Abstract
Diamond Blackfan anemia (DBA) is a lineage-selective inherited bone-marrow failure syndrome characterized primarily by anemia and physical malformations. Recent advances in identifying the genetic abnormalities underlying DBA have demonstrated involvement of genes encoding both large (RPL) and small (RPS) ribosomal subunit proteins, including mutations of RPL5, RPL11, RPL35A, RPS7, RPS10, RPS17, RPS19, RPS24, and RPS26 in 50–60% of affected patients. Despite significant progress, identification of gene abnormalities in the remaining patients remains an important question since present data suggests that mutations in other members of the ribosomal protein gene complement do not explain those cases without an identified genetic lesion in these genes. Genetic studies have also raised new questions with the recognition of substantial variability in the manifestations of DBA, ranging from ribosomal protein mutations in otherwise asymptomatic individuals to those with classic severe red-cell aplasia with characteristic malformations, at times within the same kindred. In this review, we summarize the genetic basis of DBA and discuss mechanisms by which the phenotype of DBA might be modified.
Introduction
Understanding of the genetic basis of DBA has evolved rapidly in the past decade, with the pace of identification of new DBA-related genes markedly accelerating in the past several years. Aside from the difficulties of keeping current with and assimilating a burgeoning lexicon of affected small and large ribosomal protein genes, there have been significant developments in understanding what constitutes DBA, based in part on these genetic studies. However, the rapid pace of recent gene discoveries belies a tenuous understanding of the fundamental connections between these specific genetic events and the many clinical features of DBA. Genetic discovery in DBA has progressed in a process comparable to pulling the end of a line to unroll a ball of twine: work so far has exposed a good length of twine; however, a large ball still remains, and in places there are tangled knots where none were initially obvious. In this article, we review the current literature regarding the genetics of DBA, explore current information of how genotype may influence phenotype in DBA, and review some mechanisms by which allelic and non-allelic factors may modify the phenotype in DBA.
Tugging the String: Gene Discovery in DBA
The first significant breakthrough in defining the genetic basis for DBA developed from the identification of a child with a t(X;19) balanced reciprocal translocation.1 This finding was followed by polymorphic marker linkage studies localizing a critical region in 29 multiplex families (i.e., families with multiple affected members) to 19q13 and defining a critical region based on 3 probands with microdeletions involving 19q13.2.2, 3 A ribosomal protein (r-protein) gene, RPS19, was subsequently implicated as the causative gene by positional cloning of the 19q13 breakpoint from the t(X;19) index patient and confirmed by the finding of RPS19 mutations in 10 of 40 additional patients, including 6 multiplex families in whom RPS19 mutations segregated with a clinical DBA phenotype.4 Since this initial report, the proportion of patients in whom DBA is attributable to coding sequence mutations in RPS19 has been consistently estimated at around 25% in numerous studies from case series as well as national DBA registries (Table 1).4–15 Interestingly, a lower frequency of RPS19 mutations was recently reported in a Japanese cohort of DBA patients (5/45 probands, 11%),15 suggesting the possibility of racial or ethnic differences in the frequency of DBA mutations, an area that has not yet been well explored.
Table 1.
Ribosomal protein gene involvement in DBA.
| Ribosomal Subunit | Gene | % Mutation (mutations/screened)* | 95 % CI | Mutation Types | Origin Country(ies)/Group | Reference |
|---|---|---|---|---|---|---|
| Small | RPS19 | 25% (10/40) | 14–38% | Genomic Del/rearrangement, Frame shifting In/Del, Premature stop, Splice disruption, Initiator codon disruption, a.a. substitutions | Europe and North Afica | 4 |
| 17% (4/24) | 6–36% | Northern Europe | 5 | |||
| 24% (42/172) | 18–31% | Europe and North Africa | 6 | |||
| 21%(10/48) | 15–42% | SHIP Working Group DBA (France) | 10 | |||
| 25% (5/20) | 10–47% | Czech DBA Registry | 7 | |||
| 25% (24/97) | 17–34% | AIEOP (Italy) | 11 | |||
| 15% (16/104) | 10–24% | UK DBA Registry | 13 | |||
| 24% (19/81) | 15–34% | USA and Europe | 12 | |||
| 11% (5/45) | 4–24% | Japan | 15 | |||
| RPS24 | <2% (3/215) | 0–4% | Premature stop, Splice disruption, a.a. substitution, a.a. Del, Initiator codon disruption | USA and Europe | 22 | |
| ~2%(2/92) | 0–8% | Italy | 31 | |||
| NR (0/45) | 0–9% | Japan | 15 | |||
| Patient report | - | Sweden | 24 | |||
| Patient report | - | European | 25 | |||
| RPS17 | <2% (1/24) | 0–22% | Initiator codon disruption, Frame shifting In/Del | Czech DBA Registry | 26 | |
| <1% (1/193) | 0–3% | USA and Europe | 27 | |||
| ~2% (1/45) | 0–13% | Japan | 15 | |||
| RPS26 | 10% (12/117) | 6–17% | Frame shifting In/Del, Initiator disruption, a.a. substitutions, Splice disruption | USA and Europe | 32 | |
| RPS10 | 4% (5/117) | 2–10% | Frame shifting In/Del, Premature stop, Initiator disruption | USA and Europe | 32 | |
| RPS7 | <1% (1/159) | 0–4% | Splice disruption | USA and Europe | 27 | |
| Large | RPL35A | ~3% (5/148) | 1–8% | Genomic Del, Splice disruption, a.a. substitution | USA and Europe | 29 |
| NR (0/92) | 0–5% | Italian | 31 | |||
| NR (0/45) | 0–9% | Japan | 15 | |||
| RPL5 | 9% (18/196) | 6–14% | Frame shifting In/Del, Premature stop, a.a. substitution, Initiator codon disruption, Splice disruption | USA and Europe | 27 | |
| 21% (6/28) | 10–40% | Czech DBA Registry | 30 | |||
| 13% (12/92) | 7–22% | Italy | 31 | |||
| 9% (4/45) | 3–21% | Japan | 15 | |||
| RPL11 | 7% (13/196) | 4–11% | Frame shifting In/Del, Premature stop, a.a. deletion, Splice disruption | USA and Europe | 27 | |
| 7% (2/28) | 0–24% | Czech DBA Registry | 30 | |||
| 13% (12/92) | 7–22% | Italy | 31 | |||
| ~4% (2/45) | 0–16% | Japan | 15 |
Frequency data are not included for single patient and small series reports. 95% CI - 95th percentile confidence interval by modified Wald method.
NR - None reported, a.a. - amino acid, In - insertion, Del - deletion
At the time of writing, more than 120 unique RPS19 alterations have been cataloged, ranging from genomic deletions, single base substitutions resulting in both nonsense and missense mutations, splicing consensus changes, and small insertions or deletions causing predominantly nonsense changes (www.dbagenes.unito.it, accessed January 2011).16 It was unclear how mutations or deletions of a structural constituent of a ubiquitous cellular component, the ribosome, could lead to such a distinct and fairly limited phenotype of erythroid insufficiency and physical developmental abnormalities. Since extra-ribosomal functions have been demonstrated for a number of ribosomal proteins, one hypothesis was that an unidentified erythroid-sensitive extra-ribosomal function of RPS19 might underlie the disorder. With the functional studies that would ultimately demonstrate deleterious effects on ribosomal assembly in RPS19-mutated DBA samples just underway,17–21 it was additional genetic data that suggested the ribosome was indeed the center of attention in DBA.
Another small subunit r-protein gene, RPS24, was identified as a DBA gene by sequencing of candidate r-protein genes contained in a region of chromosome 10 that was one of 3 regions identified by microarray-based linkage study of a single large multiplex family.22 As with RPS19, the informative finding from this family was confirmed by identifying additional heterozygous RPS24 mutations in unrelated probands as well as by segregation of the mutant alleles with the DBA phenotype in the index and in an unrelated family. Loss of RPS24 has since been shown to disrupt 18S ribosomal RNA (rRNA) processing in model systems and in DBA patient specimens.23 All of the mutations identified in RPS24 are nonsense truncations or disrupt the initiator codon and thus are predicted to result in functional haploinsufficiency. In contrast to RPS19, RPS24 mutations in DBA appear uncommon, with only 3 mutations initially identified from a group of 215 probands screened (about 2%) and presently only 5 unique mutations reported in the literature (Table 1).22, 24, 25 The focus on small ribosomal subunit genes intensified after a single RPS17 mutation was reported from a candidate screen for other small subunit r-protein genes in a group of 24 Czech DBA patients.26 Subsequent studies have identified 3 additional RPS17 mutations: two single mutations found in cohorts of 19827 and 45 probands15 and a case report of mutation in a single patient.28 All of the reported RPS17 mutations are heterozygous, disrupting allelic expression with 2 different mutations of the initiator codon and two small deletions that result in frame shifting mutations (Table 1).
Recognition of another cytogenetic abnormality in an unusual DBA patient led to the discovery of the first large subunit r-protein abnormality in DBA. Heterozygous mutations of RPL35A, identified as a candidate gene based on a child with a terminal deletion of 3q, were found in 3 DBA probands along with a second heterozygous 3q deletion involving RPL35A in an unrelated patient.29 Segregation of a mutated allele was demonstrated in one multiplex family, and a processing abnormality of the large subunit rRNA was demonstrated in patient samples. DBA due to RPL35A alterations is also uncommon (~3%) with 2 deletions and 3 mutations reported in the literature (Table 1). With both subunits of the ribosome now definitively involved, and more comprehensive candidate gene sequencing studies underway, the pace of gene discovery quickened, and the number of DBA-related genes has increased dramatically.
Disruption of large subunit pre-rRNA processing distinctive from that caused by RPL35A defects was shown in conjunction with patient mutations of RPL5 and RPL11.27 Mutations of these r-proteins in DBA have provided the most concrete genotype-phenotype correlation in DBA, appear at higher frequency than RPS24, RPS17 or RPL35A mutations, and have also been confirmed in additional DBA cohorts (Table 1).15, 27, 30, 31 In 2009, Gazda and colleagues reported completion of re-sequencing of 80 r-protein genes, adding mutations of three additional small subunit genes, RPS7, RPS10 and RPS26, to the growing list of DBA genes, with the latter occurring in comparatively high frequency (Table 1).32, 33 Patient samples with mutations in these genes also demonstrate disruptions in rRNA processing, demonstration of which has become a de facto requirement for confirmation of the pathogenicity of novel r-protein changes in DBA patients, particularly in sporadic cases where familial analysis is not informative.
A summary of the reported frequencies and confidence intervals of r-protein gene mutations are shown in Table 1. It should be emphasized that multiple levels of evidence associating a sequence change with a pathogenic mutation have been used in validating these DBA genes: 1) identification of recurrent gene alterations in unrelated probands, 2) segregation of sequence abnormalities in affected (both classical and non-classical) family members, 3a) demonstration of nonsense or premature stop codon mutations, and/or 3b) careful evaluations of normal r-protein gene sequences in the case of amino acid substitutions, and 5) demonstration of abnormal rRNA processing within tissues of patients harboring a sequence change. With the important caveats of cautious interpretation of the broad confidence intervals associated with frequency estimates derived from rare alterations and small sample sizes as well as recognition 7 of a paucity of representation of some racial and ethnic populations in the literature, a general pattern of mutational frequency in DBA as currently understood can be summarized: uncharacterized > RPS19 > {RPL5, RPL11, RPS26} > { RPL35A, RPS7, RPS10, RPS17, RPS24}, with r-protein gene abnormalities identifiable in about 50–60% of patients.
In contrast to cases where a pathogenic mutation can be confidently inferred from functional analysis of rRNA processing or careful evaluation of multiplex families demonstrating segregation, there are several r-protein genes in which a missense substitution or deletion of uncertain significance has been identified. These include, RPS8, RPS15, RPS27A, RPL3, RPL7, RPL9, RPL14, RPL19, RPL23A, RPL26, RPL35 and RPL36.27, 32–34 Due to the rarity of these changes, the ultimate significance of the alterations observed in these genes is currently unclear; however, with an estimated frequency of abnormalities at <1% for each individual gene, none appear likely to account for a substantial fraction of uncharacterized DBA mutations. Following the implication of haploinsufficiency of RPS14 as the causative gene in 5q− myelodysplastic syndrome (MDS), mutation screening in DBA patients failed to identify abnormalities, suggesting RPS14 does not account for a significant fraction of DBA mutations.31, 32, 35 However, the parallels between DBA and 5q− MDS are noteworthy, with refractory anemia and marrow erythroid hypoplasia as major clinical manifestations of the latter, and point to a broader role for dysfunctional ribosome synthesis in abnormal hematopoiesis and cancer predisposition. The recent identification and treatment of a patient initially diagnosed with DBA in whom a 5q− deletion was subsequently reported illustrates both the clinical overlap and importance of distinguishing these syndromes.36
Several points worth emphasis are clear from analysis of RPS19 mutations and appear to hold in all of the DBA genes subsequently identified. The definition of a mutation in most cases is based on a sequence change not seen in a cohort of normal controls, leading to a predicted change at the mRNA or protein level that could clearly lead to a deleterious effect. Not surprisingly given the scope of genes studied, the majority have been screened with focus on coding exons and splicing boundaries; with the notable exception of RPS19, detailed sequencing studies of regulatory regions, as well as reporting of synonymous coding changes and intronic single nucleotide variants (SNVs) is sparse. Based both on the substantial portion of the unexplained DBA cases and the phenotypic variability not attributable to a primary mutation, the assumptions underlying these initial screening strategies may require re-evaluation.
Importantly, all r-protein gene changes identified to date are heterozygous changes that preserve an intact allele. Bi-allelic inactivation of critical ribosomal constituents is believed to be incompatible with embryonic development in humans, and pre-implantation embryonic lethality has been demonstrated in homozygous RPS19 knock-out mice.37 A related point is that the majority of RPS19 mutations have been demonstrated,12 or are predicted, to result in functional haploinsufficiency at the mRNA level either by allelic loss, nonsense-mediated or nonstop mRNA decay.14, 38 This is true for all of the subsequently identified gene mutations. In RPS19, about 25% of mutations result in single amino acid substitutions or deletions in RPS19 protein. Some of these alterations appear to cause protein-level haploinsufficiency due to failure of translational initiation, protein instability and/or failure of nucleolar localization.18, 39, 40 The remaining amino acid substitutions cluster predominantly in functional domains important to RPS19 folding or RPS19 interaction with pre-ribosomes.41 Additionally, a potential translational regulatory function for RPS19-RPS19 mRNA binding has been reported, with alterations of this interaction seen in mutant RPS19, suggesting the possibility that in some RPS19 mutations, aberrant RPS19 protein/mRNA interactions may also disrupt translational regulation at a wild-type RPS19 allele.42 No similarly detailed structure-function relationship analysis has been reported for the remaining DBA-related r-protein genes. Unlike RPS19, though, nearly all mutations in the remaining genes cause premature termination, splicing disruption or frame shifting. Interestingly, initiator codon changes make up a disproportionate fraction of the few missense mutations identified. Of 14 unique missense mutations reported in the DBA mutation database in RPS7, RPS10, RPS17, RPS24, RPS26, RPL5, RPL11, and RPL35A, 10 result in substitutions of the initiator methionine, lending further support to the notion that functional haploinsufficiency of an r-protein gene product leads to DBA.16 In addition to mutational studies suggesting haploinsufficiency as the critical deficiency in DBA, the observed mode of inheritance in cases where a gene mutation is identifiable is consistent with autosomal dominant transmission with variable penetrance.43 Though nearly 1/3 of pedigrees from registry data as well as isolated case reports are potentially consistent with autosomal recessive inheritance, one such example is attributable to gonadal mosaicism while no instance of recessive inheritance has yet been confirmed.6, 7, 9, 11–13, 44
Based on their experiences, Gazda and colleagues estimated that about 45% of DBA cases from their cohort could not be explained by coding sequence variation in r-protein genes.32, 33 Similar results were seen in an analysis of a partially overlapping cohort of North American DBA registry patients that evaluated 77 r-protein and r-protein like genes in which 40% of patients lacked detectable r-protein coding sequence abnormalities45. What are the additional genetic lesions in DBA? It is presently unclear what extent deletions of r-protein loci may contribute to DBA. A weakness of the sequencing methods for r-protein mutations in DBA that have been reported is failure to detect copy number variation (e.g., allelic loss). Quarello and colleagues identified 1 larger (~3 Mb) and 2 small (<1 Mb) deletions in 92 RPS19 wild-type probands by screening of SNP haplotypes followed by PCR-based copy number detection.46 This suggests that methods used to detect small deletions may be important complements in genetic testing for DBA. Indeed, recent reports indicate that inclusion of strategies to detect either small (exon to gene level) or larger deletions involving confirmed DBA genes identifies a fraction, though still not all, of patients in whom mutational analysis is uninformative.34, 47, 48 Beyond the r-protein genes, based on the finding that abnormal ribosome assembly underlies all of the known DBA gene abnormalities identified, any of an estimated 200 non-ribosomal protein ribosome assembly factors could be considered potential gene candidates. Hence, gene discovery studies in DBA remain an active area of investigation.
Confronting the knots: new insights and new problems in genotype-phenotype correlations
One of the most confounding problems in DBA research has been the lack of clear correlation between genotype and phenotype. This may be partly explained by the relatively small numbers of mutations in r-protein genes other than RPS19 as well as a relatively large fraction of unknown mutations. However, with the advent of RPS19 testing, it became clear that no distinctive clinical phenotype could reliably differentiate RPS19-mutated from non-RPS19-mutated DBA.6, 13, 49 Even more surprising was the observation of substantial phenotypic diversity in patients harboring similar mutations and even within families sharing identical gene mutations. Prior to the identification of RPS19 involvement in DBA, familial DBA cases ascertained by family history were estimated to account for 10–15% of all DBA patients; currently, careful analysis for subtle hematological manifestations (elevated erythrocyte adenosine deaminase (eADA), unexplained macrocytosis and mutation testing suggest the fraction of familial cases approaches 40%.6, 13, 50
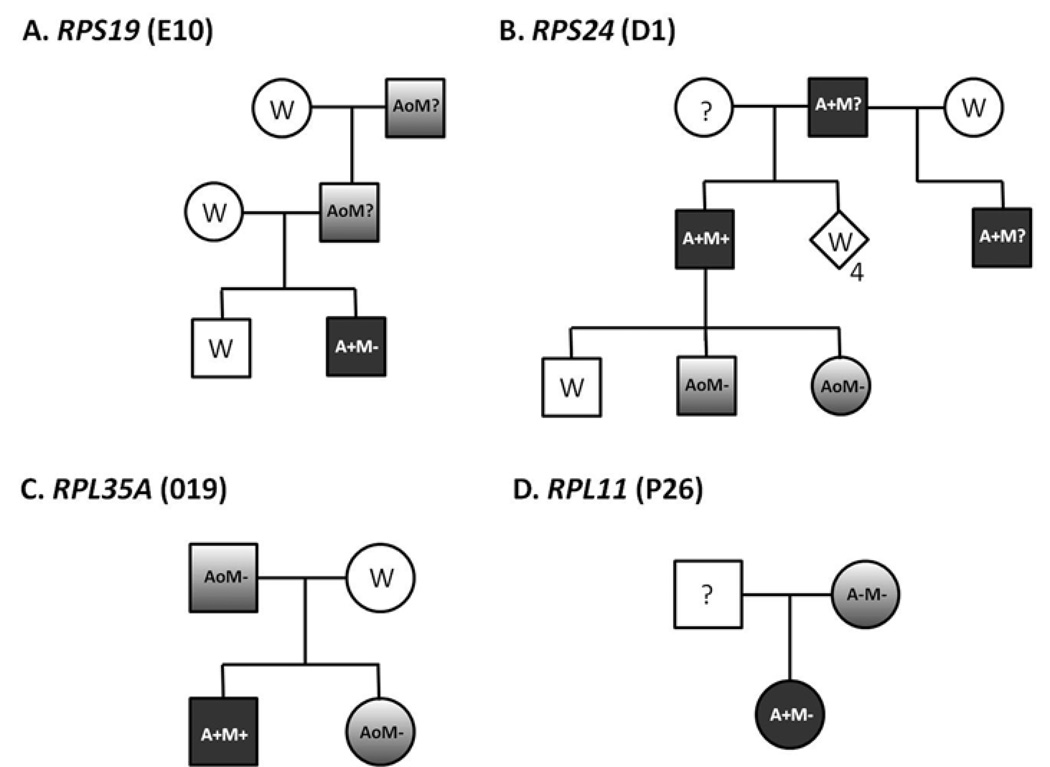
The most dramatic examples of the failure of genotype to predict phenotype are clearly evident when considering the intra-familial variability observed in family members sharing a known RPS19 mutation as illustrated in Figure 1a.5, 6, 8, 50 This phenotypic variability appears to be a general feature of r-protein mutations in DBA as similar examples of subclinical hematologic abnormalities, variable congenital abnormalities, and/or non-penetrance have been reported in RPS24, RPL35A, RPL5, and RPL11 families (Figure 1b–d). This finding strongly suggests the importance of initially unrecognized and as yet unidentified genetic modifiers to an individual's DBA phenotype.22, 27, 29 In part as a response to recognition of the substantial clinical heterogeneity and variable penetrance of r-protein mutations in DBA, recent international consensus recommendations for diagnosis and treatment of DBA suggest consideration of the diagnosis of "non-classical" DBA in any family member sharing an established DBA r-protein mutation (regardless of phenotype) or without an identifiable mutation but with a compatible physical anomaly and/or hematologic abnormalities including macrocytosis or elevated eADA.43 Table 2 presents some of the phenotypic characteristics common in classical and non-classical DBA. These findings underscore the critical importance of careful physical and hematologic evaluation of family members and genotyping when a causative mutation can be identified to provide accurate reproductive counseling and for careful donor selection when stem-cell transplantation is contemplated.
Figure 1. Examples of phenotypic variability in DBA families with r-protein mutations.
Variability in clinical expression of shared r-protein gene muations in a) RPS19,6 b) RPS24,22 c) RPL35A,29 and d) RPL11.27 The patient designation in the primary reference is indicated in parentheses. Open symbols represent phenotype- and genotype- normal individuals (W) or phenotypically normal without reported genotype data (?). Filled symbols represent affected individuals with classic DBA findings including anemia requiring transfusion or steroid therapy with or without malformation. Graded symbols represent non-classical or incomplete DBA phenotype sharing an r-protein mutation with family members. Symbol codes: A: anemia (−: none, o: only laboratory abnormalities, +: requiring treatment) M: malformation (−: none, ?: not reported, +: present). Subscript below symbols represents multiple individuals.
Table 2.
Phenotypic Features in Classic and Non-Classic DBA
| Category | Features | Classic DBA | Non-Classic DBA | |
|---|---|---|---|---|
| Anemia | Always | Variable | ||
| Age at presentation | Median 2 months, most prior to 1 year | May be incidental finding or identified during evaluation of affected relative | ||
| Elevated hematologic indices | MCV, eADA, HbF | Usually | Often, but may also be normal | |
| Steroid responsiveness | ~50% | |||
| Spontaneous Remission | ~20%52 | |||
| Physical Anomaly | Craniofacial | Hypertelorism, microcephaly, flattened nasal bridge, low hairline, microcephaly, cleft palate, micrognathia, microtia, epicanthus, congenital glaucoma | Single anomaly present in 40–50%, Multiple in ~25% | Occasionally reported |
| Musculoskeletal | Triphalayngeal or bifid thumb, thenar hypoplasia, syndactyly, webbed neck, vertebral abnormalities | |||
| Cardiovascular | Septal defects, coartcation, complex | |||
| Ophthalmalogic | Congenital glaucoma, microophthalmia | |||
| Genitourinary | Hypospadias, renal agenesis, horseshoe kidney | |||
| Cancer risk | Leukemia, lymphoma, osteogenic sarcoma, others | Elevated; under study | Unknown | |
Despite the fundamental quandary in correlating a molecular genotype with clinical phenotype, several important correlations are emerging with the accumulation of genotype data from registry-based gene discovery studies. For instance, recent reports have demonstrated interesting correlations between specific gene defects and presence or type of congenital malformations. The most striking is an association observed between the presence of oral cleft abnormalities and mutations of RPL5. Craniofacial abnormalities are among the most common congenital abnormalities in DBA, occurring in approximately 30% of patients. Oropharyngeal clefts are much less common, with reported frequencies ranging from 3–6%.51, 52 In 2008, Gazda and colleagues described the association of congenital abnormalities in 14 of 20 RPL5-mutated patients, with cleft lip and/or cleft palate identified in 9 of 14 patients with anomalies. They also noted a higher than expected proportion of physical anomalies (12 of 18) in RPL11-mutated patients.27 No oral clefts were found in 8 RPL5 mutated Czech patients, though all had congenital abnormalities.30 Six patients of 12 with RPL5 and 2 of 12 with RPL11 mutations in an Italian population had cleft palate.31 In contrast, no cleft lip/palate abnormalities have been reported in the >120 RPS19 mutated patients for whom phenotype data is reported in the DBA mutation database.16 In addition to the association of RPL5 with clefts, each of these studies demonstrate a higher overall frequency of occurrence of malformations in RPL5 and RPL11 than is typical of the DBA population.
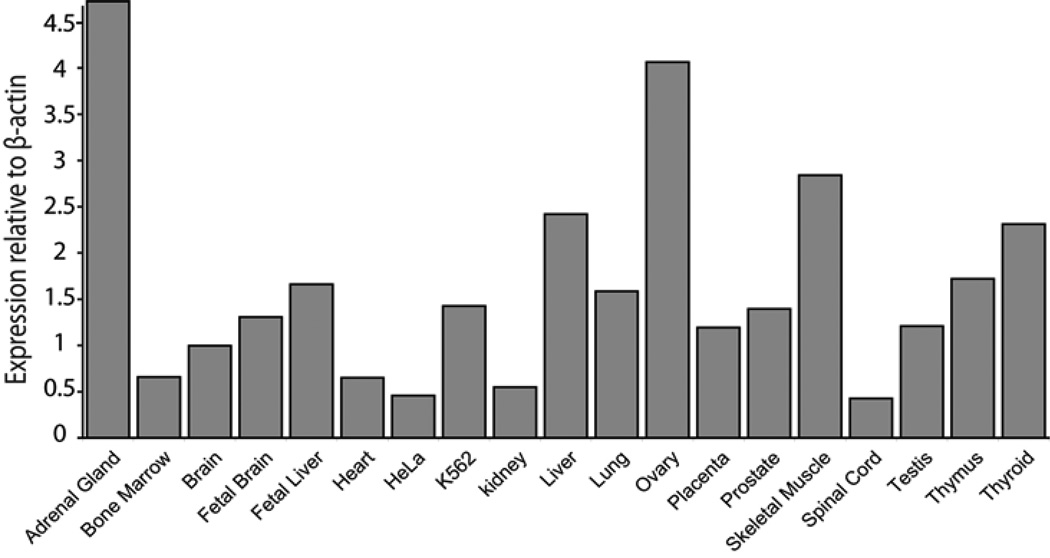
From an immediate and practical standpoint, such correlations may help guide rational selection of molecular testing for DBA.31 In addition, these differing phenotypic correlations suggest alternative pathways by which r-protein haploinsufficiency leads to specific congenital malformations, which may be distinct from the mechanism involved in other malformations and/or distinct from the mechanism of disrupted erythropoiesis. One hypothesis that may explain differential physical malformations in r-protein mutations derives from the observation that r-proteins show tissue-specific variations in both protein level and, in some cases, isoform abundance. The rate limiting step for ribosome biosynthesis, requiring equimolar amounts of r-proteins, is thus determined by the r-protein with the lowest relative expression in a tissue. A recent report shows that expression of the RPS4 paralog RPSY2 is restricted to prostate and testis.52 RPS24 was shown to have tissue-specific variability in both absolute mRNA expression levels as well as isoform distribution.22 Recent studies also suggested broad variations in RPS19 mRNA levels in different tissues (Figure 2) that appears to be independent of metabolic activity or cellular growth.54 Hypothetically, the effect of haploinsufficiency for an individual r-protein may thus be related to its expression relative to other components of the ribosome in a particular tissue.55 Much work is still required to establish the normal pattern of mature and embryologic tissue variation for r-proteins to have a better understanding of how a specific r-protein deficiency might manifest specific physical malformations.
Figure 2. Quantitative RT-PCR analysis of RPS19 on a panel of tissues and cell lines.
Quantitative RT-PCR of RPS19 mRNA corresponding to nucleotide −3 to +89 was performed in triplicate and normalized to β-actin.54 The mean RPS19 expression is related to β-actin expression.
DBA patients with chromosomal rearrangements and large deletions are a small but also potentially clinically distinctive group of DBA patients. More than 10 deletions or rearrangements have been reported at the RPS19 locus on chromosome 19.1, 3, 4, 11, 34, 46, 48, 56 In this subgroup of patients, those with larger deletions (~1Mb) are disproportionately affected by neurodevelopmental abnormalities including macrocephaly, developmental delay, psychomotor retardation or mental retardation (present in 6/7 deletions/translocations), in some cases along with additional atypical manifestations suggestive of a contiguous gene syndrome.48, 57 In comparison, neurodevelopmental abnormalities were not seen in two patients with relatively smaller (less than 1 Mb) deletions and are very rarely reported in registry data. Similarly, in two patients with 3q deletions involving RPL35A, the phenotype of one child with a large (10 Mb) deletion included developmental delay and lymphopenia atypical of classic DBA while the phenotype of a female with a smaller (4 Mb) terminal deletion appeared more typical.29 Though it is unlikely that large recurrent deletions or contiguous gene syndromes explain more than a small minority of all DBA patients, evaluation for copy loss by high resolution karyotype and/or microarray-based methods is warranted in cases with neurodevelopmental abnormalities or other atypical features. 34, 47, 48
Elucidating the Genotype/Phenotype Connection in DBA: Next Steps
It is increasingly apparent that assembling and exporting a functional ribosome is a complex and interactive process involving myriad interactions. The process does not occur in isolation, with studies demonstrating converging and diverging signaling at the level of ribosome assembly in such diverse cellular processes as nutrient availability, cell cycle control, cell size control, and apoptosis.58–62 In addition to the 4 mature rRNAs and approximately 80 structural ribosomal proteins that constitute a functional ribosome, it is estimated that at least 200 transacting factors, including helicases, nucleases, small nucleolar RNA (snoRNA), chaperones, and transporters, are involved in the process of ribosome assembly.63, 64 This large group of genes are logical candidates for evaluation based on the present understanding that all currently identified gene alterations in DBA disrupt ribosome assembly. Though the scope of such studies has heretofore been prohibitive in terms of sample availability and technical feasibility, the maturation of well-characterized patient registries, coupled with significant advances, cost-improvements and increasing availability of high-throughput sequencing technology make such studies feasible. In addition to an unexplored role as potential primary defects in a subset of DBA patients, quantitative and/or qualitative alterations in ribosome assembly factors may theoretically act as modifiers to a DBA phenotype in concert with an altered or diminished structural ribosomal constituent.
While the demonstration that different r-protein alterations cause some phenotypically distinctive features is exciting, it clearly cannot account for all of the observed heterogeneity in DBA. The most obvious example, the heterogeneity observed amongst related patients sharing a common r-protein abnormality, almost certainly reflects the influence of modifiers discrete from the primary mutation. Yet in spite of the strong evidence from which the action of modifying genes is inferred, no study has conclusively identified any modifying genetic factor with a predictable effect on the DBA phenotype. Based on current concepts of how disrupted ribosome biosynthesis may lead to DBA, many classes of modifying determinants can be theorized, with actions at the level of ribosome protein gene synthesis, ribosome assembly, or on downstream pathways activated by abortive ribosome synthesis. Proof-in-concept of a phenotypic modulator acting downstream of failed ribosome synthesis comes from studies of dark-skin mutant mice (reviewed in this issue by McGowan and Mason) where the major phenotype resulting from Rps19 mutations is abrogated when bred into a p53 mutant background.65 Co-occurrence of Li-Fraumeni syndrome and DBA has not been reported, but this observation raises the interesting question: would such an individual have anemia? Whether more common or less dramatic examples of variants that blunt the p53 response to nucleolar stress might underlie the variable phenotype in DBA currently remains an interesting, and open, question.
Modifiers acting directly at the level of ribosome synthesis might interact with deficient r-protein synthesis in a positive or negative fashion, either in exacerbating or alleviating a deficit of functional ribosomes. Again, with myriad interactions in the process, a number of interactions can be hypothesized, the majority of which are currently speculative: coding sequence amino acid changes in other small or large subunit r-proteins or assembly factors that interact with a mutant r-protein during ribosome assembly, regulatory sequence changes that enhance expression from the wild-type allele of an affected DBA gene or, conversely, hypofunctional promoter variants that exacerbate haploinsufficiency, and splicing complex variants with greater or lesser affinity for aberrant splice sites are but a few of such examples.
Emerging data have shown that the two alleles of a gene can be expressed at different levels. This may influence trait variance in heterozygotes.66, 67 Polymorphisms in regulatory cis-acting elements can mediate allele-specific effects on transcription. Hypothetically, such a transcriptional variation in wild-type alleles may contribute to the clinical variability within a family segregating an r-protein gene mutation. Targeted re-sequencing of a 20 kb genomic region spanning RPS19 in a cohort of DBA patients has revealed numerous non-transcribed genomic variants, and several of these variants coincide with predicted transcription factor binding sites of potential regulatory importance.68 Further experimental studies are now required to validate the relevance of these genomic variants on RPS19 expression in different tissues.
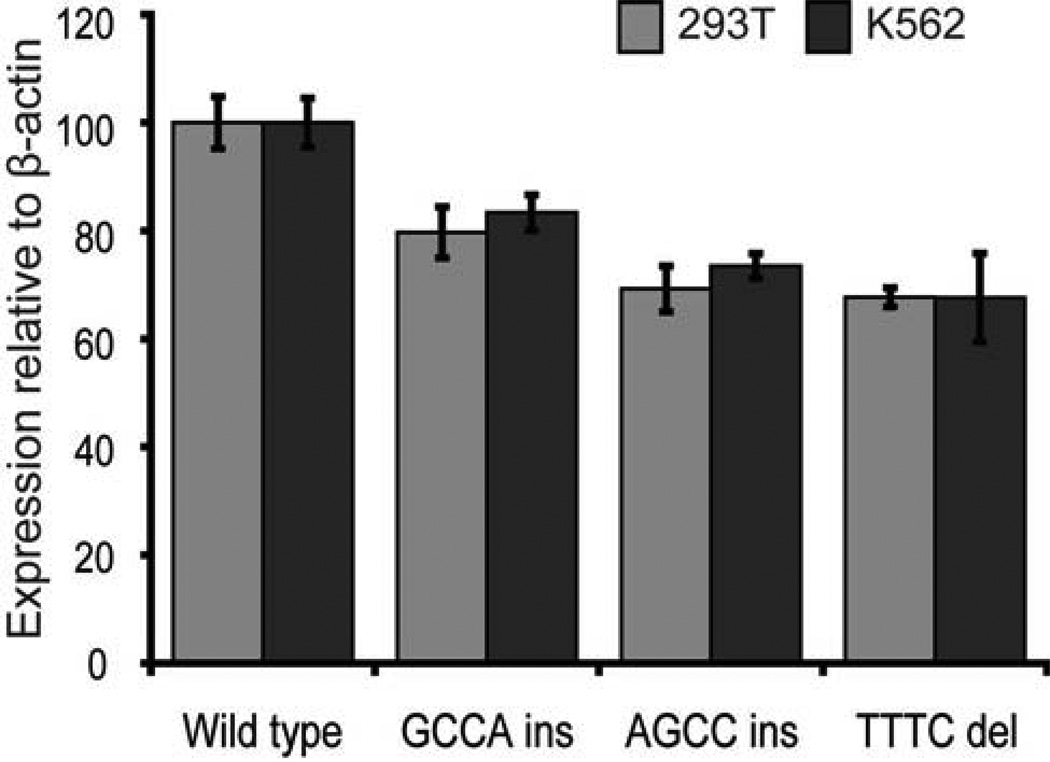
In addition to the potential effect of transcriptional-regulatory alterations, the RPS19 mRNA, like most other mRNAs encoding r-proteins, contains a 5' untranslated region sequence element, the terminal oligopyrimidine tract (5'-TOP), which is critical to post-transcriptional regulation.69, 70 Rare polymorphic sequence variants have been identified in the 5’UTR region of RPS19, some of which show an association with DBA.71–73 Recent study demonstrates that three of these 5’UTR variants result in a 20–30% reduction of RPS19 protein levels when expressed in vitro (Figure 3).54 This suggests that transcribed, non-coding polymorphic variants are of importance in the tuning of RPS19 protein levels. Analyses of the relative levels of mutant as well as non-mutant ribosomal proteins in tissues from DBA patients are needed to clarify this association with clinical expression.
Figure 3. Structural variants in the 5’UTR of RPS19 cause reduced protein levels.
Three DBA associated 5’UTR variants (c.-147_-146insGCCA, c.-147_-146insAGCC and c.- 144_-141delTTTC), were generated by site directed mutagenesis and transiently expressed in HEK293T or K562 cells from plasmid containing the complete coding sequence of RPS19 and 382 nucleotides of wild-type or variant 5’UTR. Transient expression demonstrates significantly reduced (20–30%) RPS19 protein levels for all three variants when compared to the wild-type construct (p < 0.05). Transfections were performed in triplicates and analyzed by Western blot normalized to beta-actin.54
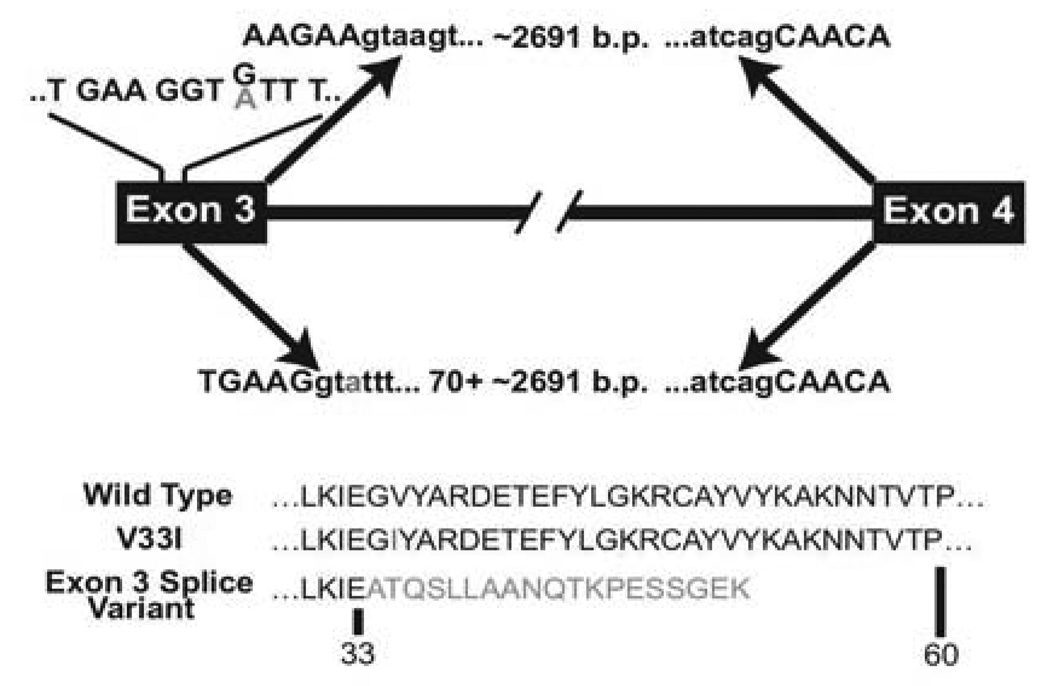
Splicing factors are another class of potential phenotypic modifiers in DBA that might modulate phenotype through differential effects at either mutant or at the counterpart wild-type r-protein. A survey of all putatively pathologic, unique r-protein variants currently reported in the DBA Gene Mutation Database demonstrates that almost 20% (41/220) disrupt splicing consensus sequences at intron boundaries. While high, this number likely underestimates the number of sequence variants with a potential to alter splicing since 1) deeper intronic sequences that harbor splicing enhancers, repressors, or branch points have not been evaluated in reported studies, 2) intronic sequence changes outside the highly conserved 4–6 base pair donor and acceptor consensus regions may not be reported, and 3) there have been no systematic analyses of missense or in-frame deletion mutations that might alter splicing patterns as a consequence of RNA sequence changes that are currently classified as resulting in amino acid substitution or deletion. One example, from a family with a mutation in RPL35A demonstrates this latter point and suggests a role for splicing in the different phenotypes observed between a classic transfusion-dependent DBA proband and his non-classic sibling and father, who share the mutation but manifest only with macrocytosis and sub-clinical anemia (Figure 1c and 4). Examination of cDNA from the proband revealed a normal product containing sequences of both the normal and mutated allele with a normal exon splicing pattern. The patient also produced a truncated mRNA, derived by an alternative splicing event within exon 3, as the result of formation of a cryptic splice donor at the mutation site. The extent to which synonymous substitutions may alter splicing is largely unexplored in DBA, as in many diseases; the large fraction of patients without identifiable mutations as currently defined, along with clear examples of aberrant splicing in DBA and other disorders74–77 suggests the need for further evaluation both as potential additional primary abnormalities as well as phenotypic modifiers in DBA.
Figure 4. Alternative splicing of RPL35A mRNA resulting in a truncated protein caused by nucleotide substitution.
Two RPL35A RNA products were amplified from the proband using RT-PCR with primers designed to amplify the full-length RPL35A message. A shorter product resulting from an alternative splicing event between exons 3 and 4 was identified along with normally-spliced mRNA containing both alleles. The arrows above the transcript diagram show the normal splicing event leading to wild-type RPL35A; sequence indicating the mutation position is shown above the schematic exon 3, with non-reference base shown in gray. The arrows below exon 3 demonstrate the abnormal splicing event resulting from activation of a cryptic splice donor site within exon 3 immediately upstream of the mutation, causing removal of 70 base pairs of 3' exon 3 coding sequence in addition to the intron. The amino acid sequence of wild-type, simple amino-acid substitution, and the splicing variant are shown below.
A relatively unique characteristic of some r-protein genes may also leave them susceptible to changes in splicing through a mechanism distinct from splice donor and acceptor sequence changes. The introns of a subset of r-protein genes, as well as other genes involved in ribosome assembly, frequently harbor snoRNAs, small non-protein coding transcripts that are involved in the process of ribosome assembly as complexes that direct post-transcriptional rRNA modifications (pseudouridylation, methylation, processing, and cleavage). SnoRNAs would seem to be poor primary DBA candidate genes since the majority found in r-protein genes are redundant in function, with multiple copy families and substantial overlap of targeted modifications. However, snoRNAs are usually co-transcribed with the host gene then processed from the intron in a manner, at least in one major class of snoRNAs, which is tightly coupled to splicing.78, 79 Could such a mechanism have relevance in DBA? A recent screening of r-protein genes in 75 North American DBA registry patients that included the majority of intron sequence and an average of 2–3 KB upstream and downstream sequence, identified 18 single nucleotide variants in 11 snoRNAs, 14 of which were novel.45 Among this group, many SNVs occurred within conserved box regions, raising the question of potential functional relevance. As with the examples outlined above, further study is required to elucidate whether such a mechanism may contribute to the phenotype in DBA.
Conclusion
The genetic landscape of DBA has changed dramatically in the past 12 years. From having no available genetic testing for DBA, more than half of DBA patients are now classifiable with a specific mutation in 1 of 9 different genes. Mutational analysis for many of these genes, initially limited to research studies, are becoming available for routine testing on a clinical basis. As a result of these observations, investigators are exploring the mechanistic connections between erythroid progenitor apoptosis, embryonic development, cancer predisposition and a new and unexpected target, the ribosome. These studies are expected to provide new avenues for exploration of more effective and less toxic therapies in patients with DBA.
These genetic discoveries have also led to significant changes in understanding the potential range of clinical manifestations of DBA. The elucidation of autosomal dominant transmission with haploinsufficient expression of r-protein genes and the associated recognition of subtle and even unaffected mutation carriers has important clinical ramifications, particularly in genetic and reproductive counseling, which underscore the importance of as yet unidentified modifying factors. Given the remarkable complexity of the ribosomal assembly process, many potentially important interactions can be hypothesized, a few of which have suggestive or circumstantial data supporting potential relevance to DBA. With the primary genetic alteration still unknown in a sizeable fraction of patients, the mechanism and contributions of potential modifiers of DBA under investigation, and an increasing understanding of how abnormalities of the ribosome may contribute to human disease, it is a reasonable expectation that the next decade of DBA research will be as active as the last.
Acknowledgments
Supported by grants from National Institutes of Health (K08 HL092224 and 5R01-HL079567-04), the Passano Foundation, the Swedish Research Council and the Daniella Maria Arturi Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jason E Farrar, Kimmel Comprehensive Cancer Center, Department of Oncology, Division of Pediatric Oncology, Johns Hopkins University, 1650 Orleans Street, CRBI-209, Baltimore, MD, 21231 Phone: 410-502-9960 Fax: 410-502-7223 jfarrar4@jhmi.edu
Niklas Dahl, Department of Genetics and Pathology, Section of Clinical Genetics, The Rudbeck Laboratory, Uppsala University Children's Hospital, S-751 85 Uppsala, Sweden Phone: +46-18 6112799 Fax: +46-18 554025 niklas.dahl@genpat.uu.se
References
- 1.Gustavsson P, Skeppner G, Johansson B, Berg T, Gordon L, Kreuger A, et al. Diamond-Blackfan anaemia in a girl with a de novo balanced reciprocal X;19 translocation. J Med Genet. 1997;34:779–782. doi: 10.1136/jmg.34.9.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gustavsson P, Willing TN, van Haeringen A, Tchernia G, Dianzani I, Donner M, et al. Diamond-Blackfan anaemia: genetic homogeneity for a gene on chromosome 19q13 restricted to 1.8 Mb. Nat Genet. 1997;16:368–371. doi: 10.1038/ng0897-368. [DOI] [PubMed] [Google Scholar]
- 3.Gustavsson P, Garelli E, Draptchinskaia N, Ball S, Willig TN, Tentler D, et al. Identification of microdeletions spanning the Diamond-Blackfan anemia locus on 19q13 and evidence for genetic heterogeneity. Am J Hum Genet. 1998;63:1388–1395. doi: 10.1086/302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 5.Matsson H, Klar J, Draptchinskaia N, Gustavsson P, Carlsson B, Bowers D, et al. Truncating ribosomal protein S19 mutations and variable clinical expression in Diamond-Blackfan anemia. Hum Genet. 1999;105:496–500. doi: 10.1007/s004399900165. [DOI] [PubMed] [Google Scholar]
- 6.Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- 7.Cmejla R, Blafkova J, Stopka T, Zavadil J, Pospisilova D, Mihal V, et al. Ribosomal protein S19 gene mutations in patients with diamond-blackfan anemia and identification of ribosomal protein S19 pseudogenes. Blood Cells Mol Dis. 2000;26:124–132. doi: 10.1006/bcmd.2000.0286. [DOI] [PubMed] [Google Scholar]
- 8.Ramenghi U, Campagnoli MF, Garelli E, Carando A, Brusco A, Bagnara GP, et al. Diamond-Blackfan anemia: report of seven further mutations in the RPS19 gene and evidence of mutation heterogeneity in the Italian population. Blood Cells Mol Dis. 2000;26:417–422. doi: 10.1006/bcmd.2000.0324. [DOI] [PubMed] [Google Scholar]
- 9.Gazda H, Lipton JM, Willig TN, Ball S, Niemeyer CM, Tchernia G, et al. Evidence for linkage of familial Diamond-Blackfan anemia to chromosome 8p23.3-p22 and for non-19q non-8p disease. Blood. 2001;97:2145–2150. doi: 10.1182/blood.v97.7.2145. [DOI] [PubMed] [Google Scholar]
- 10.Proust A, Da Costa L, Rince P, Landois A, Tamary H, Zaizov R, et al. Ten novel Diamond-Blackfan anemia mutations and three polymorphisms within the rps19 gene. Hematol J. 2003;4:132–136. doi: 10.1038/sj.thj.6200230. [DOI] [PubMed] [Google Scholar]
- 11.Campagnoli MF, Garelli E, Quarello P, Carando A, Varotto S, Nobili B, et al. Molecular basis of Diamond-Blackfan anemia: new findings from the Italian registry and a review of the literature. Haematologica. 2004;89:480–489. [PubMed] [Google Scholar]
- 12.Gazda HT, Zhong R, Long L, Niewiadomska E, Lipton JM, Ploszynska A, et al. RNA and protein evidence for haplo-insufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127:105–113. doi: 10.1111/j.1365-2141.2004.05152.x. [DOI] [PubMed] [Google Scholar]
- 13.Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br J Haematol. 2004;125:243–252. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- 14.Campagnoli MF, Ramenghi U, Armiraglio M, Quarello P, Garelli E, Carando A, et al. RPS19 mutations in patients with Diamond-Blackfan anemia. Hum Mutat. 2008;29:911–920. doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- 15.Konno Y, Toki T, Tandai S, Xu G, Wang R, Terui K, et al. Mutations in the ribosomal protein genes in Japanese patients with Diamond-Blackfan anemia. Haematologica. 2010;95:1293–1299. doi: 10.3324/haematol.2009.020826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boria I, Quarello P, Avondo F, Garelli E, Aspesi A, Carando A, et al. A new database for ribosomal protein genes which are mutated in Diamond-Blackfan Anemia. Hum Mutat. 2008;29:E263–E270. doi: 10.1002/humu.20864. [DOI] [PubMed] [Google Scholar]
- 17.Leger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, et al. Specific Role for Yeast Homologs of the Diamond Blackfan Anemia-associated Rps19 Protein in Ribosome Synthesis. J Biol Chem. 2005;280:38177–38185. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 18.Angelini M, Cannata S, Mercaldo V, Gibello L, Santoro C, Dianzani I, et al. Missense mutations associated with Diamond-Blackfan anemia affect the assembly of ribosomal protein S19 into the ribosome. Hum Mol Genet. 2007;16:1720–1727. doi: 10.1093/hmg/ddm120. [DOI] [PubMed] [Google Scholar]
- 19.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, et al. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, et al. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39:35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. American journal of human genetics. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, et al. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 24.Badhai J, Frojmark AS, Davey EJ, Schuster J, Dahl N. Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta. 2009;1792:1036–1042. doi: 10.1016/j.bbadis.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoefele J, Bertrand AM, Stehr M, Leblanc T, Tchernia G, Simansour M, et al. Disorders of sex development and Diamond-Blackfan anemia: is there an association? Pediatr Nephrol. 2010;25:1255–1261. doi: 10.1007/s00467-010-1497-y. [DOI] [PubMed] [Google Scholar]
- 26.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 27.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song MJ, Yoo EH, Lee KO, Kim GN, Kim HJ, Kim SY, et al. A novel initiation codon mutation in the ribosomal protein S17 gene (RPS17) in a patient with Diamond-Blackfan anemia. Pediatr Blood Cancer. 2010;54:629–631. doi: 10.1002/pbc.22316. [DOI] [PubMed] [Google Scholar]
- 29.Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Petrtylova K, Mihal V, et al. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat. 2009;30:321–327. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- 31.Quarello P, Garelli E, Carando A, Brusco A, Calabrese R, Dufour C, et al. Diamond-Blackfan anemia: genotype-phenotype correlation in Italian patients with RPL5 and RPL11 mutations. Haematologica. 2009;95:206–213. doi: 10.3324/haematol.2009.011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doherty L, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Clinton C, et al. Ribosomal Protein Genes RPS10 and RPS26 Are Commonly Mutated in Diamond-Blackfan Anemia. Am J Hum Genet. 2010;86:222–228. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gazda HT, Sheen MR, Doherty L, Vlachos A, Choesmel V, O'Donohue M-F, et al. Ribosomal Protein Genes S10 and S26 Are Commonly Mutated in Diamond-Blackfan Anemia. ASH Annual Meeting Abstracts. 2009;114:175. [Abstract] [Google Scholar]
- 34.Gazda H, Landowski M, Buros C, Vlachos A, Sieff CA, Newburger PE, et al. Array Comparative Genomic Hybridization of Ribosomal Protein Genes In Diamond-Blackfan Anemia Patients; Evidence for Three New DBA Genes, RPS8, RPS14 and RPL15, with Large Deletion or Duplication. ASH Annual Meeting Abstracts. 2010;116:1007. [Google Scholar]
- 35.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlachos A, Farrar J, Atsidaftos E, Muir E, Markello TC, Singh S, et al. 5q-Myelodysplastic Syndrome, In One of 23 Children Lacking a Known Ribosomal Gene Mutation, Masquerading as Diamond Blackfan Anemia (DBA) and Responding to Lenalidomide. ASH Annual Meeting Abstracts. 2010;116 LBA-2. [Google Scholar]
- 37.Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24:4032–4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatr-Aryamontri A, Angelini M, Garelli E, Tchernia G, Ramenghi U, Dianzani I, et al. Nonsense-mediated and nonstop decay of ribosomal protein S19 mRNA in Diamond-Blackfan anemia. Hum Mutat. 2004;24:526–533. doi: 10.1002/humu.20117. [DOI] [PubMed] [Google Scholar]
- 39.Da Costa L, Tchernia G, Gascard P, Lo A, Meerpohl J, Niemeyer C, et al. Nucleolar localization of RPS19 protein in normal cells and mislocalization due to mutations in the nucleolar localization signals in 2 Diamond-Blackfan anemia patients: potential insights into pathophysiology. Blood. 2003;101:5039–5045. doi: 10.1182/blood-2002-12-3878. [DOI] [PubMed] [Google Scholar]
- 40.Cretien A, Hurtaud C, Moniz H, Proust A, Marie I, Wagner-Ballon O, et al. Study of the effects of proteasome inhibitors on ribosomal protein S19 (RPS19) mutants, identified in patients with Diamond-Blackfan anemia. Haematologica. 2008;93:1627–1634. doi: 10.3324/haematol.13023. [DOI] [PubMed] [Google Scholar]
- 41.Gregory LA, Aguissa-Toure AH, Pinaud N, Legrand P, Gleizes PE, Fribourg S. Molecular basis of Diamond-Blackfan anemia: structure and function analysis of RPS19. Nucleic Acids Res. 2007;35:5913–5921. doi: 10.1093/nar/gkm626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuster J, Frojmark AS, Nilsson P, Badhai J, Virtanen A, Dahl N. Ribosomal protein S19 binds to its own mRNA with reduced affinity in Diamond-Blackfan anemia. Blood Cells Mol Dis. 2010;45:23–28. doi: 10.1016/j.bcmd.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, Ramenghi U, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourhama MH, Al-Matter ER, Aboobacker KC, Al-Humood S. Familial Diamond-Blackfan anemia. Case reports and a review of the related literature. J Trop Pediatr. 2004;50:54–56. doi: 10.1093/tropej/50.1.54. [DOI] [PubMed] [Google Scholar]
- 45.Farrar JE, Arceci RJ, Atsidaftos E, et al. Sequence abnormalities in genes encoding ribosomal proteins in patients with Diamond-Blackfan anemia: A preliminary report from the resequencing project. Presented at the 11th Annual Diamond Blackfan Anemia International Consensus Conference; 2010 March 13–14; New York, NY, USA. [Google Scholar]
- 46.Quarello P, Garelli E, Brusco A, Carando A, Pappi P, Barberis M, et al. Multiplex ligation-dependent probe amplification enhances molecular diagnosis of Diamond-Blackfan anemia due to RPS19 deficiency. Haematologica. 2008;93:1748–1750. doi: 10.3324/haematol.13423. [DOI] [PubMed] [Google Scholar]
- 47.Kuramitsu M, Morio T, Takagi M, Toki T, Terui K, Wang R, et al. New Determination Method for Extensive Gene Deletions In Diamond-Blackfan Anemia. ASH Annual Meeting Abstracts. 2010;116:4231. [Google Scholar]
- 48.Farrar JE, Vlachos A, Atsidaftos E, Carlson-Donohoe H, Ellis SR, Markello TC, et al. SNP Array Genotyping Reveals Constitutional and Mosaic Losses of Ribosomal Protein Gene Regions In Patients with Diamond Blackfan Anemia without Ribosomal Protein Gene Mutations. ASH Annual Meeting Abstracts. 2010;116:1168. [Google Scholar]
- 49.Ramenghi U, Garelli E, Valtolina S, Campagnoli MF, Timeus F, Crescenzio N, et al. Diamond-Blackfan anaemia in the Italian population. Br J Haematol. 1999;104:841–848. doi: 10.1046/j.1365-2141.1999.01267.x. [DOI] [PubMed] [Google Scholar]
- 50.Vlachos A, Klein GW, Lipton JM. The Diamond Blackfan Anemia Registry: tool for investigating the epidemiology and biology of Diamond-Blackfan anemia. J Pediatr Hematol Oncol. 2001;23:377–382. doi: 10.1097/00043426-200108000-00015. [DOI] [PubMed] [Google Scholar]
- 51.Willig TN, Niemeyer CM, Leblanc T, Tiemann C, Robert A, Budde J, et al. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. DBA group of Societe d'Hematologie et d'Immunologie Pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI) Pediatr Res. 1999;46:553–561. doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- 52.Lipton JM, Atsidaftos E, Zyskind I, Vlachos A. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr Blood Cancer. 2006;46:558–564. doi: 10.1002/pbc.20642. [DOI] [PubMed] [Google Scholar]
- 53.Lopes AM, Miguel RN, Sargent CA, Ellis PJ, Amorim A, Affara NA. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Vio. 2010;11:33. doi: 10.1186/1471-2199-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Badhai J, Schuster J, Gidlof O, Dahl N. 5' UTR variants of ribosomal protein S19 transcript determine translational efficiency: Implications for Diamond Blackfan anemia and tissue variability. PLoS ONE. 2011 doi: 10.1371/journal.pone.0017672. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellis SR, Massey AT. Diamond Blackfan anemia: A paradigm for a ribosome-based disease. Med Hypotheses. 2006;66:643–648. doi: 10.1016/j.mehy.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Cario H, Bode H, Gustavsson P, Dahl N, Kohne E. A microdeletion syndrome due to a 3-Mb deletion on 19q13.2--Diamond-Blackfan anemia associated with macrocephaly, hypotonia, and psychomotor retardation. Clin Genet. 1999;55:487–492. doi: 10.1034/j.1399-0004.1999.550616.x. [DOI] [PubMed] [Google Scholar]
- 57.Tentler D, Gustavsson P, Elinder G, Eklof O, Gordon L, Mandel A, et al. A microdeletion in 19q13.2 associated with mental retardation, skeletal malformations, and Diamond-Blackfan anaemia suggests a novel contiguous gene syndrome. J Med Genet. 2000;37:128–131. doi: 10.1136/jmg.37.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 59.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 60.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 61.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, et al. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J Biol Chem. 2006;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 64.Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 65.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pastinen T, et al. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genetic. 2010;11:533–538. doi: 10.1038/nrg2815. [DOI] [PubMed] [Google Scholar]
- 67.de la Chapelle A. Genetic predisposition to human disease: allele-specific expression and low-penetrance loci. Oncogene. 2009;28:3345–3348. doi: 10.1038/onc.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez Barrio A, Eriksson O, Badhai J, Frojmark AS, Bongcam-Rudloff E, Dahl N, et al. Targeted resequencing and analysis of the Diamond-Blackfan anemia disease locus RPS19. PloS one. 2009;4:e6172. doi: 10.1371/journal.pone.0006172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. Oligopyrimidine tract at the 5' end of mammalian ribosomal protein mRNAs is required for their translational control. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 71.Bobey NA, Carcao M, Dror Y, Freedman MH, Dahl N, Woodman RC. Sustained cyclosporine-induced erythropoietic response in identical male twins with diamond-blackfan anemia. J Pediatr Hematol Oncol. 2003;25:914–918. doi: 10.1097/00043426-200311000-00018. [DOI] [PubMed] [Google Scholar]
- 72.Cretien A, Proust A, Delaunay J, Rince P, Leblanc T, Ducrocq R, et al. Genetic variants in the noncoding region of RPS19 gene in Diamond-Blackfan anemia: potential implications for phenotypic heterogeneity. Am J Hematol. 2010;85:111–116. doi: 10.1002/ajh.21601. [DOI] [PubMed] [Google Scholar]
- 73.Huang Q, Robledo S, Wilson DB, Bessler M, Mason PJ. A four base pair insertion in exon 1 of the RPS19 gene is a common polymorphism in African-Americans. Br J Haematol. 2006;135:745–746. doi: 10.1111/j.1365-2141.2006.06368.x. [DOI] [PubMed] [Google Scholar]
- 74.Aznarez I, Chan EM, Zielenski J, Blencowe BJ, Tsui LC. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum Mol Genet. 2003;12:2031–2040. doi: 10.1093/hmg/ddg215. [DOI] [PubMed] [Google Scholar]
- 75.Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, et al. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 76.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 77.Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown JW, Marshall DF, Echeverria M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008;13:335–342. doi: 10.1016/j.tplants.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 79.Vincenti S, De Chiara V, Bozzoni I, Presutti C. The position of yeast snoRNA-coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre-mRNA. Rna. 2007;13:138–150. doi: 10.1261/rna.251907. [DOI] [PMC free article] [PubMed] [Google Scholar]