Abstract
Background
Several genome-wide association studies (GWAS) in populations of European descent have identified more than a dozen common genetic variants that are associated with prostate cancer risk.
Methods
To determine whether these variants are also associated with prostate cancer risk in the Chinese population, we evaluated 17 prostate cancer susceptibility loci in a population-based case-control study from Shanghai, including 288 prostate cancer cases and 155 population controls.
Results
After adjusting for age, two of the 17 loci were significantly associated with prostate cancer risk, while the other 15 loci were suggestively associated with prostate cancer risk in this population. The strongest associations were found for chromosome 8q24 Region 2 (rs1016343: OR=2.07, 95% CI: 1.35-3.20, P=9.4×10-4) and 8q24 Region 1 (rs10090154: OR=2.07, 95% CI: 1.31-3.28, P=0.002); additional single nucleotide polymorphisms (SNPs) assessed in these two 8q24 regions were also significant (ORRegion2=1.92-2.05, P=9.4×10-4-0.003, and ORRegion1=1.77-1.81, P=0.01 for all SNPs).
Conclusions
Our study shows that multiple prostate cancer risk loci identified in European populations using GWAS are also associated with prostate cancer risk in Chinese men, a low-risk population with mostly clinically relevant cancers. Larger studies in Chinese and Asian populations are needed to confirm these findings and the role of these risk loci in prostate cancer etiology in Asian men.
Keywords: prostate cancer, association, Asian, Chinese, 8q24
Introduction
Recent genome-wide association studies (GWAS) in 4 different study populations, all of which in European descents, have revealed more than a dozen loci associated with prostate cancer risk [1-9]. Confirmation of these risk loci among European descents has been reported extensively, but there are few studies in other racial/ethnic groups [10-19]. Most published studies in populations not of European descent were on the 8q24 loci in African American, Japanese American, Taiwanese, Latino American, Native Hawaiians, and Asian Indian populations [11-19]. Considering the fact that there are substantial differences in genetic background and lifestyles between racial groups, association studies in multiple race/ethnic populations may provide important insight into the role of genetics and gene-environmental interactions in prostate cancer etiology [20].
To date, recent GWAS have revealed 17 loci associated with prostate cancer risk in various populations worldwide [1-8,10,12-17], including 3 loci on chromosomal band 8q24 [represented by single nucleotide polymorphism (SNP) rs1447295 (Region 1), rs16901979 (Region 2), and rs6983267 (Region 3)] and 2 loci on chromosomal band 17q12 [represented by rs4430796 (Region 1) and rs11649743 (Region 2)] as well as 12 loci in other regions of the genome. To determine whether these risk loci also affect prostate cancer risk in Asian men, where prostate cancer risk is much lower but is rising steadily, we evaluated their relationship with prostate cancer risk in a population-based study conducted in Shanghai, China.
Materials and Methods
Study population
Details of this population-based case-control study have been reported elsewhere [21-23]. Briefly, cases were permanent residents of Shanghai who were newly diagnosed with prostate cancer between 1993 and 1995, identified through a rapid reporting system in 28 collaborating hospitals in urban Shanghai. The rapid reporting system captured > 95% of the cases diagnosed in urban Shanghai during the study period. Pathology material from cancer cases was reviewed by study pathologists in Shanghai and subsequently confirmed by pathologists at the Armed Forces Institute of Pathology in the USA. Based on records maintained at the Shanghai Resident Registry, male controls with no history of cancer were selected at random from the 6.5 million permanent Shanghai residents >18 years of age and frequency-matched to cancer cases by age (5-year intervals). Over 75% of the study subjects provided overnight fasting blood samples. DNA was extracted from the buffy coat fractions at the American Type Culture Collection (Manassas, VA). Only subjects with sufficient DNA available were included in the study for genotyping. In total, 288 prostate cancer cases and 155 population controls were included. There was no difference in demographic characteristics between those with and without DNA samples. This study was approved by the Institutional Review Boards at the National Cancer Institute and the Shanghai Cancer Institute and written informed consent was obtained from all study subjects.
Additional population controls
To increase the sample size of population controls for allele-based analysis, we included genotyping information from the Chinese subjects from the HapMap project in the study, including 162 Han Chinese from Beijing (CHB) and 140 Chinese from Metropolitan Denver, Colorado (CHD) [24-26]. Both CHB and CHD subjects were unrelated individuals who identified themselves as having at least three out of four Han Chinese grandparents. Data were pooled with Shanghai population controls for further analysis only if SNP allele frequencies were similar in both the Shanghai controls and HapMap subjects, as suggested by HapMap guidelines [24-26].
Selection of SNPs for evaluation and genotyping
We selected one SNPs from each of the 17 loci that were significantly associated with prostate cancer risk (P < 10-8) in four previous GWAS [1-8] and one followed-up fine mapping follow-up study [27]. These loci included three independent loci at 8q24 (Region 1 rs1447295, Region 2 rs16901979, and Region 3 rs6983267), two independent loci at 17q12 (Region 1 rs4430796 and Region 2 rs11649743), and one locus each at 2p15, 3p12, 6q25, 7p15, 7q21, 9q33, 10q11, 10q26, 11q13, 17q24.3, 19q13, and Xp11. We also selected 8 additional SNPs at 8q24 where prostate cancer associations have been reported in two fine mapping studies [15,28]. These include two additional SNPs in 8q24 Region 1 (rs4242382 and rs10090154), three additional SNPs in 8q24 Region 2 (rs1016343, rs13254738, and rs6983561), and 2 additional SNPs in 8q24 Region 3 (rs7837328 and rs7000448) as well as a SNP centromeric to the three 8q24 risk regions (rs979200).
The 25 SNPs were genotyped for all study subjects using a MassARRAY iPLEX system (Sequenom, Inc. San Diego, CA). Two duplicates and two water samples were included in each 96-well plate as PCR-negative controls. All assays were performed in a blinded fashion. The genotype call rates for these SNPs were 98% and the average concordance rate between samples was 100% among the duplicated quality control samples.
Statistical methods
Tests for Hardy-Weinberg Equilibrium (HWE) were performed for each autosomal SNP separately among control subjects, using Pearson's χ2 test. Unadjusted allelic odds ratios (ORs) and the associated 95% confidence interval (95% CI) were calculated for each risk allele (R) versus non-risk allele (N); risk alleles were defined based on European populations. Logistic regression analysis was used to test for the association between prostate cancer and the genotypes as well as for calculating ORs and 95% CI in models that were adjusted for age (4 categories: <65, 65-69, 70-74, and ≥75). Due to the limited size of the study, autosomal SNPs were modeled in a dominant mode of inheritance (NN vs NR+RR) if the risk allele frequency was less than 50% in the controls and in a recessive mode of inheritance (NN+NR vs RR) if the risk allele frequency was greater than 50% in the controls. Significant results are defined as P<0.05 (unadjusted and age-adjusted models) and P<0.002 (Bonferroni corrected for age-adjusted models). For SNPs on 8q24, pair-wise linkage disequilibrium (LD) was estimated in control subjects using Haploview [30]. Haplotype blocks were inferred using the default option of the Gabriel method [31].
Results
Table I shows selected demographic characteristics of study participants. Case and control subjects were of similar age and body size; over half of the subjects were in the normal body mass index (BMI) category (BMI of 18.5-22.9 kg/m2) as defined by the World Health Organization recommendations for Asian populations [29]. Over 55% of cases had PSA levels above 4 ng/ml. The median total PSA level for cases was 8.1 ng/mL with an interquartile range of 0.38 to 90.0 ng/mL and the median total PSA level for controls was 1.5 ng/mL with an interquartile range of 1.0 to 2.8 ng/mL. Because screening is relatively uncommon during our study period, only about 46% of the cases were localized, while over half were regional or remote. The majority of the cases (91%) were moderately or poorly differentiated.
Table I. Selected demographic characteristics of study subjects.
| Characteristic | Prostate Cancer Cases N=288 |
Controls N=155 |
||
|---|---|---|---|---|
| n | % | n | % | |
| Age (years) | ||||
| < 65 | 29 | 11.9 | 29 | 18.7 |
| 65-69 | 42 | 17.3 | 26 | 16.8 |
| 70-74 | 75 | 30.9 | 45 | 29.0 |
| ≥ 75 | 97 | 39.9 | 55 | 35.5 |
| Body mass index (BMI; kg/m2)a | ||||
| Underweight (<18.5) | 17 | 13.1 | 16 | 10.4 |
| Normal (18.5-22.9) | 67 | 51.5 | 87 | 56.5 |
| Overweight (23-27.5) | 35 | 26.9 | 38 | 24.7 |
| Obese (>27.5) | 11 | 8.5 | 13 | 8.4 |
| Prostate specific antigen (PSA; ng/ml) | ||||
| Normal (≤ 4) | 128 | 44.6 | 129 | 83.2 |
| Elevated (> 4) | 159 | 55.4 | 26 | 16.8 |
| Clinical Stage of Cancer | ||||
| Localized | 102 | 35.4 | - | - |
| Regional | 56 | 19.4 | - | - |
| Remote | 64 | 22.2 | - | - |
| Unstaged | 66 | 22.9 | - | - |
| Histological phenotype | ||||
| Well differentiated | 18 | 6.3 | - | - |
| Moderately differentiated | 76 | 26.4 | - | - |
| Poorly differentiated | 111 | 38.5 | - | - |
| Unknown | 83 | 28.8 | - | - |
BMI categories are based on the World Health Organization recommendations for Asian populations [29]
All autosomal SNPs were in HWE (P≥0.05) among control subjects (Table II). Of the 17 loci evaluated (Table II, top panel), two were significantly associated with prostate cancer in unadjusted models, including 8q24 Region 2 rs16901979 (OR=1.93, 95% CI, 1.39-2.70) and 8q24 Region 1 rs1447295 (OR=1.52, 95% CI, 1.06-2.18), while 13 other loci were suggestively associated with prostate cancer risk. Because allele frequencies for all SNPs were very similar between our controls and HapMap subjects (CHB and CHD), we combined our controls with those from HapMap and used this larger combined control group for further analysis to increase the statistical power of the study. Analysis of allele frequencies of the cases and the combined controls showed that four of the 17 loci, including the two 8q24 regions described above, 7p15 rs10486567 (P=0.009), and 10q11 rs10993994 (P=0.04), were significantly associated with prostate cancer risk. The effect of both 8q24 Regions 1 and 2 persisted after further adjustment for age using logistic regression analysis; further adjustment for BMI did not change the results. In addition, although not statistically significant, 11 other loci were suggestively associated with increased risk (ORs after age-adjustment ranged from 1.11-2.02) and 4 loci were suggestively associated with decreased risk (ORs ranged from 0.82-0.96). In general, directions of the effects as well as effect sizes for both significant and non-significant loci are the same as that seen in the European populations in almost all risk regions examined.
Table II. Association of SNPs with prostate cancer risk in a Chinese population.
| Chr | SNP | Locus | Non-risk allele (N) |
Risk allele (R)a |
Controls n=155 |
HapMapd N=302 |
Prostate Cancer Cases n=288 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk allele frequency |
Genotypesb | HWE Pc |
Risk allele frequency |
Unadjusted Allelic ORe | Genotypesb | Age-adjusted ORg | ||||||||||||||||
| NN | NR | RR | Risk allele frequency |
OR | 95% CI | P | Pf | NN | NR | RR | Model h | OR | 95% CI | P | ||||||||
| Prostate cancer risk loci identified in populations of European descent | ||||||||||||||||||||||
| 2 | rs721048 | 2p15 | G | A | 0.033 | 142 | 10 | 0 | 0.68 | 0.033 | 0.039 | 1.18 | 0.56 - 2.50 | 0.67 | 0.60 | 263 | 20 | 1 | D | 0.94 | 0.41 - 2.17 | 0.89 |
| 3 | rs2660753 | 3p12 | C | T | 0.314 | 70 | 70 | 13 | 0.44 | 0.323 | 0.310 | 0.99 | 0.74 - 1.32 | 0.92 | 0.64 | 142 | 105 | 35 | D | 0.82 | 0.54 - 1.23 | 0.34 |
| 6 | rs9364554 | 6q25 | C | T | 0.310 | 72 | 63 | 15 | 0.82 | 0.344 | 0.308 | 0.99 | 0.74 - 1.34 | 0.95 | 0.19 | 138 | 117 | 29 | D | 0.96 | 0.64 - 1.45 | 0.84 |
| 7 | rs10486567 | 7p15 | T | C | 0.128 | 116 | 35 | 2 | 0.72 | 0.126 | 0.181 | 1.47 | 1.00 - 2.17 | 0.05 | 0.009 | 192 | 73 | 14 | D | 1.53 | 0.97 - 2.43 | 0.07 |
| 7 | rs6465657 | 7q21 | T | C | 0.860 | 5 | 32 | 112 | 0.17 | 0.875 | 0.850 | 0.90 | 0.61 - 1.33 | 0.59 | 0.13 | 7 | 74 | 203 | R | 0.83 | 0.52 - 1.34 | 0.45 |
| 8 | rs16901979 | 8q24 (Region 2) | C | A | 0.235 | 85 | 52 | 8 | 0.99 | 0.264 | 0.366 | 1.93 | 1.39 - 2.70 | 1.0×10-4 | 2.0×10-4 | 110 | 139 | 34 | D | 2.03 | 1.33 - 3.10 | 0.001 |
| 8 | rs6983267 | 8q24 (Region 3) | T | G | 0.428 | 51 | 72 | 29 | 0.69 | 0.434 | 0.457 | 1.12 | 0.85 - 1.48 | 0.41 | 0.42 | 86 | 134 | 62 | D | 1.11 | 0.72 - 1.72 | 0.64 |
| 8 | rs1447295 | 8q24 (Region 1) | C | A | 0.156 | 110 | 35 | 6 | 0.15 | 0.130 | 0.222 | 1.52 | 1.06 - 2.18 | 0.02 | 3.7×10-5 | 173 | 96 | 15 | D | 1.81 | 1.16 - 2.83 | 0.009 |
| 9 | rs1571801 | 9q33 | G | T | 0.036 | 142 | 11 | 0 | 0.64 | 0.054 | 0.063 | 1.80 | 0.90 - 3.62 | 0.10 | 0.55 | 246 | 33 | 1 | D | 1.75 | 0.84 - 3.63 | 0.13 |
| 10 | rs10993994 | 10q11 | C | T | 0.434 | 43 | 77 | 24 | 0.29 | 0.443 | 0.504 | 1.32 | 0.99 - 1.75 | 0.06 | 0.04 | 75 | 130 | 77 | D | 1.12 | 0.70 - 1.77 | 0.64 |
| 10 | rs4962416 | 10q26 | A | G | 0.007 | 146 | 2 | 0 | 0.93 | 0.007 | 0.012 | 1.85 | 0.38 - 9.03 | 0.45 | 0.39 | 276 | 7 | 0 | D | 2.02 | 0.41 - 9.92 | 0.38 |
| 11 | rs10896449 | 11q13 | A | G | 0.086 | 122 | 21 | 2 | 0.33 | 0.084 | 0.111 | 1.35 | 0.82 - 2.23 | 0.24 | 0.12 | 221 | 63 | 0 | D | 1.53 | 0.89 - 2.64 | 0.13 |
| 17 | rs11649743 | 17q12 (Region 2) | A | G | 0.660 | 19 | 64 | 67 | 0.55 | 0.654 | 0.700 | 1.22 | 0.91 - 1.64 | 0.19 | 0.064 | 28 | 110 | 143 | R | 1.35 | 0.89 - 2.05 | 0.15 |
| 17 | rs4430796 | 17q12 (Region 1) | C | T | 0.720 | 11 | 63 | 78 | 0.72 | 0.724 | 0.740 | 1.14 | 0.83 - 1.56 | 0.43 | 0.42 | 17 | 110 | 155 | R | 1.18 | 0.78 - 1.78 | 0.43 |
| 17 | rs1859962 | 17q24.3 | T | G | 0.391 | 55 | 75 | 22 | 0.66 | 0.405 | 0.457 | 1.31 | 0.98 - 1.74 | 0.07 | 0.072 | 83 | 136 | 59 | D | 1.31 | 0.85 - 2.03 | 0.22 |
| 19 | rs2735839 | 19q13 | A | G | 0.580 | 24 | 79 | 47 | 0.34 | 0.596 | 0.610 | 1.14 | 0.85 - 1.52 | 0.38 | 0.69 | 46 | 131 | 107 | R | 1.15 | 0.66 - 2.00 | 0.63 |
| X | rs5945619 | Xp11 | A | G | 0.059 | 144i | 9i | 0.053 | 0.086 | 1.51 | 0.68 - 3.33 | 0.31 | 0.16 | 255i | 24i | 1.82 | 0.82 - 4.06 | 0.14 | ||||
| Additional SNPs genotyped at 8q24j | ||||||||||||||||||||||
| 8 | rs979200 | 8q24 | T | C | 0.510 | 36 | 76 | 39 | 0.93 | 0.498 | 0.459 | 0.82 | 0.62 - 1.08 | 0.16 | 0.18 | 84 | 134 | 61 | R | 0.77 | 0.47 - 1.25 | 0.29 |
| 8 | rs1016343 | 8q24 (Region 2) | C | T | 0.330 | 66 | 65 | 16 | 1.00 | 0.341 | 0.453 | 1.76 | 1.29 - 2.41 | 3.7×10-4 | 1.0×10-4 | 76 | 159 | 49 | D | 2.07 | 1.35 - 3.20 | 9.4×10-4 |
| 8 | rs13254738 | 8q24 (Region 2) | A | C | 0.697 | 11 | 70 | 71 | 0.26 | 0.303 | 0.801 | 1.82 | 1.32 - 2.56 | 0.001 | 5.9×10-4 | 9 | 92 | 176 | R | 2.05 | 1.34 - 3.13 | 9.4×10-4 |
| 8 | rs6983561 | 8q24 (Region 2) | A | C | 0.245 | 80 | 53 | 8 | 0.84 | 0.245 | 0.368 | 1.85 | 1.33 - 2.59 | 2.9×10-4 | 3.1×10-4 | 109 | 141 | 34 | D | 1.92 | 1.25 - 2.93 | 0.003 |
| 8 | rs7837328 | 8q24 (Region 3) | G | A | 0.372 | 59 | 64 | 22 | 0.50 | 0.381 | 0.417 | 1.20 | 0.90 - 1.59 | 0.22 | 0.20 | 99 | 133 | 52 | D | 1.18 | 0.77 - 1.81 | 0.45 |
| 8 | rs7000448 | 8q24 (Region 3) | C | T | 0.307 | 75 | 55 | 18 | 0.12 | 0.307 | 0.360 | 1.26 | 0.93 - 1.70 | 0.13 | 0.12 | 114 | 133 | 35 | D | 1.43 | 0.94 - 2.16 | 0.09 |
| 8 | rs4242382 | 8q24 (Region 1) | G | A | 0.168 | 108 | 37 | 7 | 0.11 | 0.140 | 0.241 | 1.56 | 1.09 - 2.22 | 0.01 | 1.0×10-5 | 163 | 105 | 16 | D | 1.77 | 1.15 - 2.75 | 0.01 |
| 8 | rs10090154 | 8q24 (Region 1) | C | T | 0.142 | 112 | 30 | 6 | 0.04 | 0.142 | 0.223 | 1.70 | 1.17 - 2.48 | 0.006 | 0.004 | 170 | 98 | 14 | D | 2.07 | 1.31 - 3.28 | 0.0019 |
Risk alleles are defined based on European populations
NN=homozygous non-risk alleles; NR=heterozygous; RR=homozygous risk alleles
HWE=Hardy-Weinberg Equilibrium
Chinese subjects from the HapMap Project: CHB (N=162) & CHD (N=140)
Based on analysis of Shanghai prostate cancer cases versus Shanghai controls; OR=Odds Ratio; CI=confidence interval
Based on analysis of Shanghai prostate cancer cases versus all controls (Shanghai controls, CHB and CHD; N=457)
Based on logistic regression analysis of Shanghai prostate cancer cases versus Shanghai controls adjusting for age (4 categories: <65, 65-69, 70-74, and ≥75)
D=dominant mode of inheritance; R=recessive mode of inheritance
Haploid
Additional SNPs were selected to be genotyped from previous fine mapping studies of 8q24 to provide better coverage of this region. These include two additional SNPs at Region 1, three additional SNPs at region 2, two additional SNPs at Region 3, and 1 SNP centromeric to the risk regions.
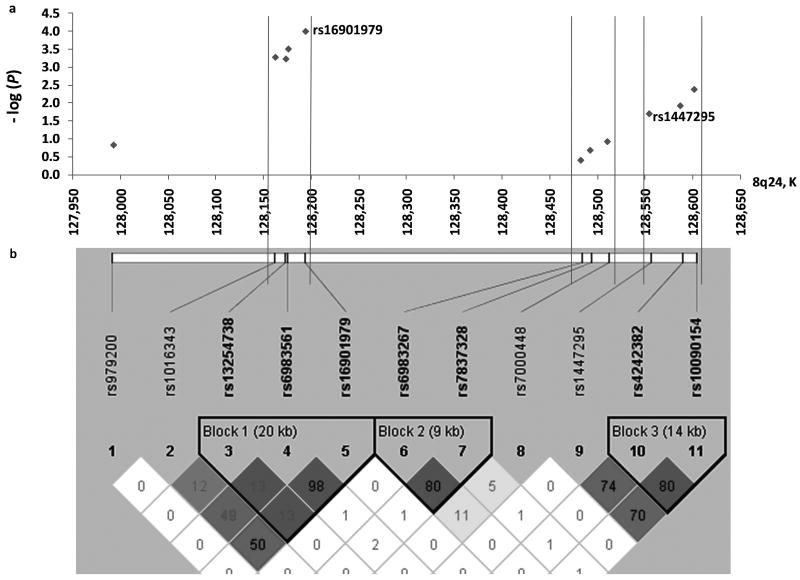
Additional SNPs genotyped in the three 8q24 risk regions (Table II, bottom panel and Figure 1a) show all SNPs in 8q24 Regions 1 and 2 were significantly associated with prostate cancer risk (P<0.05) in both unadjusted and adjusted models. The strongest associations were found for the additional three SNPs in Region 2 (rs1016343, rs13254738, and rs6983561; Padjusted ranged from 9.4×10-4-0.003), followed by the additional two SNPs in Region 1 (rs4242382, and rs10090154; Padjusted ranged from 0.002-0.01). The additional SNPs at region 3 (rs7837328, and rs7000448) were not significantly associated with prostate cancer similar to rs6983267, nor was rs979200 at the centromeric boundary of 8q24 (Padjusted>0.05). All SNPs within the same region were in significant LD with one another (Figure 1b). Among all 25 SNPs typed, three SNPs in the 8q24 Region 2 (rs16901979, rs1016343, and rs13254738) as well as one SNP in 8q24 Region 1 (rs10090154) remained significant after Bonferroni correction (P<0.002) in adjusted models.
Figure 1.
A schematic view of genetic association between SNPs at 8q24 and prostate cancer risk. (Upper panel) Allelic association tests for SNPs at 8q24 (127,950,000-128,600,000) and prostate cancer risk in the Shanghai population. (Lower panel) Pair-wise LD and inferred haplotype blocks of SNPs at this region based on the control subjects in the Shanghai population. The color of each square represents the pair-wise D′; the darker dhases corresponding to stronger D′, with the brightest red representing D′=1 and pure white represents D′=0. The number in the square represents r2.
Discussion
In this population-based study of prostate cancer in Chinese men, we systematically evaluated 17 reported prostate cancer risk loci identified through GWAS in populations of European descent. We found that two of the 17 loci (8q24 Regions 1 and 2) were significantly associated with prostate cancer risk in Chinese men in all statistical models. These results suggest that some prostate cancer risk variants identified in populations of European descent are also relevant for Chinese men, a low-risk population with mostly clinically relevant cancers.
Evaluation of additional SNPs at the 3 risk loci on 8q24 in this Chinese population showed that SNPs in Regions 1 and 2 were significantly associated with prostate cancer risk while associations for SNPs in Region 3 were mostly null, although this result could be due to limited power related to minor allele frequencies of SNPs on Region 3. These results are consistent with those from three previous studies of 8q24 in Asian populations [12,16,17], which found significant associations with Regions 1 and 2 while results for Region 3 were mixed. Taken together, these results suggest that risk Regions 1 and 2 of 8q24 may be more important than Region 3 of 8q24 in relation to prostate cancer risk in Asian populations, although larger studies are needed to confirm these findings.
There are several notable advantages to replicating GWAS findings from populations of European descent in other racial/ethnic groups, including minority populations. First, most of the cases had aggressive prostate tumors, since PSA screening in China was relatively uncommon during the early 1990s in China when the parent study was conducted. Second, this is an efficient and cost-effective approach for identifying high-risk variants in minority populations. Targeted evaluations of a dozen high-risk loci for prostate cancer, identified from GWAS in European populations, in minority populations are feasible and efficient because smaller numbers of SNPs need to be typed and evaluated, thus incurring a lower cost. The identification of several regions associated with prostate cancer risk in this small Chinese study population successfully demonstrated the benefit. Third, genetic association studies in multi-ethnic populations provide important information in distinguishing true from false associations. For example, confirming the same risk loci in populations that have substantially different genetic backgrounds and lifestyle provides further support for the observed associations. For the SNPs that fail to be confirmed in other racial/ethnic groups given sufficient power, the substantially different genetic backgrounds and environmental exposures between the populations may provide a unique opportunity to study gene-gene and gene-environmental interaction in the disease risk [20].
Limitations of the study should be noted. The statistical power of this small study was limited. For example, we had only 80% power to detect SNPs with allelic OR ≥ 1.6 and minor allele frequency (MAF) ≥ 20% in this study population [32]. Therefore, caution should be exercised in interpreting these results (i.e., we could not exclude the possibility that some of the non-significant SNPs in this study may still be modestly associated with prostate cancer risk in the Chinese population). False positive associations due to multiple testing are likely. After adjustment for 25 tests, only four SNP in 8q24 remained significant after Bonferroni correction. Finally, although our cases and controls came from a relatively homogenous Chinese population in Shanghai, we could not rule out the possibility that some observed associations in the study may have been subject to potential population stratification, since the genetic background (i.e., ancestry informative markers) in our subjects had not been measured and controlled for.
Conclusions
In summary, we found that multiple prostate cancer risk-associated loci identified in populations of European descent using GWAS may be associated with prostate cancer risk in the Chinese population. Our study demonstrated an efficient and cost-effective approach to confirming in a Chinese population the genetic variants implicated in other study populations. Larger studies in the Chinese population are needed to confirm these findings and further evaluate additional genetic variants, especially those with a modest effect. Identification of variants associated with prostate cancer may improve our understanding of the disease etiology and have potential implications for the early detection, diagnosis, and treatment of prostate cancer.
Acknowledgments
We thank J.Cheng and the staff of the Shanghai Cancer Institute for specimen collection and processing; collaborating hospitals and urologists for data collection; and local pathologists for pathology review; S. Niwa and A. Truelove of Westat and G.Yuan of Information Management Systems for data management and preparation; and J. Koci of the Scientific Applications International Corporation for management of the biologic samples. This research was supported in part by the Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, and by National Cancer Institute CA105055, CA106523, and CA95052 to J.X. L.W.C. is supported by the Cancer Prevention Fellowship Program, Office of Preventive Oncology, National Cancer Institute.
References
- 1.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 3.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 5.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Balter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Gronberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindstrom S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common sequence variants on 2p15 and Xp11. 22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 9.Chu LW, Reichardt JK, Hsing AW. Androgens and the molecular epidemiology of prostate cancer. Curr Opin Endocrinol Diabetes Obes. 2008;15:261–270. doi: 10.1097/MED.0b013e3282febcf6. [DOI] [PubMed] [Google Scholar]
- 10.Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, Ingles SA, Schaid D, Thibodeau S, Dork T, Neal D, Cox A, Maier C, Vogel W, Guy M, Muir K, Lophatananon A, Kedda MA, Spurdle A, Steginga S, John EM, Giles G, Hopper J, Chappuis PO, Hutter P, Foulkes WD, Hamel N, Salinas CA, Koopmeiners JS, Karyadi DM, Johanneson B, Wahlfors T, Tammela TL, Stern MC, Corral R, McDonnell SK, Schurmann P, Meyer A, Kuefer R, Leongamornlert DA, Tymrakiewicz M, Liu JF, O'Mara T, Gardiner RA, Aitken J, Joshi AD, Severi G, English DR, Southey M, Edwards SM, Al Olama AA, Eeles RA. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waters KM, Le Marchand L, Kolonel LN, Monroe KR, Stram DO, Henderson BE, Haiman CA. Generalizability of Associations from Prostate Cancer Genome-Wide Association Studies in Multiple Populations. Cancer Epidemiol Biomarkers Prev. 2009;18:1285–1289. doi: 10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher FR, Feigelson HS, Cox DG, Haiman CA, Albanes D, Buring J, Calle EE, Chanock SJ, Colditz GA, Diver WR, Dunning AM, Freedman ML, Gaziano JM, Giovannucci E, Hankinson SE, Hayes RB, Henderson BE, Hoover RN, Kaaks R, Key T, Kolonel LN, Kraft P, Le Marchand L, Ma J, Pike MC, Riboli E, Stampfer MJ, Stram DO, Thomas G, Thun MJ, Travis R, Virtamo J, Andriole G, Gelmann E, Willett WC, Hunter DJ. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 14.Robbins C, Torres JB, Hooker S, Bonilla C, Hernandez W, Candreva A, Ahaghotu C, Kittles R, Carpten J. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salinas CA, Kwon E, Carlson CS, Koopmeiners JS, Feng Z, Karyadi DM, Ostrander EA, Stanford JL. Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1203–1213. doi: 10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- 16.Terada N, Tsuchiya N, Ma Z, Shimizu Y, Kobayashi T, Nakamura E, Kamoto T, Habuchi T, Ogawa O. Association of genetic polymorphisms at 8q24 with the risk of prostate cancer in a Japanese population. Prostate. 2008;68:1689–1695. doi: 10.1002/pros.20831. [DOI] [PubMed] [Google Scholar]
- 17.Tan YC, Zeigler-Johnson C, Mittal RD, Mandhani A, Mital B, Rebbeck TR, Rennert H. Common 8q24 sequence variations are associated with Asian Indian advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:2431–2435. doi: 10.1158/1055-9965.EPI-07-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Kibel AS, Hu JJ, Turner AR, Pruett K, Zheng SL, Sun J, Isaacs SD, Wiley KE, Kim ST, Hsu FC, Wu W, Torti FM, Walsh PC, Chang BL, Isaacs WB. Prostate Cancer Risk Associated Loci in African Americans. Cancer Epidemiol Biomarkers Prev. 2009:1055–9965. doi: 10.1158/1055-9965.EPI-09-0091. EPI-1009-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M, Huang YC, Ko Il, Yang S, Chang YH, Huang WJS, Chen YMA. The rs1447295 at 8q24 Is a Risk Variant for Prostate Cancer in Taiwanese Men. Urology. doi: 10.1016/j.urology.2009.02.035. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 20.Tang H. Confronting ethnicity-specific disease risk. Nat Genet. 2006;38:13–15. doi: 10.1038/ng0106-13. [DOI] [PubMed] [Google Scholar]
- 21.Hsing AW, Chua S, Jr, Gao YT, Gentzschein E, Chang L, Deng J, Stanczyk FZ. Prostate cancer risk and serum levels of insulin and leptin: a population-based study. J Natl Cancer Inst. 2001;93:783–789. doi: 10.1093/jnci/93.10.783. [DOI] [PubMed] [Google Scholar]
- 22.Hsing AW, Deng J, Sesterhenn IA, Mostofi FK, Stanczyk FZ, Benichou J, Xie T, Gao YT. Body size and prostate cancer: a population-based case-control study in China. Cancer Epidemiol Biomarkers Prev. 2000;9:1335–1341. [PubMed] [Google Scholar]
- 23.Hsing AW, Gao YT, Wu G, Wang X, Deng J, Chen YL, Sesterhenn IA, Mostofi FK, Benichou J, Chang C. Polymorphic CAG and GGN repeat lengths in the androgen receptor gene and prostate cancer risk: a population-based case-control study in China. Cancer Res. 2000;60:5111–5116. [PubMed] [Google Scholar]
- 24.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Zheng SL, Wiklund F, Isaacs SD, Purcell LD, Gao Z, Hsu FC, Kim ST, Liu W, Zhu Y, Stattin P, Adami HO, Wiley KE, Dimitrov L, Sun J, Li T, Turner AR, Adams TS, Adolfsson J, Johansson JE, Lowey J, Trock BJ, Partin AW, Walsh PC, Trent JM, Duggan D, Carpten J, Chang BL, Gronberg H, Isaacs WB, Xu J. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Lange EM, Isaacs SD, Liu W, Wiley KE, Lange L, Gronberg H, Duggan D, Carpten JD, Walsh PC, Xu J, Chang BL, Isaacs WB, Zheng SL. Chromosome 8q24 risk variants in hereditary and non-hereditary prostate cancer patients. Prostate. 2008;68:489–497. doi: 10.1002/pros.20695. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organisation Expert Consultation Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363:157–163. doi: 10.1016/S0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 32.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]