-bearing DHL-4 cells. Apparently in DHL-4 cells, lacking the transcription-dependent activities of wt p53 may in part, account for the drug resistance.
Fig. 3.
Growth curve (a) and cell death ELISA analysis (b) of lymphoma cells (DoHH2 and DHL-4) after doxorubicin (D, 200 ng/ml) treatment. c Relative levels of complex I and II activities. DoHH2 and DHL-4 cells were treated with doxorubicin (D, 200 ng/ml) for 24 h. Mitochondrial fractions were then prepared for the activity assays. *P < 0.05 compared with its own control (C, saline treatment for 24 h) group; #P < 0.05 compared with the D/24h group
In addition, the transcription-independent activities of mitochondrial p53 may also play an important role in this event. Compared with early mitochondrial translocation of p53 (at 6 h) shown in Fig. 2, apoptosis (Fig. 3b) did not reach a significant level until 24 h after doxorubicin treatment, suggesting that p53 mitochondrial translocation precedes apoptotic cell death. Consistent with the growth curve, the level of apoptosis in DoHH2 cells was twice (was two times the level seen in DHL-4 cells) of that seen in DHL-4 cells. Similarly, mutant p53-bearing Raji cells also showed resistance to doxorubicin treatment compared to DoHH2 cells (data not shown).
Decreases in mitochondrial complex I and complex II activities were associated with translocation of wt p53
As electron respiration chain components, mitochondrial complex I and II activities were examined in isolated mitochondria from DoHH2 and DHL-4 cells after doxorubicin treatment for 24 h. Mitochondrial samples (protein concentrations were adjusted to 1 μg/μl) were subjected to three fast freeze–thaw cycles in hypotonic buffer before the assay.
As shown in Fig. 3c, there were significant decreases (approximately 50%) in both Complex I and Complex II activities in wt p53-bearing DoHH2 cells at 24 h post doxorubicin treatment; whereas, in mutant p53-containing DHL-4 cells, the activities were decreased slightly (approximately 15%, which was not statistically significant).
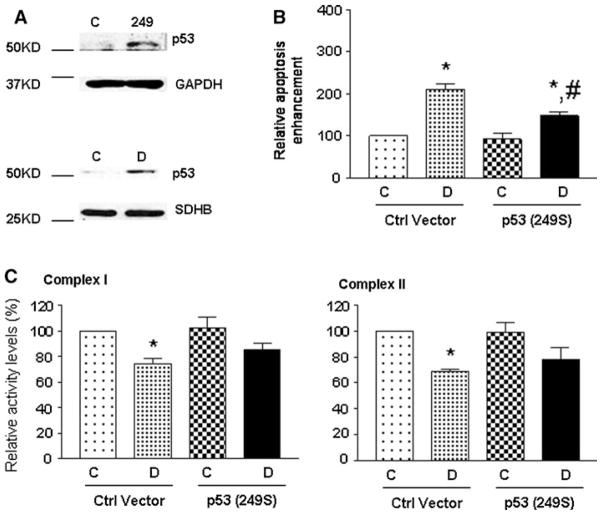
Expression of mutant p53 (p53 249S) endowed Ramos cells resistant to doxorubicin-induced cell death and decreased complex activities
To verify the role of mutant p53 in mitochondrial Complex activities, p53 deficient Ramos cells were transfected with mutant p53 (p53 249S). The expression of mutant p53 was confirmed by Western blot analysis (anti-p53 antibody: FL393), as shown in Fig. 4a. This mutant form of p53 could also translocate to mitochondria after doxorubicin treatment (Fig. 4a, lower panel). Upon doxorubicin treatment, the mutant p53 (249S) transfected cells showed approximately 30% less apoptotic cell death compared to the control vector transfected cells (Fig. 4b). In addition, doxorubicin treatment caused significant decreases in the activities of Complex I and II in the control vector transfected cells, but not in the p53 (249S) transfected cells (Fig. 4c).
Fig. 4.
Mutant p53 (p53 249S) caused chemoresistance in p53-deficient Ramos cells. Western blot analysis of the total cellular and mitochondrial p53 (a). Top panel (total cell lysate): C control (GFP) vector transfected samples; 249: p53 (249S) vector transfected samples. Lower panel (mitochondrial fractions): Control (C), Saline treatment; doxorubicin (D): 200 ng/ml for 24 h. Both were from p53 (249S) transfected samples. (b) Cell death ELISA analysis. Control (C): saline treatment; doxorubicin (D): 200 ng/ml for 24 h. *P < 0.05 compared with own control (C, saline treatment for 24 h) Group; #P < 0.05 compared with the doxorubicin (D) Group. (c) Relative levels of complex I and II activities. Mitochondrial fractions were prepared for the activity assays. Control (C): saline treatment; doxorubicin (D): 200 ng/ml for 24 h. *P < 0.05 compared with own control Group
A small molecule, ellipticine, reactivated mutant p53
Ellipticine has been shown to reactivate mutant p53 functions in a number of cancer cells [23]. Raji cells were treated with ellipticine and/or doxorubicin for 8 h (Raji cells were included in this study based on the fact that the mutated sites of p53 are known [24]). No significant amount of apoptotic cell death was detected in Raji cells after doxorubicin treatment for 8 h, therefore, this time point was chosen to test whether ellipticine might enhance chemotherapy by reactivating mutant p53. Both total cell lysate and mitochondrial fractions were prepared for the study.
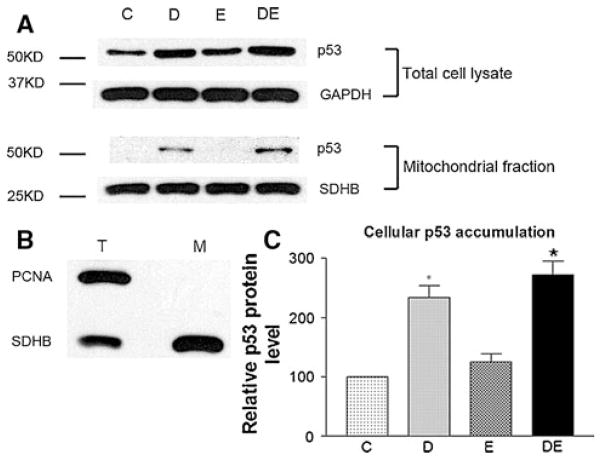
As shown in Fig. 5a, neither total cellular nor mitochondrial p53 was significantly increased after 2.5 μM ellipticine treatment for 8 h. Next, the mitochondrial p53 levels were determined. The purity of the mitochondrial fraction was demonstrated in Fig. 5b, in which no nuclear contamination was detected using PCNA as a nuclear marker. Figure 5c demonstrated that ellipticine alone did not induce a significant increase of the total cellular levels of p53 after doxorubicin treatment; however, it enhanced mitochondrial p53 translocation (Fig. 5a, lower panel).
Fig. 5.
Western blot analysis of p53 in p53-mutant Raji cells after doxorubicin and ellipticine treatment. Control (C): DMSO treatment (ellipticine was dissolved in DMSO); doxorubicin (D): 200 ng/ml; ellipticine (E): 2.5 μM; doxorubicin 200 ng/ml and ellipticine 2.5 μM (DE). Cells were treated for 8 h. A representative result was shown. a p53 levels in total cell lysate and mitochondrial fractions. b Detection of mitochondria purity (revealed by the nuclear marker PCNA and the mitochondrial marker SDHB). GAPDH and SDHB served as the loading controls for total cell lysate (T) and mitochondrial (M) fractions, respectively. c Quantification of p53 expression levels in total cell lysate. *P < 0.01 compared with the control
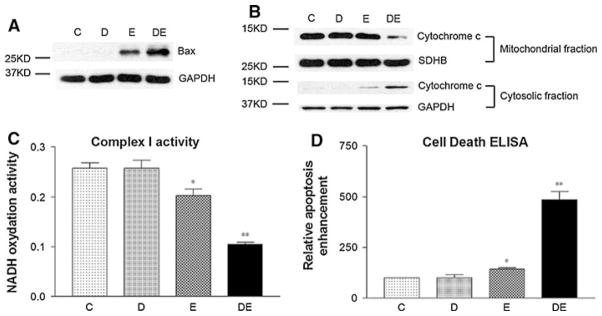
Does ellipticine reactivate mutant p53 in Raji cells? As a p53 target gene, Bax was induced by ellipticine alone, as shown in Fig. 6a. In addition, a slight release of cytochrome c (Fig. 6b), a significant decrease in Complex I activity (Fig. 6c), and a significant increase in apoptosis (Fig. 6d) were all observed by ellipticine treatment, suggesting the reactivation of mutant p53. Moreover, ellipticine sensitized chemoresistant Raji cells to doxorubicin treatment, as demonstrated by a greater increase in Bax expression (Fig. 6a) and cytochrome c release (Fig. 6b), a greater decrease in Complex I activity, and a greater increase in apoptosis (Fig. 6d). As the control, p53 deficient Ramos cells were also treated with the same doses of ellipticine and doxorubicin, and no Bax protein expression was detected by the treatment of ellipticine alone or in combination with doxorubicin (data not shown).
Fig. 6.
Ellipticine reactivated mutant p53 in Raji cells. Control (C): DMSO treatment; doxorubicin (D): 200 ng/ml; ellipticine (E): 2.5 μM; doxorubicin 200 ng/ml and ellipticine 2.5 μM (DE). Cells were treated for 8 h. A representative result was shown. a Bax expression in total cell lysate. b Cytochrome c detected in both mitochondrial and cytosolic fractions. GAPDH and SDHB served as loading controls. c Mitochondrial complex I activity assay. d Apoptosis levels revealed by cell death ELISA. *P < 0.05 compared with the control. **P < 0.001 compared with the control
Discussion
Mitochondrial translocation of wt p53 induced by genotoxic and non-genotoxic reagents has been recognized. Since 50% of human cancers carry mutant p53 [2], whether mutant p53 translocates to mitochondria, thereafter, affecting mitochondrial functions has not been well studied. Solving this puzzle may help in understanding the features of p53 mutations in human cancers, as well as, the role of mitochondria during cancer therapy. There are a large number of established human lymphoma cells, and the status of p53 is known in some of these cell lines, making these cells suitable models for such studies. As a chemotherapeutic drug with a wide anticancer spectrum, doxorubicin is known to induce mitochondrial translocation of wt p53 both in vitro and in vivo. Importantly, it is also used to treat lymphoma. In the mitochondria, wt p53 has been demonstrated to affect mitochondrial energization [25], and regulate mitochondrial respiration [26]. Whether mutant p53 preserves these activities is a very important question to address.
Our studies in human lymphoma cells with doxorubicin treatment demonstrated that p53 could be induced and translocated to mitochondria in lymphoma cells carrying both wt (JOK-1 and DoHH2) and mutant (DHL-4 and Raji) p53. These results suggest that the status of p53 may not affect its mitochondria translocation. In addition, we have observed that γ irradiation emitted from Co60 also induced p53 mitochondrial translocation in JOK-1 cells, whereas no p53 was detected in Ramos cells after γ irradiation (data not shown).
For the next step, more studies are needed to thoroughly demonstrate the role of mutant p53 in the mitochondria. For instance, clinical tissue samples after chemo or radiation-therapy could be obtained to examine the mitochondrial translocation. It will also be important to detect p53 mitochondrial translocation in other human cancers with high p53 mutation rates, e.g., breast and colon cancer [27].
p53 has been shown to regulate mitochondrial respiration through its target genes [26]. In addition, in the mitochondria, wt p53 has been suggested to physically interact with a number of (mitochondrial) proteins including Bcl-2/Bcl-xL [11], DNA polymerase γ [28], nuclear factor A [29], and MnSOD [11]. Our future studies will examine whether mutations in p53 abolish the interaction between p53 and its mitochondrial targets, which may serve as a potential mechanism of how mutant p53 affects mitochondrial functions differently than wt p53.
Mutations in p53 could attenuate or abolish the transcriptional activity of wt p53. Evidence has shown that conformation of the DNA binding domain is flexible and that conformational changes in mutant p53 are reversible, at least to some extent. The best example is the temperature-sensitive p53 mutations that can keep a wt p53 conformation at a permissive temperature, which is mostly lower than the normal body temperature. When the temperature goes up, the wt conformation of the mutant p53 is disrupted [30]. Another supportive phenomenon is that C-terminal peptides and antibodies against the C-terminus can restore wt activity to mutant p53 at least in some cases [31]. Unlike wt p53, mutant p53 is normally expressed at high levels in tumor cells, most likely due to the inability to induce components of p53 negative-feedback loops such as MDM2, Cop1, or Pirh2. Thus, restoration of p53 wt function to the highly accumulated mutant p53 in tumor cells may possibly result in considerable therapeutic responses and has been a long-time effort for treatment of these cancers. The National Cancer Institute has screened a small-molecule library and ellipticine is one of the compounds that show the ability to reactivate p53 target genes and apoptosis [32]. Ellipticine is first known as an inhibitor of the topoisomerase II enzyme [33], and studies have demonstrated that ellipticine could form covalent DNA adducts and reactivate mutant p53 to become transcriptionally active in human cervical, breast, and prostate cancer cells [34, 35]. Altering the mutant p53 conformation by ellipticine has been suggested as a possible mechanism [34]. In our studies, ellipticine alone did not alter the expression levels of p53 (Fig. 5), but induced Bax protein expression and apoptosis in mutant p53-bearing Raji cells (Fig. 6), which is consistent with the previous notion [34].
More importantly, we report a novel finding that, in mutant-p53 lymphoma cells, the combination of ellipticine with a chemotherapeutic agent (e.g., doxorubicin) can induce a more intense apoptotic response than ellipticine alone. Normally, in a cell, the p53 protein is maintained at a low concentration in cytoplasm. In response to various stress signals, p53 is activated and stabilized through post-transcriptional modifications followed by translocation to both nucleus and mitochondria. In the nucleus, p53 functions as a transcription factor and increases the expression of target genes such as Bax, PUMA, Noxa, FAS and Pidd that promote apoptosis. While in the mitochondria, p53 directly interacts with Bcl-2 family members leading to increased cytochrome c release. Thus, the multiple p53 apoptotic actions depend on both correct conformation and localization of p53. Herein, we hypothesize that, in mutant-p53 lymphoma cells, doxorubicin treatment induces p53 cellular accumulation and mitochondrial translocation, whereas, ellipticine shows no apparent effect on p53 protein expression and localization. Given the fact that ellipticine-induced apoptosis is significantly enhanced by doxorubicin co-treatment, we hypothesize, in mutant p53-bearing cells, doxorubicin-based stress signals direct mutant p53 to the exact subcellular position, such as the nucleus and mitochondria, where ellipticine reactivates the mutant p53 activities. It is well known that these two reagents exert their effects on cancer cells via two major mechanisms which result in DNA disruption: (1) intercalation between the bases of DNA and blocks DNA synthesis and transcription; (2) inhibiting the activity of topoisomerase II leading to breaks in the genomic DNA [33–37]. However, in our case, these two reagents showed a cooperative effect and worked synergistically to fully activate the p53-mediated apoptotic pathway. In mutant p53-bearing cells, we did not observe any interactions between mutant p53 and mitochondrial protein such as Bcl-2 and Bax in the presence of ellipticine or doxorubicin (data not shown). The mechanism by which ellipticine alters mutant p53 conformation, as well as, reactivates mutant p53 activities is yet uncertain. Potentially, ellipticine may alter p53’s post-transcriptional modification status, e.g., phosphorylation, via its roles as an inhibitor of Cdc2, topoisomerase II, and cascein kinase 2 [38, 39]. These possibilities need to be explored in future studies.
In summary, our studies demonstrated that in lymphoma cells: (1) doxorubicin treatment increased the protein levels of both wt and mutant p53; (2) both wt and mutant p53 translocated to mitochondria; (3) mutant p53-contaning cells were more resistant to doxorubicin treatment; (4) in wt p53-bearing cells, mitochondrial Complex I and II activities were greatly decreased by doxorubicin treatment, whereas mutant p53-containing cells were not affected significantly; (5) transfection of mutant p53 caused resistance to doxorubicin treatment in p53 deficient Ramos cells; and (6) for the preclinical application of our studies, a small molecule ellipticine reactivated mutant p53 and sensitized chemoresistant lymphoma cells to doxorubicin treatment. These results suggest that mutant p53 can translocate to mitochondria similar to wt p53; however, the effects on mitochondrial functions could be different from wt p53. The data generated from this study may provide novel information regarding the role of mutant p53 in mitochondria, which may contribute to the understanding of the relationship between p53 mutations and drug resistance, helping to design a better strategy for p53-mediated cancer therapy.
Acknowledgments
The authors would like to thank Dr. Mitchell Smith at Fox Chase Cancer Center, Philadelphia, PA, for providing us with human lymphoma B-cell lines DoHH2; and Dr. Heinz Kohler at the University of Kentucky for providing Raji, DHL-4, Romas, and JOK-1 cells. We also would like to thank Rachael Walton (C.E. Byrd High School, Shreveport, LA) and Adrienne Parker (Wiley College, Marshall, TX) for their technical support. This work was supported by Grant Number R03CA128077 from the National Cancer Institute and Grant Number NSF(2010)-PFUND-199 from the Louisiana Board of Regents.
Contributor Information
Fei Wang, Department of Pharmacology, Toxicology & Neuroscience, LSU Health Sciences Center in Shreveport, Shreveport, LA 71130, USA. College of Life Science, Jilin University, Changchun, China.
Jianfeng Liu, Department of Pharmacology, Toxicology & Neuroscience, LSU Health Sciences Center in Shreveport, Shreveport, LA 71130, USA.
Delira Robbins, Department of Pharmacology, Toxicology & Neuroscience, LSU Health Sciences Center in Shreveport, Shreveport, LA 71130, USA.
Kerri Morris, College of Arts and Sciences, University of Louisiana at Monroe, Monroe, LA 71209, USA.
Amos Sit, Department of Pharmacology, Toxicology & Neuroscience, LSU Health Sciences Center in Shreveport, Shreveport, LA 71130, USA.
Yong-Yu Liu, Department of Basic Pharmaceutics Sciences, University of Louisiana at Monroe, Monroe, LA 71209, USA.
Yunfeng Zhao, Email: yzhao1@lsuhsc.edu, Department of Pharmacology, Toxicology & Neuroscience, LSU Health Sciences Center in Shreveport, Shreveport, LA 71130, USA.
References
- 1.Levine JJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, et al. Datebase of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 5.Jia HP. Controversial Chinese gene-therapy drug entering unfamiliar territory. Nat Rev Drug Discov. 2006;4:269–270. doi: 10.1038/nrd2017. [DOI] [PubMed] [Google Scholar]
- 6.Tokino T, Nakamura Y. The role of p53-target genes in human cancer. Crit Rev Oncol Hematol. 2000;33:1–6. doi: 10.1016/s1040-8428(99)00051-7. [DOI] [PubMed] [Google Scholar]
- 7.Moll UM, Zaika A. Nuclear and mitochondrial apoptotic pathways of p53. FEBS Lett. 2001;493:65–69. doi: 10.1016/s0014-5793(01)02284-0. [DOI] [PubMed] [Google Scholar]
- 8.Li PF, Dietz R, von Harsdorf R. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 1999;18:6027–6036. doi: 10.1093/emboj/18.21.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchenko ND, Zaika AI, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 10.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, et al. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 12.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11(3):577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 13.Heyne K, Schmitt K, Mueller D, Armbruester V, Mestres P, et al. Resistance of mitochondrial p53 to dominant inhibition. Mol Cancer. 2008;7:54–70. doi: 10.1186/1476-4598-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, et al. CP-31398 reactivates mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117:3753–3764. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahyar-Roemer M, Fritzsche C, Wagner S, Laue M, Roemer K. Mitochondrial p53 levels parallel total p53 levels independent of stress response in human colorectal carcinoma and glioblastoma cells. Oncogene. 2004;23:6226–6236. doi: 10.1038/sj.onc.1207637. [DOI] [PubMed] [Google Scholar]
- 16.Allman R, Errington RJ, Smith PJ. Delayed expression of apoptosis in human lymphoma cells undergoing low-dose taxol-induced mitotic stress. Br J Cancer. 2003;88:1649–1658. doi: 10.1038/sj.bjc.6600905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao C, Nakajima T, Taya Y, Tsuchida N. Activation of p53 in MDM2-overexpressing cells through phosphorylation. Biochem Biophys Res Commun. 1999;264:860–864. doi: 10.1006/bbrc.1999.1611. [DOI] [PubMed] [Google Scholar]
- 18.Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 1991;88:5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiman KG, Magnusson KP, Ramqvist T, Klein G. Mutant p53 detected in a majority of Burkitt lymphoma cell lines by monoclonal antibody PAb240. Oncogene. 1991;6:1633–1639. [PubMed] [Google Scholar]
- 20.Karpova MB, Sanmun D, Henter JI, Smirnov AF, Fadeel B. Betulinic acid, a natural cytotoxic agent, fails to trigger apoptosis in human Burkitt’s lymphoma-derived B-cell lines. Int J Cancer. 2006;118:246–252. doi: 10.1002/ijc.21311. [DOI] [PubMed] [Google Scholar]
- 21.Yen HC, Oberley TD, Vichitbandha S, Ho YS, St Clair DK. The protective role of manganese superoxide dismutase against adriamycin-induced acute cardiac toxicity in transgenic mice. J Clin Invest. 1996;98:1253–1260. doi: 10.1172/JCI118909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Wang LM, Chaiswing L, Yen HC, Oberley TD, et al. Tamoxifen protects against acute tumor necrosis factor alpha-induced cardiac injury via improving mitochondrial functions. Free Radic Bio Med. 2006;40:1234–1241. doi: 10.1016/j.freeradbiomed.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Bykov VJ, Selivanova G, Wiman KG. Small molecules that reactivate mutant p53. Eur J Cancer. 2003;39:1828–1834. doi: 10.1016/s0959-8049(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 24.Duthu A, Debuire B, Romano J, Ehrhart JC, Fiscella M, et al. p53 mutations in raji cells–characterization and localization relative to other burkitt’s lymphomas. Oncogene. 1992;7:2161–2167. [PubMed] [Google Scholar]
- 25.Donahue RJ, Razmara M, Hoek JB, Knudsen TB. Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. FASEB J. 2001;15:635–644. doi: 10.1096/fj.00-0262com. [DOI] [PubMed] [Google Scholar]
- 26.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 27.Soussi T. p53 alterations in human cancer: more questions than answers. Oncogene. 2007;26:2145–2156. doi: 10.1038/sj.onc.1210280. [DOI] [PubMed] [Google Scholar]
- 28.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, et al. p53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 30.Brown CR, Hong-Brown LQ, Welch WJ. Correcting temperature-sensitive protein folding defects. J Clin Invest. 1997;99:1432–1444. doi: 10.1172/JCI119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selivanova G, Kawasaki T, Ryabchenko L, Wiman KG. Reactivation of mutant p53: a new strategy for cancer therapy. Semin Cancer Biol. 1998;8:369–378. doi: 10.1006/scbi.1998.0099. [DOI] [PubMed] [Google Scholar]
- 32.Sugikawa E, Hosoi T. Mutant p53 mediated induction of cell cycle arrest and apoptosis at G1 phase by 9-hydroxyellipticine. Anticancer Res. 1999;19:3099–3108. [PubMed] [Google Scholar]
- 33.Huff AC, Kreuzer KN. Evidence for a common mechanism of action for antitumor and antibacterial agents that inhibit type II DNA topoisomerases. J Biol Chem. 1990;265:20496–20505. [PubMed] [Google Scholar]
- 34.Peng Y, Li C, Chen L, Sebti S, Chen J. Rescue of mutant p53 transcription function by ellipticine. Oncogene. 2003;22:4478–4487. doi: 10.1038/sj.onc.1206777. [DOI] [PubMed] [Google Scholar]
- 35.Stiborová M, Sejbal J, Borek-Dohalská L, Aimová D, Poljaková J, et al. The anticancer drug ellipticine forms covalent DNA adducts, mediated by human cytochromes P450, through metabolism to 13-hydroxyellipticine and ellipticine N2-oxide. Cancer Res. 2004;64:8374–8380. doi: 10.1158/0008-5472.CAN-04-2202. [DOI] [PubMed] [Google Scholar]
- 36.Fornari FA, Randolph JK, Yalowich JC, Ritke MK, Gewirtz DA. Interference by doxorubicin with DNA unwinding in MCF-7 breast tumor cells. Mol Pharmacol. 1994;45:649–656. [PubMed] [Google Scholar]
- 37.Pigram WJ, Fuller W, Hamilton LD. Stereochemistry of intercalation: interaction of daunomycin with DNA. Nature New Biol. 1972;235:17–19. doi: 10.1038/newbio235017a0. [DOI] [PubMed] [Google Scholar]
- 38.Ohashi M, Sugikawa E, Nakanishi N. Inhibition of p53 protein phosphorylation by 9-hydroxyellipticine: a possible anticancer mechanism. Jpn J Cancer Res. 1995;86:819–827. doi: 10.1111/j.1349-7006.1995.tb03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubert A, Paris S, Piret JP, Ninane N, Raes M, et al. Casein kinase 2 inhibition decreases hypoxia-inducible factor-1 activity under hypoxia through elevated p53 protein level. J Cell Sci. 2006;119:3351–3362. doi: 10.1242/jcs.03069. [DOI] [PubMed] [Google Scholar]