Abstract
Background
Identifying viral and host determinants of HIV-1 elite control may help inform novel therapeutic and/or vaccination strategies. Previously, we observed decreased replication capacity in controller-derived viruses suggesting that fitness consequences of HLA class I-associated escape mutations in Gag may contribute to this phenotype. This study examines whether similar functional defects occur in Pol proteins of elite controllers.
Methods
Recombinant NL4-3 viruses encoding plasma RNA-derived Reverse Transcriptase (RT)-Integrase sequences from 58 elite controllers and 50 untreated chronic progressors were constructed and replication capacity measured in vitro using a GFP reporter T-cell assay. Sequences were analyzed for drug resistance and HLA-associated viral polymorphisms.
Results
Controller-derived viruses displayed significantly lower replication capacity compared to those from progressors (p<0.0001). Among controllers, the most attenuated viruses were generated from individuals expressing HLA-B*57 or B*51. In viruses from B*57+ progressors (N=8), a significant inverse correlation was observed between B*57-associated RT-Integrase escape mutations and replication capacity (R=−0.89; p=0.003); a similar trend was observed in B*57+ controller-derived viruses (N=20, R=−0.36; p=0.08).
Conclusions
HIV-1 Pol function appeared to be compromised in elite controllers. As observed previously for Gag, HLA-associated immune pressure in Pol may contribute to viral attenuation and subsequent control of viremia.
Keywords: HIV-1, elite controller, viral replication capacity, HLA class I, Pol, Cytotoxic T-lymphocyte (CTL), immune escape
Introduction
Elite controllers are a rare group of HIV-1 infected individuals who spontaneously maintain plasma viremia below the limit of standard clinical detection (< 50 viral RNA copies/ml) without antiviral therapy1. Elucidating the mechanisms responsible for this phenotype may reveal host and viral factors that may be modulated for prophylactic or therapeutic intervention.
HIV-1 replication capacity (RC) likely plays an important role in pathogenesis2, 3, but its relevance to the elite controller phenotype remains unclear. Although a recent examination of a large number of elite controller-derived HIV sequences revealed no evidence of gross mutational defects, large insertions or deletions, nor shared ancestry4, virus function may nevertheless be compromised. Indeed, previous studies have reported lower RC in viruses isolated from viremic long-term non-progressors compared to those from chronic infected individuals2, 3, 5, and reduced entry efficiency has been observed for elite controller-derived envelopes compared to those from chronic infected individuals6.
Although RC is determined in large part by the founder virus acquired at transmission7, 8, it can change over time as host and other selective pressures drive intra-host HIV-1 evolution9–11. In vitro fitness costs of HLA-restricted cytotoxic T-lymphocyte (CTL) escape mutations in Gag12–17 and Nef18 have been demonstrated, and evidence suggests that immune-mediated fitness defects may be relevant to the controller phenotype19, 20. A recent case report described reduced RC of virus isolated from a B*27/B*57-expressing elite controller compared to the transmitted donor virus19. Furthermore, we have previously described reduced in vitro RC of recombinant viruses expressing Gag-Protease from elite controllers compared to progressors in chronic20 and acute/early21 infection, an observation attributable at least in part to immune selection20, 22. A biologically relevant role for immune-mediated fitness defects is supported by relative early viremia control in individuals who acquire HIV-1 harboring escape mutations from donors expressing protective HLA alleles7, 8. Moreover, evidence for sequential reductions in RC as a result of the accumulation of HLA-restricted CTL escape mutations has been reported in Gag16, 23.
However, comparably little is known about the consequence of mutations outside of the Gag gene on viral RC in elite controllers, and what relevance this may have to the controller phenotype. Since mutations in the Pol gene that emerge under antiretroviral drug selection pressures can affect fitness24–28, we therefore examined whether elite controller viruses exhibited functional defects in this gene. To do this, recombinant viruses encoding plasma RNA-derived Reverse Transcriptase (RT)-Integrase sequences were generated in an NL4-3 virus backbone from 58 elite controllers and 50 untreated chronic progressors and their in vitro RC was examined using a GFP reporter T-cell assay20, 29. Similar to previous observations for Gag-Protease20, 21 and Envelope6, we observed reduced function of controller-derived RT-Integrase, indicating that differences in Pol activity may contribute to HIV-1 control.
Methods
Study participants
Fifty-eight elite controllers (all <50 copies RNA/ml; median CD4 = 799 [IQR 593–1037] cells/mm3) and 50 untreated chronic progressors (median viral load 4.98 [IQR 4.51–5.35] log10 copies RNA/ml; median CD4 = 318 [IQR 61–476] cells/mm3) were included. Characteristics of this elite controller cohort have been described previously30. In addition, 76 of these 108 (70%) patients were previously evaluated for Gag-Protease function20. HLA class I typing was performed using standard sequence-based methods. This study was approved by the institutional review board at Massachusetts General Hospital; written informed consent was obtained from all participants.
Generation of recombinant RT-Integrase viruses
a) Bulk (quasispecies) method
For elite controllers, RT-PCR products spanning Pol were generated as described4. For progressors, HIV-1 Pol was RT-PCR amplified from extracted plasma RNA using sequence-specific primers. Second round PCR was performed using PAGE-purified “recombination primers” designed to match the NL4-3 sequence directly upstream of RT (forward; 100bp) and downstream of Integrase (reverse; 98bp). Primer sequences are available upon request.
Plasmid pNL4-3DRT-Integrase was developed by inserting unique restriction enzyme sites for BstEII at the 5’ end of RT and the 3’ end of Integrase using the QuikChange XL kit (Stratagene) followed by deletion of the intervening region by BstEII digestion (New England Biolabs). This plasmid was maintained using Stbl3 E. coli cells (Invitrogen). To generate recombinant viruses, 10µg of BstEII-linearized plasmid plus 50µl of 2nd round amplicon (approximately 5µg) were mixed with 2.0×106 cells of a Tat-driven GFP reporter T-cell line (GXR 25 cells29) in 800µl of R10+ medium (RPMI 1640 containing 10% FCS, 2mM L-glutamine, 100 units/mL penicillin, and 100µg/mL streptomycin), and transfected by electroporation using a BioRad GenePulser Xcell (exponential protocol: 300V, 500µF). Following transfection, cells were rested for 45 min at room temperature, transferred to 25 cm2 flasks in 5 mL of R10+ medium, and fed with 5 mL R10+ medium on day 5. GFP expression was monitored daily by flow cytometry (FACScalibur, BD Biosciences) starting on day 10. Once GFP+ expression reached ~15% among viable cells, representing the early phase of exponential spread, culture supernatants containing the recombinant viruses were harvested and aliquots stored at −80°C.
b) Clonal method
In addition, RT-integrase sequences from a random subset of 14 controllers and 10 progressors were cloned (TOPO-TA cloning kit; Invitrogen), purified and used as starting material to generate recombinant viruses as described above. All clones were resequenced to confirm patient origin.
Replication capacity assays
Virus titers and replication assays were performed as described12, 20, 29. Replication assays were initiated at MOI=0.003, and included six negative (uninfected cells only) and six NL4-3 controls. For each virus, the natural log slope of the percent (%) of GFP+ cells was calculated during the exponential phase of spread (days 3–6). This value was divided by the mean rate of spread of NL4-3 to generate a normalized, quantitative measure of replication capacity (RC). An RC of 1.0 indicates a rate of spread equal to NL4-3, while RC <1.0 and >1.0 indicate rates of spread that are slower than or faster than NL4-3, respectively. All assays were performed in triplicate in independent experiments and replication rates were averaged. The experimental procedure is illustrated in Supplemental Figure 1.
Re-sequencing of recombinant viral stocks
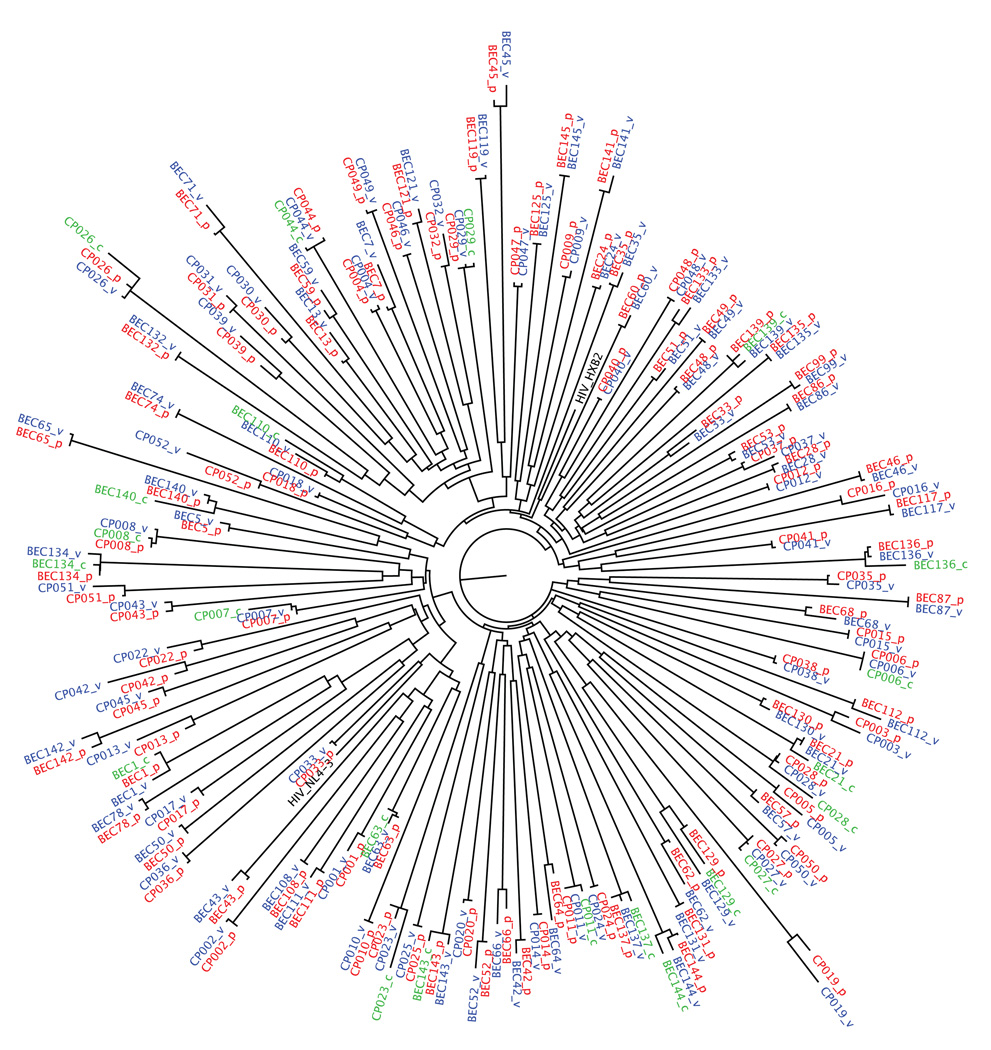
For all bulk (quasispecies-containing) recombinant viruses, HIV-1 RNA was extracted from viral culture supernatant (QIAamp viral RNA kit; Qiagen), amplified and sequenced as described in4, and compared to the original plasma HIV-1 RNA sequences (Figure 1). All viruses were confirmed as subtype B using RIP (http://www.hiv.lanl.gov). Nucleotide alignments were performed using GeneCutter and maximum-likelihood phylogenetic trees were generated using PHYml31; both available at http://www.hiv.lanl.gov. Trees were visualized using Figtree v.1.2.2 (http://tree.bio.ed.ac.uk/software/figtree/). Resistance mutations were identified using the Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu/). Most controller-derived RT-Integrase sequences were previously deposited in Genbank4. Accession numbers for the remaining sequences from controllers and progressors are GQ284657-GQ284730.
Figure 1. Phylogenetic tree illustrating the relationship between HIV-1 RT-Integrase sequences from plasma and recombinant viruses from study subjects.
Prefixes “BEC” and “CP” discriminate sequences from controllers and progressors, respectively. Red (suffix “p”) Blue (suffix “v”) and Green (suffix “c”) sequences indicate bulk plasma, bulk recombinant and clonal recombinant virus sequences, respectively. The sequences of HIV-1 subtype B reference strains HXB2 and NL43 are included for comparison.
Statistical analysis
Student’s T-test was used to compare differences in RC between groups (e.g. controllers/progressors; presence/absence of HLA alleles, etc). Spearman’s and Pearson’s correlation was used to investigate the relationship between clinical parameters (CD4/pVL) and the presence of HLA-associated escape mutations, respectively, and viral RC. In an exploratory analysis, the Mann-Whitney U-Test was used to identify specific amino acids in RT-Integrase associated with RC; here, q-values were used to address multiple tests32.
Results
Generation of recombinant viruses expressing RT-Integrase sequences from controllers and progressors
Recombinant viruses were generated using bulk patient plasma-derived PCR amplicons containing Reverse Transcriptase (RT) and Integrase sequences as described in Methods, and stocks harvested during the early phase of exponential viral spread. For elite controllers (N=58), the median time to harvest was 25 days (Interquartile Range [IQR] 21–28) while for progressors (N=50) it was 19 days [IQR 15–21]. This difference was statistically significant (p<0.0001). The RT-Integrase sequences of the recombinant virus and the original plasma HIV RNA were highly concordant (Figure 1). The median number of full amino acid differences between these sequences was 1 [IQR 0–2] out of a total of 849 codons spanning RT-Integrase, a similarity of 99.9%. In a more conservative analysis where amino acid mixtures were considered full differences, the median number of differences was 6 [IQR 3–9] (99.3% similarity); values that are comparable to the average inter-laboratory nucleotide concordance of sequence-based genotypic drug resistance assays (99.4%)33. These data indicate that our approach did not result in substantial in vitro selection and that at least some quasi-species diversity was maintained in the recombinant viral stocks.
Reduced RC of RT-Integrase viruses derived from elite controllers
RC of bulk (quasispecies-containing) recombinants was assayed in three independent experiments and results reported as the mean. Concordance between replicates was high (R=0.77, p<0.0001; with a median difference between replicates of 8.03% [IQR 3.76–16.4%]).
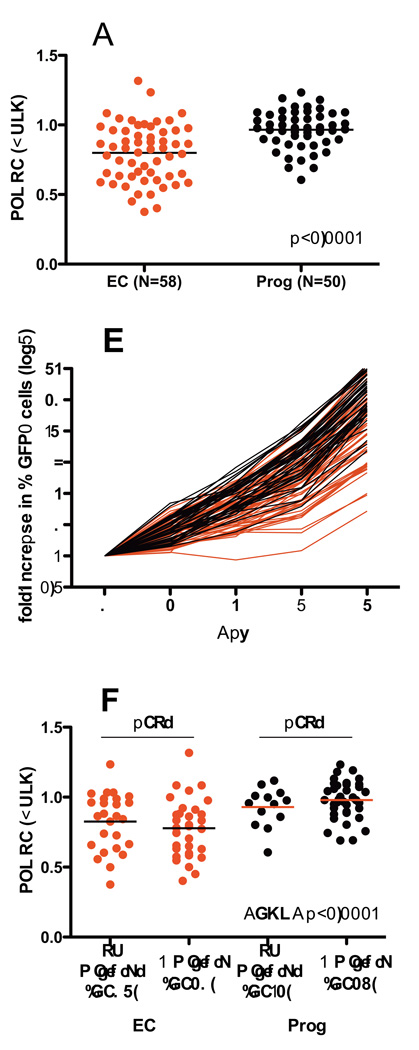
Controller-derived RT-Integrase recombinant viruses constructed using bulk (quasispecies-containing) methods displayed significantly reduced RC compared to those derived from progressors (Figures 2A and 2B). The median RC of controller viruses was 0.83 [IQR 0.63–0.96] compared to 0.98 [0.89–1.07] for progressor viruses (p<0.0001). To assess whether this result may be affected by the diversity of the recombinant quasispecies, we stratified recombinant virus sequences based on the presence or absence of amino acid mixtures in the RT-Integrase region, but observed no significant differences in RC between the two in either controllers or progressors (Figure 2C). However, the overall differences in RC between controllers and progressors remained highly statistically significant regardless of the presence or absence of amino acid mixtures (ANOVA p<0.0001).
Figure 2. Replication Capacities of bulk RT-Integrase recombinant viruses derived from elite controller and chronic progressor sequences.
Panel A: RC of bulk RT-Integrase recombinant viruses from controllers (EC) and Progressors (Prog) are shown. RC values are normalized to the mean RC of NL4-3, such that an RC of 1.0 indicates equivalent growth to NL4-3, while RC >1 and RC <1 indicate faster or slower growth compared to NL4-3, respectively. Results represent the average of three independent experiments. Horizontal bars indicate median values for each group. Panel B: The rate of viral spread in culture (expressed as fold-increase in % GFP+ cells over the assay period) is shown for controller-derived (red lines) vs. progressor-derived (black lines) viruses. Panel C: RC results of bulk RT-Integrase recombinant viruses are stratified by the absence or presence of amino acid mixtures in the chimeric viral stocks. No significant difference in RC was observed between non-mixture-containng vs. mixture containing recombinant viruses derived from controllers or progressors.
In order to further rule out potential bias due to the quantity and/or diversity of the initial bulk PCR inoculum between controllers and progressors, and the potential existence of minority quasispecies in the recombinant virus stocks, we generated clonal recombinant viruses from a randomly-selected subset of 14 controllers and 10 progressors. Consistent with the original findings, median RC of clonal controller viruses was 0.73 [IQR 0.61–0.98] compared to 0.97 [0.86–1.08] for clonal progressor viruses in this subset (p=0.028; Supplementary Figure 2A). In addition, robust concordance was observed between RC of independently constructed clones (N=10 pairs, R=0.84, p=0.002, not shown) as well as between the RC of quasispecies-containing versus clonal recombinant viruses (R=0.7, p=0.0003, Supplementary Figure 2B).
Taken together, the RT-Integrase region from most controllers exhibited reduced function compared to progressors, regardless of whether quasispecies-containing or clonal recombinants were evaluated.
Reduced RC is not due to resistance mutations and does not correlate with Gag-Protease function
Resistance mutations can affect viral RC24–28. Although all patients were untreated at the time of sample collection, 8 of 58 (14%) controller-derived and 8 of 50 (16%) progressor-derived viruses harbored at least one major resistance mutation in RT (p=0.8), most frequently at codons 215 (N=9) and 219 (N=6), K70R (N=5), D67N (N=4), and others. Of these 16 sequences, 10 encoded ≥2 resistance mutations. After exclusion of the 16 resistant viruses from analysis, the difference in RC between controller and progressor-derived viruses remained statistically significant (p<0.0001, not shown). Furthermore, no significant differences were observed between resistant and nonresistant viruses within each patient group (p>0.05), suggesting that decreased RC in controllers is not due to drug resistance mutations.
Reduced RC does not correlate with clinical parameters, or Gag/protease function, in controllers or progressors
We next addressed whether RT-Integrase function correlated with clinical parameters. Among controllers, all pVL were <50 copies/ml and the median CD4 count was 799 [IQR 593–1037] cells/mm3. Among progressors, the median pVL was 4.98 [IQR 4.51–5.35] log10 HIV RNA copies/ml and the median CD4 count was 318 [IQR 61–476] cells/mm3. In an analysis stratified by patient group, we observed no correlation between RC and CD4 count in controllers (Spearman R=−0.03, p=0.8) or progressors (R=0.06, p=0.7), nor between replication and pVL in progressors (R=0.05, p=0.7) (not shown).
We also evaluated whether RT-Integrase RC correlated with functional RC data for Gag-Protease previously obtained on a subset of these individuals (reported in20). Although both the previous and present study report reduced RC among controller-derived viruses, we observed no correlation between RC of Gag-Protease viruses and RT-Integrase viruses in an analysis stratified by patient group (controllers [N=38, R=−0.09, p=0.6]; progressors [N=38, R=0.13, p=0.4]), suggesting that these observations are largely independent.
Associations between RC and HLA class I expression
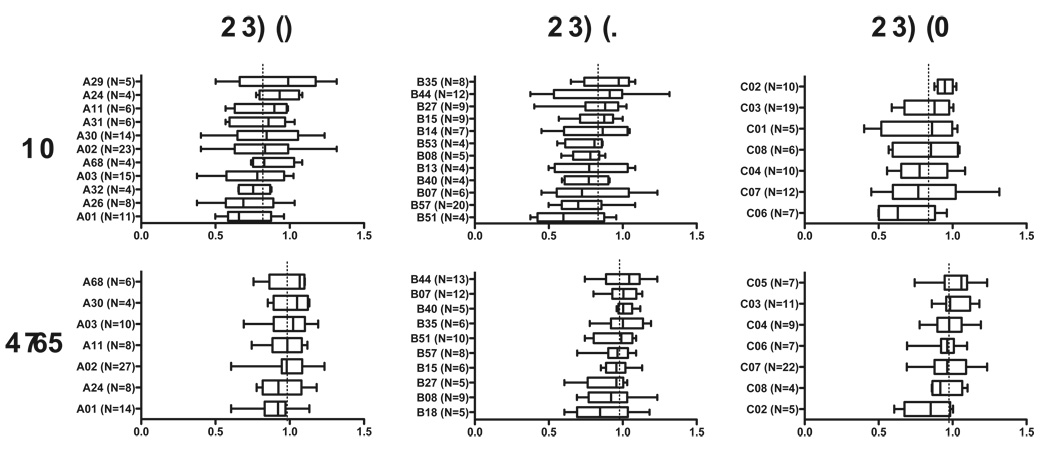
We next investigated associations between RC and HLA class I allele expression, stratified by patient group (Figure 3). In controllers, no statistically significant associations were observed, however the poorest replicating viruses originated from HLA-B*51- and B*57-expressing individuals (p=0.09 and p=0.08, respectively). Among progressors, viruses from A*01- and C*02-expressing individuals displayed significantly lower RC (p=0.016 and p=0.02, respectively).
Figure 3. Associations between HLA class I allele expression and Replication Capacities of RT-Integrase recombinant viruses.
Box and whisker plots indicate the median (line), interquartile range (box) and range (whiskers) of viral RC, stratified by HLA class I alleles expressed. Asterisks indicate observations that trend higher or lower than the population mean, with p<0.1 (Wilcoxon Rank-sum test). Dotted vertical line indicates median replication capacity for each patient group. HLA class I alleles with a minimum of N=4 observations are shown.
Correlation between replication and HLA-associated sequence polymorphisms
To further assess the impact of immune selection pressure on RT-Integrase function, we investigated potential correlations between the presence of HLA-associated polymorphisms (defined according to a population-based analysis of >1500 subtype-B infected individuals worldwide34) and RC. For each patient, we determined the total possible number of HLA-associated polymorphic sites in RT-Integrase according to their HLA class I profile, as well as the number of these sites that exhibited a known HLA-associated polymorphism in the recombinant virus sequence. No significant correlation was observed between RC and the overall burden of HLA-A, B and C-associated polymorphisms in RT-Integrase, assessed in absolute terms (“number of escaped sites”) or relative terms (“proportion of escaped sites”) in either controllers or progressors (all p>0.05, not shown).
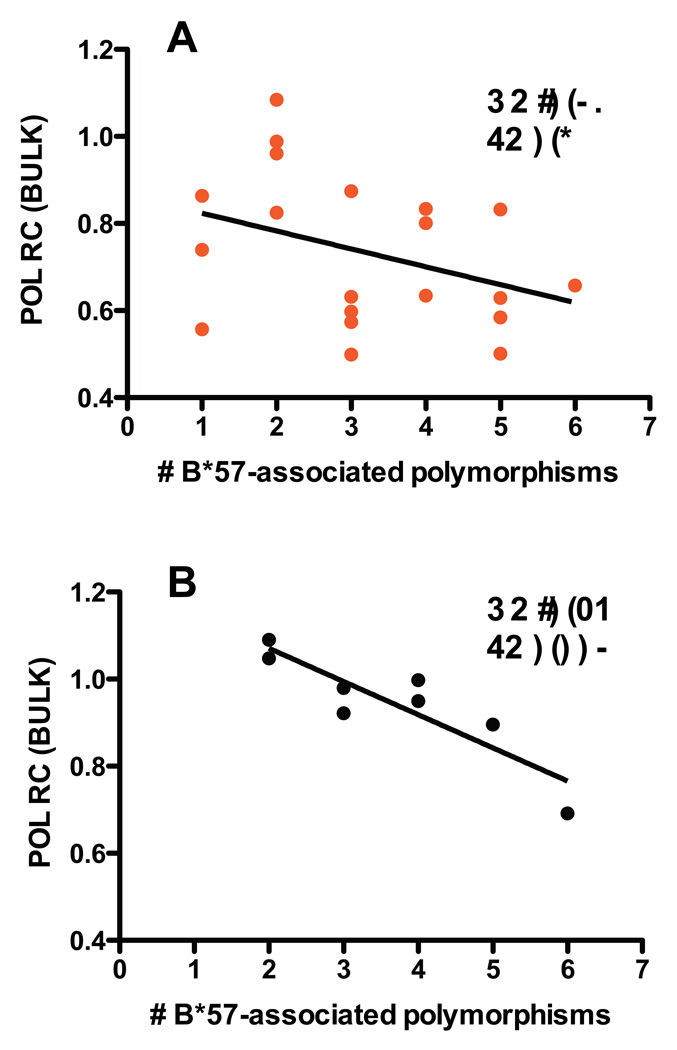
We next investigated the relationship between HLA-associated polymorphisms and RC in an allele-specific manner, for all alleles with frequencies ≥5. Although no correlations achieved statistical significance in controllers, an inverse relationship was observed between the number of B*57-associated polymorphisms and lower RC in B*57-expressing controllers (N=20; R=−0.36, p=0.1) (Figure 4A). In B*57-expressing progressors, a strong inverse relationship was observed between the number of B*57-associated RT-Integrase polymorphisms and RC (N=8, R=−0.89, p=0.003); no other significant correlations were observed (Figure 4B). Therefore, although the average RC of viruses derived from B*57-expressing progressors did not significantly differ from the population average, the strong dose-dependent relationship between B*57-associated polymorphisms and decreasing RC suggests that these polymorphisms negatively influence RT-Integrase function in a cumulative manner.
Figure 4. Relationship between the number of HLA-B*57-associated escape mutations in RT-Integrase and viral Replication Capacity.
In viruses from the 20 elite controllers (panel A) and 8 progressors who expressed HLA-B*57 (panel B), the relationship between the number of HLA-B*57-associated polymorphisms in RT-Integrase and viral RC was characterized using Pearson’s correlation. A regression line is drawn to highlight the trend.
No association between replication and transmitted HLA-associated polymorphisms
Immune-mediated fitness defects have been reported in individuals acquiring HIV-1 from donors who express protective HLA alleles7, 8. Therefore, we investigated whether RC correlated with protective allele-associated viral polymorphisms in individuals not expressing these alleles. No significant difference was observed in the average number of polymorphisms associated with protective HLA alleles (defined as HLA-B*13, B*27, B*51, B*57, B*5801 and B*8135–40) in controllers (N=19; mean protective allele-associated polymorphisms/sequence=8.6) versus progressors (N=22; mean polymorphisms/sequence=7.6; p=0.25) not expressing these alleles. Moreover, no correlation was observed between the number of protective HLA-associated polymorphisms and RC in controllers (R=0.23, p=0.34) or progressors (R=0.34, p=0.13) not expressing these alleles. An analysis restricted to B*57-associated mutations in controllers and progressors not expressing this allele also failed to demonstrate significant differences in polymorphism frequency between groups, nor correlations with RC (all p>0.1, not shown). Altogether, these results suggest that the observed reductions in RC seen in controllers were not likely due to transmitted immune escape mutations.
Associations between RC and specific amino acid residues in RT-Integrase
In an exploratory analysis, the Mann-Whitney U-test was used to examine associations between RT-Integrase amino acid variation and RC. This analysis was performed on the entire dataset, as well as stratified by patient group. In the combined analysis of all polymorphisms occurring at frequencies ≥5, 28 residues (20 in RT, 8 in Integrase) were associated with RC at p<0.05 (corresponding q-values 0 to 0.77; Table 1). The strongest association was observed for Integrase codon 265, where the consensus Alanine was associated with significantly higher RC than the polymorphism Valine (p<0.0001, q=0). Stratification by patient group also identified the codon 265 association as significant in controllers (p<0.01, q=0.4) but not in progressors (not shown).
Table 1. Exploratory analysis of RT-Integrase residues associated with viral replication capacity.
Results limited to those observed in a minimum of 5 sequences, p<0.05. Putative HLA class I alleles are described in 34; optimally-described HLA-restricted epitopes from (http://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html).
| Protein | Codon | Residue | N with |
N without |
MEDIAN replication with |
MEDIAN replication without |
p-value | q-value | Putative HLA/epitope |
|---|---|---|---|---|---|---|---|---|---|
| RT | 211 | R | 59 | 49 | 0.86 | 0.98 | 0.003 | 0.41 | A32, A68, B15, B44 |
| 211 | K | 44 | 64 | 0.98 | 0.87 | 0.004 | 0.41 | A32, A68, B15, B44 | |
| 245 | V | 69 | 39 | 0.95 | 0.87 | 0.049 | 0.77 | A29, B57, B58 B57-IW9 | |
| 294 | P | 99 | 9 | 0.89 | 1.01 | 0.024 | 0.68 | ||
| 294 | T | 5 | 103 | 1.10 | 0.90 | 0.047 | 0.77 | ||
| 329 | I | 87 | 21 | 0.88 | 0.96 | 0.036 | 0.71 | C05 | |
| 345 | P | 101 | 7 | 0.88 | 1.04 | 0.010 | 0.48 | A11, A11-IK10 | |
| 366 | K | 96 | 12 | 0.89 | 0.99 | 0.041 | 0.74 | B13, A03-RK11 | |
| 366 | R | 12 | 96 | 0.99 | 0.89 | 0.041 | 0.74 | B13, A03-RK11 | |
| 369 | T | 96 | 12 | 0.89 | 1.00 | 0.039 | 0.74 | B13 | |
| 376 | S | 6 | 102 | 0.63 | 0.92 | 0.036 | 0.71 | B53, B58, C12, B58-IW9 | |
| 381 | V | 102 | 6 | 0.92 | 0.60 | 0.015 | 0.48 | C12, B58-IW9 | |
| 381 | I | 6 | 102 | 0.60 | 0.92 | 0.015 | 0.48 | C12, B58-IW9 | |
| 390 | R | 51 | 57 | 0.96 | 0.86 | 0.032 | 0.71 | B58 | |
| 390 | K | 58 | 50 | 0.87 | 0.97 | 0.033 | 0.71 | B58 | |
| 431 | T | 6 | 102 | 1.04 | 0.89 | 0.024 | 0.68 | ||
| 431 | K | 101 | 7 | 0.88 | 1.03 | 0.027 | 0.68 | ||
| 449 | D | 5 | 103 | 1.00 | 0.89 | 0.028 | 0.68 | A26-EY9 | |
| 452 | I | 12 | 96 | 0.81 | 0.93 | 0.028 | 0.68 | B44, A26-EY9 | |
| 517 | L | 89 | 19 | 0.88 | 0.98 | 0.027 | 0.68 | ||
| INT | 20 | K | 6 | 102 | 1.03 | 0.89 | 0.011 | 0.48 | |
| 119 | S | 80 | 28 | 0.88 | 0.96 | 0.028 | 0.68 | A01, B39, C05 | |
| 156 | K | 99 | 9 | 0.88 | 1.04 | 0.005 | 0.41 | B50 | |
| 156 | N | 9 | 99 | 1.04 | 0.88 | 0.005 | 0.41 | B50 | |
| 201 | I | 34 | 74 | 0.98 | 0.88 | 0.022 | 0.68 | ||
| 201 | V | 75 | 33 | 0.88 | 0.99 | 0.025 | 0.68 | ||
| 265 | A | 81 | 27 | 0.95 | 0.73 | 0.00008 | 0 | B42-VI9, B15-RY9 | |
| 265 | V | 27 | 81 | 0.73 | 0.95 | 0.00013 | 0 | B42-VI9, B15-RY9 | |
Discussion
We recently reported that elite controllers display defects in Gag-Protease function due in part to immune selection by protective HLA alleles, most notably B*5720, thus further supporting an impact of Gag escape mutations on viral fitness and HIV disease12–17, 19, 23. However, comparatively little is known about potential immune-mediated attenuation of other HIV-1 proteins and whether this might be relevant to the controller phenotype.
Here, we extend our previous findings and demonstrate that recombinant viruses encoding RT-Integrase sequences derived from elite controllers displayed significantly reduced RC compared to viruses derived from untreated chronic progressors. This was true regardless of whether recombinant viruses contained quasispecies diversity, or whether they were generated from cloned sequences. The observed defects in controller-derived viruses were not likely due to the presence of drug-resistance mutations24–28 or the presence of putative transmitted immune escape mutations7, 8, although the possibility that such mutations were transmitted but then subsequently reverted cannot be ruled out. The presence of RT drug resistance mutations in a minority of elite controller samples merits mention: previous treatment is not an exclusion criterion for the International HIV controllers study (http://www.hivcontrollers.org/). Alternatively, these mutations may represent transmitted resistance mutations.
Instead, reduced RC of controller-derived recombinant RT-Integrase viruses may be explained, at least in part, by the selection of fitness-attenuating mutations that result from effective HLA-restricted CTL responses to the incoming virus. We observed that viruses generated from B*51 and B*57-expressing controllers exhibited the most profound RC defects. Both of these alleles restrict strong CTL responses against epitopes in Pol41, 42 and have been identified as “protective” with respect to HIV disease progression36, 43–45. Furthermore, among B*57-expressing individuals, the number of B*57-associated polymorphisms correlated inversely with RC, suggesting that the accumulation of B*57-associated mutations in Pol can negatively impact viral RC in a dose-dependent manner. Similar results have been reported previously for HLA-B*5703-selected mutations in the Gag p24/capsid protein16. Notably, although both B*57+ controllers and progressors harbored B*57-associated escape mutations, viruses from B*57-expressing controllers displayed further reduced RC than their progressor counterparts, suggesting the presence of additional functional defects in controller sequences beyond those associated with commonly-observed primary escape mutations. Previous studies have described rare and/or unique Gag escape mutations associated with fitness costs in controllers22, although non-immune mechanisms cannot be ruled out. The selection of compensatory mutations in progressors could also contribute to observed differences, as has been reported in Gag13. Observed associations between HLA-A*01 and C*02 and viral RC are also notable and merit further study.
Substantial overlap in the RC distribution of controller- and progressor-derived viruses indicates that reduced RT-Integrase function is not common to all controllers. Similarly, viruses generated from B*51 and B*57-expressing progressors did not exhibit significantly reduced RC compared to their non-B*51/B*57-derived counterparts. This underscores the observation that expression of a protective allele does not guarantee viral attenuation, and that analysis at the individual sequence level is necessary to elucidate relationships between viral polymorphisms and fitness (indeed, analysis at the sequence-level revealed a significant dose-dependent relationship between the B*57-associaed escape mutations and RC in viruses from these individuals). Furthermore, more than half of controllers expressed neither B*51 nor B*57, yet they still harbored viruses with reduced RC compared to progressors, indicating that immune selection pressure by these two alleles does not solely account for the observed effects. We cannot rule out transmission of attenuated viruses in at least some controllers, nor the selection of unique mutations, immune-mediated or otherwise, that incur fitness costs.
The inherent challenges associated with identifying elite controllers and extracting HIV RNA from individuals with undetectable plasma viral loads limit the size and thus the power to comprehensively evaluate associations between RC and specific HLA alleles or viral polymorphisms. Despite this, we were able to identify a number of Pol codons that may be associated with reduced RC, including, among others, the B*57-associated V245E mutation in RT (residue 2 of B*57 ISW9 epitope46, 47) that is selected relatively rapidly following infection in B*57 (and B*58)-expressing individuals48. The observation that V245E reverts following transmission to non-B*57/B*58-expressing individuals48, 49 also supports a fitness cost. Integrase 265V was identified as the strongest correlate of lower RC, but mechanisms for this are unclear. This residue lies within described B*15 and B*42 epitopes; however, to our knowledge no HLA-associated polymorphisms have been reported. Although candidate residues were identified in this exploratory analysis, a comprehensive identification of codons associated with Pol RC will require a larger dataset followed by in vitro validation.
In summary, we observed significantly reduced RC of recombinant NL4-3 viruses encoding RT-Integrase from elite controllers compared to those from untreated chronic progressors, regardless of whether recombinant viruses contained quasispecies mixtures or were generated from cloned sequences. Controller-derived viruses from individuals expressing “protective” HLA-B*51 and B*57 alleles exhibited the lowest overall RC, and a dose-dependent inverse relationship was observed between the number of B*57-associated mutations and viral replication in both controllers and progressors. Although it is well-established that envelope is a major determinant of fitness2, 50, a growing body of evidence indicates that mutations outside envelope mediated by immune14, 16, 20, 23 or other27 selective pressures may also result in fitness defects. Results are consistent with functional defects in viruses isolated from HIV-1 elite controllers6, 20, 21, which may arise as a result of immune selection pressures that reduce viral replication capacity20.
Supplementary Material
Panel A: RT-Integrase recombinant viruses were generated from cloned sequences for a random panel of N=14 controllers and N=10 progressors. Consistent with results from bulk (quasispecies-containing) recombinant stocks, controller-derived viruses encoding clonal RT-Integrase sequences exhibited significantly lower RC than those derived from progressors (p=0.028). Panel B: Robust agreement is observed between the original RC of bulk (quasispecies-containing) and clonal recombinant virus stocks from the same individual (R=0.70; p=0.0003).
Acknowledgments
We gratefully acknowledge the efforts of the clinical and laboratory staff of the International HIV Controllers Study (IHCS), as well as the healthcare professionals and researchers who have referred HIV controllers or contributed samples to the IHCS. A complete list of contributors and collaborators can be found at http://www.hivcontrollers.org. We also thank the dedicated individuals who have participated in this study, without whom this research would not be possible. We also thank Alexander Filiposki at Simon Fraser University for laboratory assistance.
Sources of Support: This work was supported in part by grants AI028568 and AI030914 from the NIAID-NIH, the Howard Hughes Medical Institute, the Harvard University Center for AIDS Research, the Bill and Melinda Gates Foundation, and a gift from the Mark and Lisa Schwartz Foundation (to BDW), as well as an Operating Grant from the Canadian Institutes of Health Research (CIHR) and a Jim Gray Seed Grant from Microsoft Research (to MAB and ZLB). ZLB is supported by a CIHR New Investigator Award. CJB is supported by a NSERC Julie Payette Scholarship. The authors declare no conflicts of interest related to this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented in part as: Li C, Brumme ZL, Miura T, Rosato P, Sela J, Brumme CJ, Heckerman D, Pereyra F, Walker BD, Brockman MA. Reduced replication capacity of NL4-3 chimeric viruses encoding RT-Integrase sequences from HIV-1 elite controllers. Abstract P09-11, AIDS Vaccine 2009.
Disclosure: Funding for this study was received from NIH, HHMI, CIHR and other sources listed above.
References
- 1.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007 Sep;27(3):406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Quinones-Mateu ME, Ball SC, Marozsan AJ, et al. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol. 2000 Oct;74(19):9222–9233. doi: 10.1128/jvi.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaak H, Brouwer M, Ran LJ, de Wolf F, Schuitemaker H. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J Infect Dis. 1998 Mar;177(3):600–610. doi: 10.1086/514219. [DOI] [PubMed] [Google Scholar]
- 4.Miura T, Brockman MA, Brumme CJ, et al. Genetic Characterization of Human Immunodeficiency Virus type 1 in Elite Controllers: Lack of gross genetic defects or common amino acid changes. J Virol. 2008 Jun 18;82(17):8422–8430. doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navis M, Schellens I, van Baarle D, et al. Viral replication capacity as a correlate of HLA B57/B5801-associated nonprogressive HIV-1 infection. J Immunol. 2007 Sep 1;179(5):3133–3143. doi: 10.4049/jimmunol.179.5.3133. [DOI] [PubMed] [Google Scholar]
- 6.Lassen KG, Lobritz MA, Bailey JR, et al. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 2009 Apr;5(4):e1000377. doi: 10.1371/journal.ppat.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopera DR, Woodman Z, Mlisana K, et al. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 2008 Mar 7;4(3):e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goepfert PA, Lumm W, Farmer P, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008 Apr 21; doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999 Dec;73(12):10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troyer RM, Collins KR, Abraha A, et al. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol. 2005 Jul;79(14):9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arien KK, Vanham G, Arts EJ. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol. 2007 Feb;5(2):141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockman MA, Schneidewind A, Lahaie M, et al. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J Virol. 2007 Nov;81(22):12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneidewind A, Brockman MA, Yang R, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007 Nov;81(22):12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Picado J, Prado JG, Fry EE, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006 Apr;80(7):3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford H, Prado JG, Leslie A, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007 Aug;81(15):8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford H, Lumm W, Leslie A, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009 Apr 13;206(4):909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutwell CL, Rowley CF, Essex M. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J Virol. 2009 Mar;83(6):2460–2468. doi: 10.1128/JVI.01970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueno T, Motozono C, Dohki S, et al. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J Immunol. 2008 Jan 15;180(2):1107–1116. doi: 10.4049/jimmunol.180.2.1107. [DOI] [PubMed] [Google Scholar]
- 19.Bailey JR, O'Connell K, Yang HC, et al. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J Virol. 2008 Aug;82(15):7395–7410. doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura T, Brockman MA, Brumme ZL, et al. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J Virol. 2009 Jan;83(1):140–149. doi: 10.1128/JVI.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura T, Brumme ZL, Brockman MA, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J Virol. 2010 Aug;84(15):7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura T, Brockman MA, Schneidewind A, et al. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphotye recognition. J Virol. 2009 Mar;83(6):2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troyer RM, McNevin J, Liu Y, et al. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 2009 Apr;5(4):e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrigan PR, Bloor S, Larder BA. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998 May;72(5):3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber J, Chakraborty B, Weberova J, Miller MD, Quinones-Mateu ME. Diminished replicative fitness of primary human immunodeficiency virus type 1 isolates harboring the K65R mutation. J Clin Microbiol. 2005 Mar;43(3):1395–1400. doi: 10.1128/JCM.43.3.1395-1400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paredes R, Sagar M, Marconi VC, et al. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J Virol. 2009 Feb;83(4):2038–2043. doi: 10.1128/JVI.02154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wainberg MA. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev Anti Infect Ther. 2004 Feb;2(1):147–151. doi: 10.1586/14787210.2.1.147. [DOI] [PubMed] [Google Scholar]
- 28.Gandhi RT, Wurcel A, Rosenberg ES, et al. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin Infect Dis. 2003 Dec 15;37(12):1693–1698. doi: 10.1086/379773. [DOI] [PubMed] [Google Scholar]
- 29.Brockman MA, Tanzi GO, Walker BD, Allen TM. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J Virol Methods. 2006 Feb;131(2):134–142. doi: 10.1016/j.jviromet.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Pereyra F, Addo MM, Kaufmann DE, et al. Genetic and Immunologic Heterogeneity among Persons Who Control HIV Infection in the Absence of Therapy. J Infect Dis. 2008 Feb 15;197(4):563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 31.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003 Oct;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 32.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galli RA, Sattha B, Wynhoven B, O'Shaughnessy MV, Harrigan PR. Sources and magnitude of intralaboratory variability in a sequence-based genotypic assay for human immunodeficiency virus type 1 drug resistance. J Clin Microbiol. 2003 Jul;41(7):2900–2907. doi: 10.1128/JCM.41.7.2900-2907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brumme ZL, John M, Carlson JM, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef Proteins. PloS ONE. 2009 Aug 19;4(8):e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honeyborne I, Prendergast A, Pereyra F, et al. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007 Apr;81(7):3667–3672. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 37.Kawashima Y, Pfafferott K, Frater J, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009 Feb 25;458(7238):641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goulder PJ, Phillips RE, Colbert RA, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997 Feb;3(2):212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 39.Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004 Aug;4(8):630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 40.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004 Dec 9;432(7018):769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 41.Altfeld M, Kalife ET, Qi Y, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006 Oct;3(10):e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomiyama H, Sakaguchi T, Miwa K, et al. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum Immunol. 1999 Mar;60(3):177–186. doi: 10.1016/s0198-8859(98)00113-x. [DOI] [PubMed] [Google Scholar]
- 43.Kawashima Y, Kuse N, Gatanaga H, et al. Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. J Virol. 2010 Jul;84(14):7151–7160. doi: 10.1128/JVI.00171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996 Apr;2(4):405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 46.Klein MR, van der Burg SH, Hovenkamp E, et al. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. J Gen Virol. 1998 Sep;79(Pt 9):2191–2201. doi: 10.1099/0022-1317-79-9-2191. [DOI] [PubMed] [Google Scholar]
- 47.Allen TM, Altfeld M, Geer SC, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005 Nov;79(21):13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brumme ZL, Brumme CJ, Carlson J, et al. Marked epitope and allele-specific differences in rates of mutation in HIV-1 Gag, Pol and Nef CTL epitopes in acute/early HIV-1 infection. J Virol. 2008 Jul 9;82(18):9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Gladden AD, Altfeld M, et al. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J Virol. 2007 Jan;81(1):193–201. doi: 10.1128/JVI.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lobritz MA, Marozsan AJ, Troyer RM, Arts EJ. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J Virol. 2007 Aug;81(15):8258–8269. doi: 10.1128/JVI.02739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panel A: RT-Integrase recombinant viruses were generated from cloned sequences for a random panel of N=14 controllers and N=10 progressors. Consistent with results from bulk (quasispecies-containing) recombinant stocks, controller-derived viruses encoding clonal RT-Integrase sequences exhibited significantly lower RC than those derived from progressors (p=0.028). Panel B: Robust agreement is observed between the original RC of bulk (quasispecies-containing) and clonal recombinant virus stocks from the same individual (R=0.70; p=0.0003).