Abstract
Heparanase (HPSE) is a potent pro-tumorigenic, pro-angiogenic and pro-metastatic enzyme that is overexpressed in brain metastatic breast cancer (BMBC). However, little is known about the regulation of this potential therapeutic target in BMBC, which remains very poorly managed in the clinic. We hypothesized HPSE gene expression might be regulated by microRNA that might be exploited therapeutically. Using miRanda and RNAhybrid, we identified miR-1258 as a candidate microRNA that may directly target HPSE and suppress BMBC. In support of our hypothesis, we found that miR-1258 levels inversely correlated with heparanase expression, enzymatic activity, and cancer cell metastatic propensities, being lowest in highly aggressive BMBC cell variants compared to either non-tumorigenic or non-metastatic human mammary epithelial cells. These findings were validated by analyses of miR-1258 and heparanase content in paired clinical specimens of normal mammary gland versus invasive ductal carcinoma, and primary breast cancer versus BMBC. In regulatory experiments, miR-1258 inhibited the expression and activity of heparanase in BMBC cells, whereas modulating heparanase blocked the phenotypic effects of miR-1258. In functional experiments, stable expression of miR-1258 in BMBC cells inhibited heparanase, in vitro cell invasion, and experimental brain metastasis. Together, our findings illustrate how microRNA mechanisms are linked to brain metastatic breast cancer through heparanase control, and they offer a strong rationale to develop heparanase-based therapeutics for treatment of cancer patients with brain metastases, BMBC in particular.
Keywords: MicroRNA, Heparanase, LNA-ISH, Brain metastasis, Breast cancer
Introduction
Brain metastases are the most common malignancy of the central nervous system and their incidence is increasing. This is particularly valid in brain metastatic breast cancer (BMBC), whose patients have, even with best available treatments, only a low (20%) one-year survival rate with considerable morbidity (1). Despite its devastating consequences in patient survival, knowledge of the biology regulating brain metastasis remains fragmentary. Understanding its mechanisms will prompt the development of new therapies to combat this disease.
Heparanase is the dominant endoglycosidase (endo-β-D-glucuronidase) in mammals, cleaving heparan sulfate (HS) to fragments retaining biological activity (2, 3). An established role for heparanase is to release multiple HS/heparin-binding growth and angiogenic factors which are stored in the extracellular matrix, and to regulate their levels and overall potency. Heparanase intervention can thus initiate broad effects that dramatically alter the microenvironment and stimulate tumor cell growth and metastasis (2, 3). Our previous work has implicated heparanase as a molecular determinant of brain metastasis (4–7), with highest levels in brain metastatic cell variants, and also produced by brain endothelial and glial cells, likely working in the latter through feed-back loops to foster tumor cell invasion (5). We have demonstrated that heparanase is a downstream target of EGFR/HER2 signaling in human BMBC cells, translocates to the nucleolus following EGF treatment, and affects the activity of DNA Topoisomerase I and cell proliferation (4). Therefore, altering heparanase expression, localization, and action can provide the opportunity to block multiple pathways which are crucial for tumor cell colonization and their subsequent growth in the brain microenvironment. However, there are gaps in the knowledge of how the molecule is precisely regulated at the gene level.
A growing number of reports indicate that microRNAs (miRNAs) affect tumor progression as oncogenes or tumor suppressor genes (8–12). MiRNAs are single-stranded, noncoding RNA molecules consisting of ~ 22 nucleotides that result from the sequential processing of putative miRNA transcripts by the RNaseIII enzymes Drosha and Dicer (8). A number of miRNAs have been shown to be differentially expressed between normal mammalian gland and breast carcinoma tissues (8–10), however, roles of miRNAs in BMBC have not been deciphered.
The objective of this study was to investigate microRNA - mediated mechanisms that alter invasive and metastatic modalities of BMBC through the downregulation of heparanase. We provide evidence that a specific microRNA - miR-1258 - targets the heparanase gene and has profound effects on heparanase gene expression and function of this molecule in BMBC cell invasion and metastatic profiles.
Materials and Methods
Antibodies and reagents
Rabbit anti-human heparanase polyclonal antibody (1453) was provided by Dr. Israel Vlodavsky (The Bruce Rappaport Faculty of Medicine, Technion, Israel); mouse anti-human heparanase monoclonal antibody was obtained from Cedarlane Laboratories (Burlington, NC); secondary antibodies (goat anti-rabbit IgG [H+L] and goat anti-mouse IgG [H+L]) used for immunohistochemistry and Western blot analyses were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cell invasion assay kits were obtained from Cell Biolabs, Inc. (San Diego, CA). MicroRNA isolation/detection kits were purchased from Ambion (Austin, TX). MicroRNA-1258 mimic and inhibitor (anti-miR-1258) were purchased from Dharmacon (Lafayette, CO). MiRIDAN microRNA mimic is a double-stranded synthetic RNA synthesized and chemically modified using proprietary procedures while the microRNA inhibitor was engineered by proprietary synthetic hairpins to generate an antisense RNA to the mature miR-1258. Both mimic and inhibitor were designed based on miRNA sequences available in miRBase databases (microrna.sanger.ac.uk).
Cell lines and tissue culture
All cell lines were obtained between late 2009 – beginning 2010, and freshly recovered from liquid nitrogen before they were used for indicated experiments (< than 6 months). Brain metastasis-selected MDA-MB-231BR cell variants (231BR1 and 231BR3) were obtained from Dr. Janet Price (Department of Cancer Biology, The University of Texas MD Anderson Cancer Center, Houston, TX). The 231BR clones are the result of sequential cycles of injection of 231 parental cells into the internal carotid artery of nude mice with increased propensities to form brain metastasis in these animals (231BR3 > 231BR2 > 231BR1) (13). These clones were recently obtained at early passage and tested (2010) for consistent in vivo abilities to metastasize to brain. Cells were cultured in Dulbecco’s Modified Eagle Medium plus F12 (DMEM/F12) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). HMEC and MCF-10A cells (obtained in 2010 from Dr. Thomas Westbrook and the Cell Core facility of the Department of Molecular and Cellular Biology – BCM, respectively) were cultured in mammary epithelial basal medium (MEBM) containing 10% FBS and 10 ng/ml of human EGF, 5 μg/ml of insulin, and 1 μg/ml of hydrocortisone. SUM-225 and SUM-149 cells (obtained in 2010 from Drs. Daniel Medina and Robert Weinberg’s laboratories, respectively) were cultured in Ham’s/F12 medium supplemented with 5% calf serum, 5 μg/ml of insulin and 1 μg/ml of hydrocortisone. All cells were cultured in a humidified, 5% CO2 incubator at 37°C, tested every month for Mycoplasma contamination, and used only at low passage and if Mycoplasma negative (2009 – 2010). All cell lines were regularly examined by microscopy for phenotypic changes before their use.
Human BMBC tissues
Thirteen consecutive cases of paired (normal mammary gland/invasive ductal carcinoma, and primary/brain metastatic breast carcinoma) human tissue specimens were employed for miR-1258 and HPSE expression analyses. These cases were identified from 1991 to 2009 at a single institution (UCLA Medical Center, Los Angeles, CA). The presence of adjacent benign mammary tissue, primary breast carcinoma, and BMBC was confirmed by review of the hematoxylin and eosin (H & E) stained slides by a breast pathologist (PS). All patients had brain metastases morphologically similar to their breast primary and no additional cancer history. All primary and metastatic tumors were invasive ductal carcinomas characterized morphologically by cohesive sheets and nests of malignant epithelial cells, some with occasional tubule formation, moderate to severe nuclear pleomorphism, conspicuous mitoses, and scant to moderate eosinophilic cytoplasm. Adjacent brain parenchymal tissue was distinguished from BMBC by the presence of glial cells with fine elongated cytoplasmic processes and indistinct cell boundaries within a delicate fibrillary background. Benign breast parenchyma was distinguished from primary breast carcinoma by the presence of morphologically bland columnar cells with an underlying myoepithelial cell layer confined to intact terminal duct lobular units and larger ductal structures. Archived, formalin-fixed, and paraffin-embedded tissue blocks were serially sectioned at 4 micron intervals and placed on glass slides for miR-1258 and HPSE expression analyses. An additional H&E slide was prepared so that the presence of invasive carcinoma, adjacent benign mammary tissue, metastatic carcinoma, and adjacent brain parenchyma could be verified.
Prediction of miR-1258 target site of human heparanase 3′-UTR
Using the miRanda software (14) for target prediction (www.microrna.org) and sequence analyses, we identified hsa-miR-1258 as a miRNA that targets HPSE 3′-UTR. The sequence of miR-1258 was extracted from miRBase 14.0 and folded in the context of the target site identified by miRanda including flanking 3 nts upstream and 15 nts downstream. MiR-1258 was found to bind HPSE with a minimum free energy of < - 21.4 kcal/mol, predicting the generation of a highly stable duplex.
LNA-based miRNA in situ hybridization
Locked nucleic acid - in situ hybridization (LNA-ISH) is a novel technology for the detection of miRNA levels in formalin-fixed, paraffin-embedded human cancer tissues, possessing a superior sensitivity and excellent specificity [Allen Gene Expression Atlas Development (www.brainmap.org)] (15, 16). MiR-1258 probes for LNA-ISH were synthesized by Exiqon (Odense M, Denmark). Probes include: 1) specific hsa-miR-1258 (5′-TTCCACGACCTAATCCTAACT-3′); 2) positive control U6 (5′-CACGAATTTGCGTGTCATCCTT-3′); 3) negative control scrambled-miR (5′-GTGTAACACGTCTATACGCCCA-3′). MiR-1258 levels were examined and compared between paired normal human mammary gland versus invasive ductal carcinoma, and primary versus brain metastatic breast cancer tissues. Hybridized, complementary probes were detected by a catalyzed reporter deposition using biotinylated tyramide followed by colorimetric detection of biotin with an avidin-alkaline phosphatase conjugate (16). The latter yields a dark blue precipitate whose abundance is directly proportional to the number of detected miRNA molecules. Relative microRNA levels were determined using the Celldetekt protocol (16) whose software assigns a value of 9 for strong (indicated by red color), 4 for moderate (blue), and 1 for weak (yellow) signal staining intensity, respectively. Composite values were then normalized on the basis of total signal in scanned fields and data were analyzed for statistical significance. LNA-ISH assays and data interpretation were performed by the RNA Core Facility at Baylor College of Medicine (Houston, TX).
Luciferase reporter assays
The plasmids containing wild-type (Cat. # HmiT000974-MT01) or mutant (Cat. # CS-HmiT000974-MT01) Luc-HPSE 3′-UTR were specifically synthesized (GeneCopoeia, Germantown, MD) and used in luciferase reporter assays. These plasmids contain Firefly luciferase that is fused to the 3′-UTR of human HPSE, and Renilla luciferase which functions as a tracking gene. Luciferase activity assays were performed following manufacturer’s protocols. Briefly, 293T cells were seeded in 6-well plates, co-transduced with the lentiviral constructs containing miR-1258 and wild-type or mutated HPSE 3′-UTR. Following transduction, cells were split into 12-well plates, harvested after 24 hr, then Firefly and Renilla luciferase activities were measured sequentially using the dual luciferase assay kit (GeneCopoeia). Results were expressed as relative luciferase activity units by a plate reader (POLAR Star Omega, BMG LabTech, Chicago, IL). The activities were normalized with Renilla luciferase. Results represent three independent experiments, and each performed in triplicate.
TaqMan miRNA Assays
Quantitative real-time PCR was performed to assess miR-1258 levels using TaqMan Real-Time PCR Assay (Applied Biosystems Inc., Foster City, CA). Specific probes for hsa-miR-1258 (I.D. # 4427975, ABI) and endogenous control RNU44 (I.D. # 4373384, ABI) were used for PCR-based detection of mature miR-1258. Real-time RT-PCR based detection was achieved using a mirVana microRNA detection kit (Ambion, Austin, TX) following kit instructions. In brief, the assay includes two steps: stem-loop RT and real-time PCR. MicroRNAs were isolated using the mirVana miRNA kit (Ambion, Austin, TX). Stem-loop RT primers bind to at the 3′ portion of miRNA molecules and are transcribed with reverse transcriptase using a kit (Cat # 4366596, ABI). Then, the product was quantified using conventional TaqMan PCR that included a miR-1258 specific forward primer: (5′-CTGCGAGTCCCTGGAGTTAG-3′); reverse primer (5′-CGGTCCCCTA-ACTACCCATT-3′); and SYBR – labeled TaqMan probes for miR-1258 and RNU44 control levels. PCR assays were performed using an I-Cycler detector (Bio-Rad Laboratories Inc., Hercules, CA). Parameters were: 48°C for 30 min, then 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec, and 60°C for 60 sec. Each assay was performed in triplicate. PCR results were recorded as threshold cycle numbers (Ct), normalized against RNU44, and expressed as fold change.
Immunohistochemistry (IHC)
Heparanase expression was examined by IHC using formalin-fixed, paraffin-embedded human and mouse brain tissues. Anti-HPSE monoclonal antibody (Cedarlane) was used at 1:3,000 dilution, and incubated overnight (16 hr) at 4°C, followed by incubation with biotinylated anti-mouse IgG (H+L) for 30 min at room temperature (25°C). Staining was performed in parallel and by using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA), per manufacturer’s instructions. Then, stained and coded sections were reviewed by pathologists blinded to study groups. Staining of heparanase was distributed through a 0 – 3+ intensity scoring scale: 0 corresponded to background negative staining; 1+, weak staining; 2+, moderate staining; 3+, strongest staining. Heparanase positive cases were those that possessed an intensity score of ≥ 1, as pathologically assessed (Table 1, supplement).
Transduction, transient transfection, and rescue assays
Lentiviral constructs containing miR-1258 (Cat. # HmiR0620-MR01) or scrambled miRNA (Cat. # CmiR0001-MR01) were purchased from GeneCopoeia. Precursor miRNA expression constructs were prepared in a feline immunodeficiency virus (FIV) based lentiviral vector system (pEZX-MR01) with the neomycin selection gene under the control of the CMV promoter. 293T cells at 70–80% confluency were infected with lentiviral constructs, and then particles were collected, and transduced into BMBC 231BR3 cells at a multiplicity of infection equal to one, per manufacturer’s protocol. Transient transfections were performed as previously described (4). Rescue experiments were accomplished using OmicsLinks™ expression clone vector expressing heparanase without 3′-UTR (EX-Z6097-M09, GeneCopoeia™) co-transfected with miR-1258 or scrambled contructs into 231BR3 cells. Clones of EX-Z6097-M09 expressing heparanase or empty vector were first transfected into 231BR3 cells, and then miR-1258 or scrambled constructs were introduced into cells 48 hour later. Transfection efficiency was monitored by GFP imaging and examined by Western blotting. Selected clones for cell invasion and experimental metastasis assays carrying miR-1258 or scrambled and HPSE or vector were then used in chemoinvasion assays, and injected into mice as described below. After 6 weeks, animals were sacrificed and mouse brains examined for metastases and HPSE content.
Heparanase expression and activity assays
Cells were serum-starved overnight (16 hr), then transfected (miR-1258 mimic and/or inhibitor) or transduced (lentiviral constructs containing either miR-1258 or scrambled-miRNA) following manufacturer’s protocols. Cell lysates were collected and examined for heparanase expression by Western blot analyses, and heparanase activity was examined using the heparan sulfate degrading enzyme assay kit (TakaRa Mirus.Bio, Madison, WI), as previously reported (4).
Cell invasion assays
Chemoinvasion analyses were performed following indicated treatments and using corresponding cell invasion kits (Cell Biolabs Inc., San Diego, CA), (4).
Experimental in vivo BMBC assays
Female athymic nude mice (nu/nu, 4–5 weeks old) were purchased from Harlan Sprague Dawley, Inc. (Indianapolis, IN), and maintained at the accredited animal facility of Baylor College of Medicine (BCM). All studies were conducted according to NIH animal use guidelines and a protocol approved by the BCM Animal Care Committee. To obtain stable cell lines expressing miR-1258 or scrambled-miRNA, lentiviral constructs were prepared in 293T cells and transduced into BMBC 231BR3 cells. After 48 hr of cell transduction, heparanase expression status was examined by Western blot analyses. 231BR3 cells transduced with lentiviral constructs carrying either miR-1258 or scrambled control (n = 30 mice per treatment group) were injected intracardiacally into nude mice (0.5 × 106 cells per mouse) after they were anesthesized with isofluorane. Mice were euthanized by CO2 asphyxiation when they showed signs of neurological impairment (usually 5–6 weeks), the whole lungs and brains removed, and fixed in Bouin’s solution. Serial sections, obtained by cutting every 300 μm of tissue, were analyzed by H&E staining for presence of brain metastatic lesions. Presence of brain metastasis was confirmed by pathologists blinded to experimental groups. Brain metastases were then counted under a Zeiss microscope (100 – 400× magnification). Three independent experiments were performed, data were pooled and analyzed for statistical significance.
Statistical analyses
All data were analyzed using ANOVA or Student’s t test, and represent the mean ± standard deviation. A p value less than 0.05 was considered statistically significant. Statistical tests were performed with SAS statistical software (version 9.1; SAS Institute, Cary, NC).
Results
MiR-1258 directly targets the heparanase gene and downregulates heparanase protein
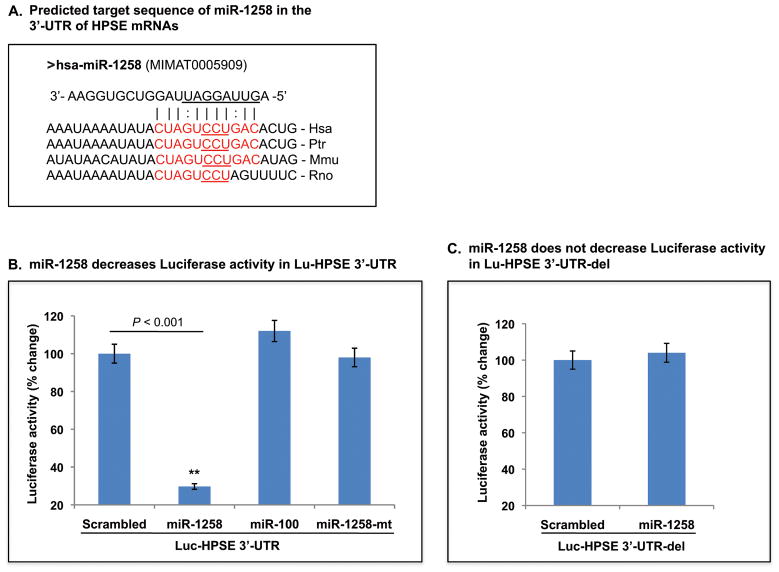
MiR-1258 maps on human chromosome 2 (180433516 - 180434172 - www.genome.ucsc.edu) within an intron of the zinc finger protein 385B isoform 1 gene. MiR-1258 is transcribed in the antisense orientation to the gene. The 3′-UTR of the human heparanase gene (hpse) is unusually short extending only 27 nucleotides following the stop codon. Consequently, there is a dearth of miRNA binding sites that can be targeted for post-transcriptional regulation. We identified miR-1258 as a miRNA candidate predicted to target HPSE using the miRanda software and RNAhybrid programs (14). The putative miR-1258 target site in the 3′-UTR of HPSE shows a near-perfect match to nucleotides 2–12 of miR-1258, including the complete 5′-seed region. The miR-1258 target site is also highly conserved between human (Hsa), chimpanzee (Ptr) and mouse (Mmu) (Fig. 1A). To determine if miR-1258 acts directly on HPSE 3′-UTR, we conducted a series of luciferase reporter assays by co-transfection of miR-1258 (wild type/mutant) and luciferase constructs containing HPSE 3′-UTR (wild type/mutant). Luciferase activity was decreased by miR-1258 approximately 70% compared to scrambled control, and neither a miR-1258 mutant or an alternate microRNA (miR-100) decreased activity in the Luc-HPSE 3′-UTR constructs (Fig. 1B). Moreover, co-transduction of miR-1258 with a luciferase construct containing the 5′-CCU-3′ deletion mutant (Luc-HPSE 3′-UTR-del) failed to decrease luciferase activity (Fig. 1C).
Figure 1. MiR-1258 directly targets heparanase.
A, Alignment of miR-1258 with HPSE 3′-UTRs. Complementary sequences of miR-1258 to mammalian HPSE 3′-UTRs are shaded red. The underlined sequence (5′-CCU-3′) in the human HPSE mRNA and common to other mammalian heparanases, denotes a deletion in the construct carrying Luc-HPSE 3′-UTR-del. Seed sequences of miR-1258 are underlined. Hsa = human; Ptr = pan troglotydes; Mmu = mus musculus; Rno = rat. B and C, Effect of miR-1258 (wild type and mutant) on HPSE 3′-UTR luciferase reporters. Constructs carrying Luc-HPSE 3′-UTR (B) or Luc-HPSE 3′-UTR-del (C) were transfected in 293T cells with indicated miRNAs. MiR-100 expressed in a lentiviral construct was used as an additional control for miR-1258 specificity. After transfections (24 hr), cells were harvested and luciferase activity assays were performed. Bars represent the mean and standard deviation of three independent experiments. See Materials and Methods for additional details.
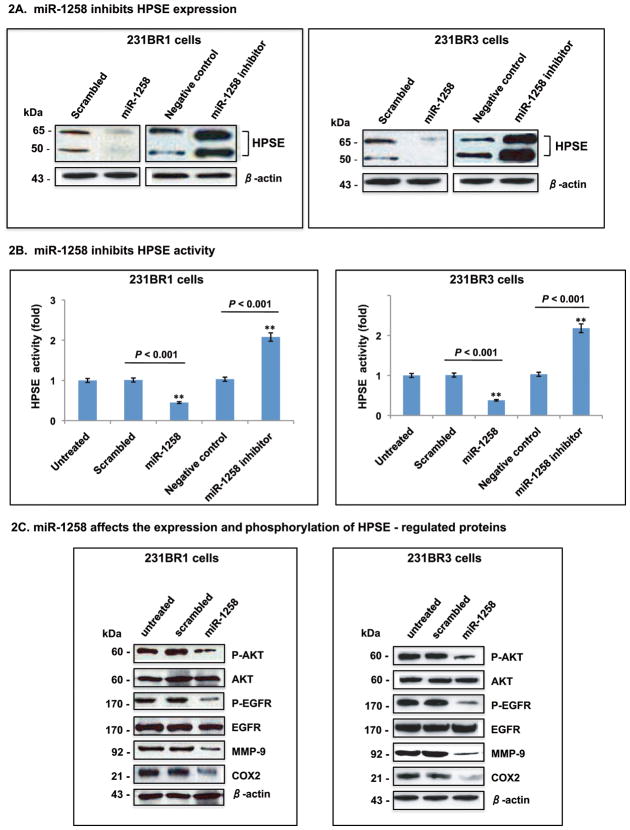
Next, to determine whether miR-1258 functionally affected endogenous HPSE expression in BMBC cells, we transiently transfected miR-1258 mimic, miR-1258 inhibitor, or negative control oligo into 231BR3 and SUM-149 cells, and studied heparanase expression and activity which were reduced by miR-1258 mimic, but augmented by miR-1258 inhibitor (Figs. 2A and 2B). Findings were further verified by using lentiviral constructs containing miR-1258 or scrambled control (Figs. 2A and 2B). In addition, we investigated whether the miR-1258 mediated inhibition of HPSE affected heparanase pathways. We focused on Akt signaling and key heparanase regulated targets - MMP-9, COX2, and EGFR (3, 17, 18). Following miR-1258 cell transduction, phosphorylation levels of Akt and EGFR were greatly reduced, along with MMP-9 and COX2 protein expression levels (Fig. 2C). Thus, we conclude that miR-1258 directly targets heparanase and may affect BMBC cell invasion and metastases through HPSE – targeted proteins and Akt signaling.
Figure 2. MiR-1258 inhibits heparanase expression and activity in BMBC cells.
BMBC cells (231BR3 and 231BR1) were transiently transfected with anti-miR-1258 and negative control (NC), or stably transduced with lentiviral constructs expressing wild type miR-1258 or scrambled miR (scrambled). After 48 hr, cell lysates were prepared and examined simultaneously using Western blotting and the TakaRa heparan sulfate degrading enzyme assay kit (4). HPSE protein and enzyme activity in 231BR1 and 231BR3 cells were shown as (A) and (B). β-actin was used as a loading control. Bars represent the mean and standard deviation of three independent experiments. C, Effect of miR-1258 - mediated inhibition of HPSE on heparanase - regulated proteins. Expression clones carrying miR-1258 or scrambled-miR (scrambled) were transduced into 231BR3 or 231BR1 cells using same procedures as (A) and (B), then cell lysates were examined for protein expression using antibodies indicated. Three independent experiments were performed, and representative analyses are shown.
MiR-1258 expression is attenuated in human breast cancer cells and patient tissues
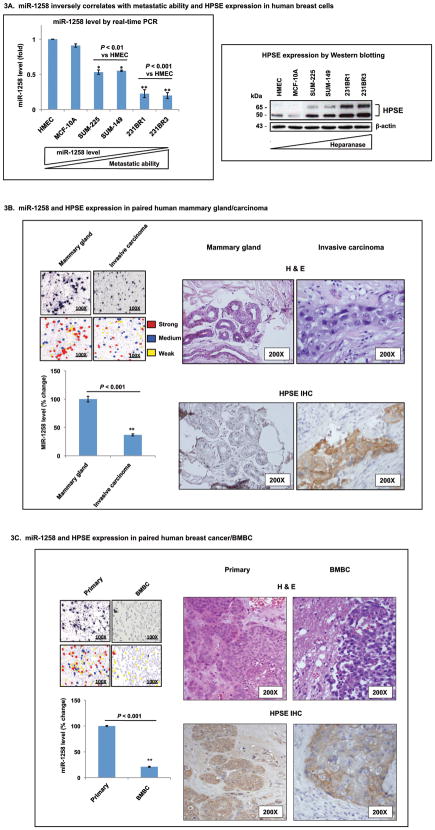
To examine patterns of miR-1258 expression in human breast cancer cells, we selected six human breast-derived cell lines that were classified into three groups according to their metastatic propensities: 1) non-tumorigenic HMEC and MCF-10A; 2) tumorigenic but non-metastatic SUM-149 and SUM-225; 3) BMBC 231BR1 and 231BR3 cells. Basal expression levels of miR-1258 were significantly reduced in BMBC cell lines compared to either non-metastatic (but tumorigenic) or immortalized (but non-tumorigenic) cells. Of note, 231BR1 and 231BR3 cells exhibited approximately 8-fold reduction of miR-1258 by TaqMan PCR compared to primary human mammary epithelial cells (HMEC) (Fig. 3A, top). To assess whether a reduced miR-1258 level correlates with heparanase expression, we examined HPSE levels in these cell lines and detected that heparanase expression was inversely related with miR-1258, with highest levels in brain metastatic 231BR3 and 231BR1 cells (Fig. 3A, bottom). Next, we investigated whether mechanisms involving miR-1258 play a role in BMBC progression. First, we designed LNA-miR-1258 probes for in situ hybridization (LNA-ISH) in paired clinical tissues - normal mammary gland versus invasive ductal breast carcinoma, or primary versus BMBC. Subsequent to LNA-ISH, we detected a reduction of miR-1258 levels by approximately 60% in invasive ductal carcinoma, and 80% in BMBC, respectively, compared to levels in corresponding normal or primary tissues (Figs. 3B and 3C). Then, we performed heparanase staining (by IHC) in both tumor and adjacent tissues in paired tissues. We detected the presence of HPSE staining positivity in neoplastic epithelial cells while the mesenchymal stroma of the tumor and adjacent non-neoplastic tissues in the mammary gland were both negative. Images of IHC and corresponding H&E staining of these areas are shown (Figs. 3B and 3C). While we were able to determine the generality of progressive downregulation of miR-1258 expression from normal, invasive primary, brain metastatic tissues, and statistical significance, we could not achieve a complete correlation with the up-regulation of HPSE expression in same tissues (Figs. 3B and 3C). Our data are however still of significance because they align well with several reports demonstrating that heparanase can also affect primary breast cancer growth as a potent pro-tumorigenic in addition to a pro-metastatic function (19).
Figure 3. MiR-1258 levels inversely correlate with metastatic ability of human breast cell lines and patient tissues.
A, miR-1258 analyses in breast cell lines with differing metastatic propensities. (Top) miR-1258 levels were examined in six breast cell lines by TaqMan RT-PCR and data shown by fold change compared to human mammary epithelial cells (HMEC). RNU44 was used as endogenous control. (Bottom) Differential HPSE expression in same cell lines was examined by Western blotting. Based on their tumorigenic and metastatic potential, cell lines were classified into three groups: 1) non-tumorigenic HMEC and MCF-10A; 2) tumorigenic but non-metastatic SUM-149 and SUM-225; 3) BMBC 231BR1 and 231BR3 cells. β-actin was used as a control for equal loading. B and C, miR-1258 analyses in paired mammary gland versus invasive ductal carcinoma (n = 5 paired cases), or primary versus BMBC (n = 8 paired cases) patient tissues were completed using locked nucleic acid in situ hybridization (LNA-ISH). Hybridized, complementary probes were detected by a catalyzed reporter deposition using biotinylated tyramide followed by colorimetric detection of biotin with an avidin-alkaline phosphatase conjugate (Left) Representative LNA-ISH images and quantification of miR-1258 signal intensity in the normal mammary gland versus invasive ductal carcinoma, and primary versus brain metastatic breast cancer (BMBC) were shown. Graph depicts relative miR-1258 expression in indicated tissues. While high miR-1258 levels were detected in the epithelial component of normal mammary gland. Each group represents the average of total signals in examined fields (n = 10) (right). Heparanase expression in paired cases of normal mammary gland/invasive ductal carcinoma, and primary/BMBC was examined by IHC (HPSE IHC) (left). See Materials and Methods for additional details.
MiR-1258 inhibits BMBC cell invasion and brain metastasis by directly targeting HPSE
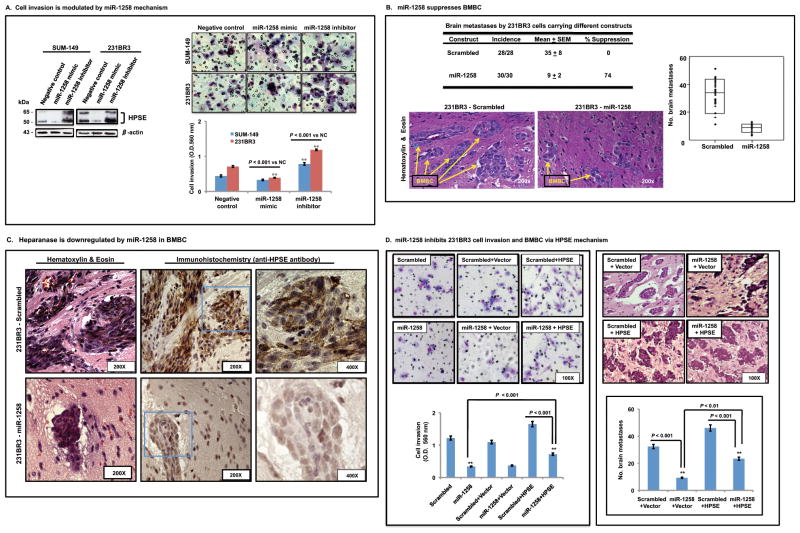
To evaluate miR-1258 functionality, we examined effects of miR-1258 on HPSE content and cell invasion. Downregulation of heparanase expression (Fig. 4A, left) was reflected by a corresponding modulation of cell invasion: decreased by miR-1258 and increased by miR-1258 inhibitor in both SUM-149 and 231BR3 cells. Significant changes (miR-1258 inhibitor vs. negative control, NC) were detected in both SUM-149 and 231BR3 cell lines, especially in highly brain metastatic 231BR3 cells overexpressing HPSE (Fig 4A). Moreover, stable miR-1258 expression significantly reduced the number of BMBC by more than 3-fold, as well as size of metastatic tumors compared to scrambled controls (Figs. 4B and 4C). Neoplastic cells that metastasize to the brain were identified by H&E staining, and HPSE expression levels in tumors were determined by IHC analyses (Fig. 4C). Brain H&E sections from all mice used in this study were examined for presence and number of brain metastases using a standard microscopy (Zeiss microscope, 200× magnification). Representative images are shown (Fig. 4B).
Figure 4. MiR-1258 inhibits BMBC cell invasion and brain metastasis by HPSE mechanism.
A, miR-1258 downregulates heparanase. Cells (SUM-149, 231BR3) were transfected with miR-1258, anti-miR-1258 (inhibitor), and negative controls (NC) followed by Western blotting to detect HPSE expression (left), and miR-1258 inhibits BMBC cell invasion (right). Chemoinvasion analyses using Matrigel™ chambers were performed in parallel with Western blotting analyses for HPSE expression. Representative images of chamber inserts are shown. Cell invasive values were quantified (O.D. 560 nm; n = 10) following manufacturer’s instruction as described in Materials and Methods. B, miR-1258 suppresses brain metastasis. 231BR3 cells stably expressing miR-1258 or scrambled-miR (scrambled) were injected intracardiacally into female nude mice (0.5 × 106 cells per mouse, 30 mice per group). After 6 weeks, mice were sacrificed, brains harvested, and analyzed for metastatic tumors. (Top) Incidence, and mean number of brain metastases in each group. (Middle) Hematoxylin and eosin (H&E) visualization of BMBC and its suppression by miR-1258 compared to scrambeled. (Bottom) Combined data (n = 3 assays) are shown graphically with dots representing the number of brain metastases from each mouse; the box represents the 10th and 90th percentile; and the black line in each box is the mean for each group. C, Heparanase expression was examined by IHC in experimental BMBC, and representatives IHC and H&E sections are shown (n = 4). D, Rescue experiments show that introduction of HPSE-expressing clone blocked miR-1258 – mediated effects on BMBC in vitro cell invasion and in vivo brain metastases. Shown are representative images visualizing brain metastases in animals (H & E staining; n = 10 mice per group, 3 group per assay). Bars represent the mean and standard deviation of two independent experiments. See Materials and Methods for additional details.
Since HPSE is functionally modulated by miR-1258, we performed rescue experiments to confirm that HPSE is required for miR-1258 effects on BMBC cell invasion and metastasis. MiR-1258-mediated changes in phenotype (decreased invasion or metastasis) could be reversed by the introduction of HPSE- expressing clones (without 3′-UTR), compared to control empty vector (Fig. 4D). Furthermore, we found that an enhanced invasive phenotype by HPSE could be reversed by miR-1258 treatment, and accompanied with morphology changes in BMBC cells by invasion assays, and a significant modulation of the number of brain metastases (Fig. 4D). These data further support that miR-1258 directly targets HPSE, which in turn cause the suppression of cell invasion and brain metastasis.
Discussion
We provide first-time evidence demonstrating that the ectopic miR-1258 negatively regulates heparanase expression and activity, and results in the inhibition of BMBC cell invasion and onset of brain metastasis. Our findings indicate that miR-1258 is a suppressor of brain metastatic breast cancer.
The notion that heparanase plays important roles in cancer progression at multiple steps, such as cell proliferation, angiogenesis, invasion, and metastasis, is well established (2–7). However, precise molecular mechanisms responsible for heparanase regulation have not been fully elucidated. Although heparanase modulation using RNAi or antisense-mediated suppression has been previously reported (2, 3, 20), it is challenging to maintain functional concentrations of the siRNA or antisense in vivo since the cells would try to recover the HPSE status. Therefore, the development of miRNA-based approaches regulating heparanase is critical and of potential therapeutic value.
In present study, we show that miR-1258 exerts its roles in BMBC cells by inhibiting heparanase expression and activity, and does so by directly targeting HPSE 3′-UTR (Figs. 1 and 2). To determine whether miR-1258 plays a role in breast cancer metastases, notably to brain, six human breast cell lines were selected and examined for miR-1258 and heparanase expression. An inverse correlation was demonstrated: that decrease of miR-1258 associated with an increase of HPSE content and correlated with metastatic abilities of these cell lines (Fig. 3A). Moreover, miR-1258 downregulated heparanase expression and activity (Figs. 2 and 4), resulting in inhibition of cell invasion (Fig. 4A) and brain metastases in xenografts (Figs. 4B–D).
To investigate the clinical relevance of these findings, we extended the above studies to clinical patient samples by using paired - normal mammary gland versus invasive ductal carcinoma, and primary versus BMBC tissues. MiR-1258 expression was assessed by using unique locked-nucleic acid (LNA) miR-1258 probes which were specifically designed to detect discriminatory microRNA levels in formalin-fixed paraffin-embedded tissues by subsequent LNA-miR-1258 in situ hybridization (15). MiR-1258 levels were significantly reduced in both of human primary and brain metastatic breast cancer tissues (Figs. 3B and 3C, left panels), which imply that miR-1258 may be involved in breast cancer progression. Interestingly, we found that heparanase staining positivity was strong in some primary or invasive carcinomas with HPSE IHC intensity comparable to cases of brain metastatic breast cancer (Figs. 3B and 3C, right panels). These results were not entirely unexpected, considering that several reports have demonstrated that heparanase can also affect primary breast cancer growth (19). The discrepancy observed between miR-1258 and HPSE expression in primary tissues can be explained by the extensive tumor heterogeneity observed in these tissues, and the likely presence of tumor cells which may have poor, if not absent, invasive capabilities. Our focus was directed towards miR-1258 functional studies since it is a novel miRNA involved in cancer progression, particular in miR-1258 effects on mechanisms of brain metastasis, beyond its incidence (13). The decreased number of brain metastasis by miR-1258 treatment, but same incidence, can be explained by mechanisms of cross-talk between tumor and normal cells of the brain microenvironment in response to a brain metastatic insult; one of which is the production of glia heparanase (5) not inhibited by miR-1258 since targeting only HPSE endogenously expressed in neoplastic cells. Others may involve factors, pathways and proteases acting independently from the miR-1258/HPSE axis, and implicated in breast cancer brain colonization (1).
We investigated mechanistic insights by which miR-1258 - mediated HPSE affect BMBC cell invasion and brain metastasis. It has been reported that HPSE regulates the expression of many molecules keenly involved in angiogenesis and metastasis, including MMP-9, COX2, and EGFR (3, 17, 18). We have shown that treatment of miR-1258 resulted in changes of MMP-9, COX2, and EGFR protein expression (Figs. 2C). Moreover, by transducing lentiviral vectors expressing miR-1258 or scrambled control into 231BR3 cells, HPSE inhibition by miR-1258 not only decreased MMP-9 and COX2 protein levels (immunofluorescence assays confirmed these results; not shown), but also the phosphorylation of Akt and EGFR (Fig. 2C). To validate that HPSE is required for miR-1258 roles in vitro and in vivo, we performed rescue experiments which show that decreased cell invasive values and numbers of brain metastasis by miR-1258 treatment could be reversed by employing an expression vector containing human HPSE. However, this did not completely restore cell invasion and number of brain metastasis (Fig. 4D), implying that other targets might contribute to BMBC onset and progression (1). Based upon our analyses using the miRanda program, potential additional targets by miR-1258 include MMP-16, FOXO3, and JUN, all involved in neoplastic events. Although these targets showed weaker affinities and lower complementarities in respective seed sequences, they might play roles, at least partially, in regulatory networks leading to BMBC.
In summary, we have identified a link between miR-1258 and heparanase, which represents a novel mechanism of endogenous regulation of this molecule known to support metastasis. Our investigations introduce new concepts that brain metastasis might be attacked by microRNA-based strategies, and can have profound implications for the development of heparanase-based therapeutics in brain metastasis in general, brain metastatic breast cancer in particular.
Supplementary Material
Acknowledgments
We would like to thank Drs. Israel Vlodavsky (The Bruce Rappaport Faculty of Medicine, Technion, Israel) for the gift of rabbit anti-HPSE polyclonal antibody, Janet Price (University of Texas M.D. Anderson Cancer Center, Houston, TX) for providing MDA-MB-231BR1 and MDA-MB-231BR3 cell lines, Robert A. Weinberg (MIT Ludwig Center for Molecular Oncology, Cambridge, MA) for providing SUM-149 cell lines, Thomas Westbrook (Department of Biochemistry and Molecular Biology - BCM) for HMEC cells, and Daniel Medina (Department of Molecular and Cellular Biology -BCM) for the SUM-225 cell line. We thank Drs. Martin Matzuk and Zhifeng Yu for providing the miR-100 lentiviral construct and valuable suggestions. We also thank Drs. Dandan He and Diego Florentin (Department of Pathology and Immunology - BCM) for their assistance in the animal surgeries and tissue preparation. Finally, we thank Dr. Anita Chandler (Department of Molecular Physiology and Biophysics - BCM) and Michael Wetzel of our laboaratory for their expert editorial assistance, and Dr. Joel Neilson (Department of Molecular Physiology and Biophysics - BCM) for valuable suggestions in microRNA technologies and sequence information.
This study was supported by NIH grant 2R01 CA086832 – 07 to D.M.
Abbreviations
- BMBC
brain metastatic breast cancer
- COX2
cyclooxigenase2
- GFP
green fluorescent protein
- HPSE
heparanase
- miRNA
microRNA
- HS
heparan sulfate
- LNA-ISH
locked nucleic acid - in situ hybridization
- MMP-9
matrix metalloprotease-9
- RT-PCR
reverse transcriptase - polymerase chain reaction
- 3′-UTR
3′-untranslated region
Footnotes
Author contributions: L.Z. designed and performed experiments, analyzed data, and wrote the manuscript; P.S.S. provided patient samples and clinical information. J.G. provided pathologic assessment of tumor metastasis in pre-clinical and clinical models, and quantitative analysis for HPSE expression in human tissues examined. P.G. identified the miR-1258 targets and provided microRNA expertise to this project and revised the manuscript. D.M. designed experiments, reviewed the manuscript, and supervised the research as principal investigator. Authors disclosed no conflicts of interest.
Authors declare no competing conflicts of interests.
References
- 1.Palmieri D, Chambers AF, Felding-Habermann B, Huang S, Steeg P. The biology of metastasis to a sanctuary site. Clin Cancer Res. 2007;13(6):1656–62. doi: 10.1158/1078-0432.CCR-06-2659. [DOI] [PubMed] [Google Scholar]
- 2.Ilan N, Elkin M, Vlodavsky I. Regulation, function, and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38 (12):2018–39. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Zcharia E, Jia J, Zhang S, Baraz L, Lindahl U, Peretz T, Vlodavsky I, Li JP. Newly generated heparanase knockout mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS ONE. 2008;4(4):e5181, 1–13. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Sullivan P, Suyama J, Marchetti D. EGF - induced heparanase nucleolar localization augments DNA Topoisomerase I activity in brain metastatic breast cancer. Molecular Cancer Res. 2010;8(2):278–90. doi: 10.1158/1541-7786.MCR-09-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchetti D, Li J, Shen R. Astrocytes contribute to the brain-metastatic specificity of melanoma cell by producing heparanase. Cancer Res. 2000;60:4767–70. [PubMed] [Google Scholar]
- 6.Murry BP, Blust BE, Singh A, Foster TP, Marchetti D. Heparanase mechanisms of melanoma metastatic to the brain: Development and use of a brain slice model. J Cell Biochem. 2006;97:217–25. doi: 10.1002/jcb.20714. [DOI] [PubMed] [Google Scholar]
- 7.Marchetti D, Nicolson G. Human heparanase: A molecular determinant of brain metastasis. Adv Enzyme Reg. 2001;41:343–59. doi: 10.1016/s0065-2571(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. New Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 9.Tavazoie SF, Alarco C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Richardson A, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin1. Cancer Res. 2010;70(1):378–87. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM, Anderson ML. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70(5):1906–15. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim LS, Huang S, Lu W, Chelouche D, Price J. Vascular endothelial growth factor expression promotes the growth of breast cancer brain metastasis in nude mice. Clin & Exp Met. 2004;21:107–18. doi: 10.1023/b:clin.0000024761.00373.55. [DOI] [PubMed] [Google Scholar]
- 14.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNA in animal embryos using LNA - modified oligonucleotide probes. Nat Methods. 2006;3(1):27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 16.Carson JP, Eichele G, Chiu W. A method for automated detection of gene expression required for the establishment of a digital transcriptome-wide gene expression atlas. J Microsc. 2005;217:275–81. doi: 10.1111/j.1365-2818.2005.01450.x. [DOI] [PubMed] [Google Scholar]
- 17.Imada T, Matsuoka J, Nobuhisa T, et al. COX-2 induction by heparanase in the progression of breast cancer. Int J Mol Med. 2006;17(2):221–28. [PubMed] [Google Scholar]
- 18.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, IIan N. Heparanase auguments epidermal growth factor receptor phosphorylation: Correlation with head and neck tumor progression. Cancer Res. 2008;68 (24):10077–85. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen I, Pappo O, Elkin M, et al. Heparanase prompts growth, angiogenesis, and survival of primary breast tumors. Int J Cancer. 2006;118:1609–17. doi: 10.1002/ijc.21552. [DOI] [PubMed] [Google Scholar]
- 20.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–30. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.