Abstract
The discovery of Suv39h1, the first SET domain-containing histone lysine methyltransferase (HKMT), was reported in 2000. Since then, research on histone methylation has progressed rapidly. Among the identified HKMTs in mammals, G9a and GLP are the primary enzymes for mono- and dimethylation at Lys 9 of histone H3 (H3K9me1 and H3K9me2), and exist predominantly as a G9a–GLP heteromeric complex that appears to be a functional H3K9 methyltransferase in vivo. Recently, many important studies have reported that G9a and GLP play critical roles in various biological processes. The physiological relevance of G9a/GLP-mediated epigenetic gene regulation is discussed.
Keywords: G9a, GLP, H3K9, histone methylation
The role of histone post-translational modifications, especially histone lysine methylation, in diverse biological processes has been a rapidly growing research subject in the last decade. Since the Su(var)3-9–Enhancer of zeste–Trithorax (SET) domain of Suv39h1/KMT1a was demonstrated to be the catalytic domain for lysine methylation (Rea et al. 2000), many SET domain-containing molecules have been found to be novel histone lysine methyltransferases (HKMTs) in various species, including viruses, bacteria, yeast, fungi, and other multicellular organisms (Qian and Zhou 2006; Pontvianne et al. 2010). With the sole exception of Dot1/Dot1L, which methylates Lys 79 of histone H3, all of the identified HKMTs are SET domain-containing molecules. Furthermore, since two classes of histone lysine demethylases (HKDMs) have been discovered (Shi et al. 2004; Tsukada et al. 2006), it is now apparent that methylation of histone lysine residues is dynamically regulated by specific HKMTs and HKDMs (Klose and Zhang 2007; Mosammaparast and Shi 2010). Lysine demethylase 1 (LSD1, also known as KDM1) is a flavin adenine dinucleotide-containing enzyme that removes mono/dimethylation. The Jumonji C-terminal domain (JmjC) family of HKDMs uses Fe2+ and α-ketoglutarate as cofactors to remove all methylation states. In addition to histones, nonhistone substrates also have been reported for various HKMTs (SET7/SET9, G9a, GLP, PR-SET7/SET8, SMYD2, and SMYD3) (Sampath et al. 2007; Huang and Berger 2008) and HKDMs (LSD1) (Nicholson and Chen 2009). The biological significance of nonhistone lysine methylation currently is not well understood, and is also a subject of growing interest.
G9a/KMT1C and GLP/Eu-HMTase1/KMT1D are members of the Suv39h subgroup of SET domain-containing molecules, and together they are the key HKMTs for H3K9me1 and H3K9me2 (Jenuwein et al. 1998; Tachibana et al. 2001, 2002, 2005). Among the mammalian HKMTs, G9a and GLP have been extensively investigated at the biochemical and physiological levels, and some specific inhibitors have been developed. Recently, multiple studies have identified the critical roles that G9a and GLP play in diverse biological processes. This review introduces the biochemical properties of G9a and GLP and specific inhibitors, describes the biological significance of these enzymes, and discusses the unresolved aspects of G9a/GLP-mediated epigenetic gene regulation.
Biochemical properties of G9a and GLP
Lysine methyltransferase activity
G9a was reported as the second HKMT and can methylate histone H1 and H3 (K9 and K27) in vitro (Tachibana et al. 2001). Biochemically, both Suv39h1 and G9a can catalyze mono-, di-, and trimethylation reactions on H3K9 (Collins et al. 2005; Kubicek et al. 2007). Investigations of G9a-deficient and Suv39h1/h2-double-deficient cells demonstrated that Suv39h1/h2 are crucial HKMTs for H3K9me3 on pericentromeric heterochromatin, and that G9a is a major H3K9me1 and H3K9me2 HKMT of euchromatin (Tachibana et al. 2002; Peters et al. 2003; Rice et al. 2003). G9a is also involved in H3K9me3 modification in vivo (Yokochi et al. 2009), and it has been shown recently that G9a is indeed involved in H1 and H3K27 methylation in vivo (Trojer et al. 2009; Weiss et al. 2010; Wu et al. 2011).
GLP was originally described as a gene encoding a G9a-like protein. Since then, various biochemical studies have demonstrated that GLP and G9a possess the same substrate specificities on histones (Ogawa et al. 2002; Tachibana et al. 2005; Weiss et al. 2010). Although G9a and GLP independently exert HKMT activities and have the same histone substrate specificities in vitro, the levels of H3K9me1 and H3K9me2 are severely reduced by either G9a or GLP knockout. Moreover, the G9a and GLP double knockout does not further reduce H3K9me1 and H3K9me2 levels (Tachibana et al. 2005). Thus, G9a generally cannot compensate for the loss of GLP HKMT function in vivo, and vice versa.
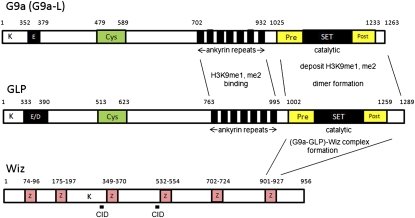
Biochemical characterization of G9a and GLP showed that G9a and GLP can form a homomeric and heteromeric complex via their SET domains; however, endogenous molecules exist exclusively as the stoichiometric G9a–GLP heteromeric complex in various human and mouse cells (Tachibana et al. 2005). Therefore, the G9a–GLP heteromeric complex seems to be the functional H3K9 HKMT in vivo (Tachibana et al. 2005, 2008). Furthermore, complementation experiments using methyltransferase-defective mutants have shown that the enzymatic activity of G9a is more important for the in vivo HKMT function than that of GLP (Tachibana et al. 2008). Although it is not yet clear why G9a or GLP generally cannot function alone as an HKMT in vivo, there are some possible mechanistic explanations for the dominant existence of the G9a–GLP heterocomplex inside cells, even though G9a and GLP can also form homodimers. First, G9a forms a stoichiometric complex not only with GLP but also with Wiz, a multi-zinc finger-containing molecule. Knockout or knockdown of GLP or Wiz concomitantly reduces G9a protein levels (Tachibana et al. 2005; Ueda et al. 2006). Second, Wiz seems to recognize and bind the homodimer or heterodimer structure of G9a and GLP SET domains, but most stably interacts with the G9a–GLP heterodimer (Ueda et al. 2006). Therefore, we speculate that the G9a–GLP(–Wiz) complex is the most stable form, and therefore exists as the dominant intracellular form. The domain organization of the core G9a complex components G9a, GLP, and Wiz is shown in Figure 1.
Figure 1.
Domain organization of the core G9a complex components: G9a, GLP, and Wiz. Amino acid sequences of mouse (m) G9a-L (9QZ148), mGLP (A2AIS4), and mWiz (O88286-2). (K) Potential methylation sites by G9a or GLP (K167 and K239 in G9a-L, K154 and K206 in GLP, and K467 in Wiz) (Sampath et al. 2007; Rathert et al. 2008); (E) Glu-rich region; (E/D) Glu/Asp-rich region; (Cys) Cys-rich region; (Pre) pre-SET domain; (SET) SET domain; (Post) post-SET domain; (Z) zinc finger motif; (CID) CtBP-interacting region (Ueda et al. 2006).
Link between H3K9 methylation and DNA methylation
In Neurospora, H3K9 methylation controls DNA methylation (Tamaru and Selker 2001; Rountree and Selker 2010). The DNA methyltransferase DIM-2 forms a complex with HP1. The DIM-5 HKMT methylates H3K9 to form H3K9me3. Subsequently, the DIM-2–HP1 complex is recruited to H3K9me3-positive nucleosome loci via HP1 binding to H3K9me3 and induces DNA methylation. In Arabidopsis, DNA methylation and H3K9 methylation are functionally interdependent (Jackson et al. 2002; Malagnac et al. 2002; Johnson et al. 2007; Tamaru 2010). The SET- or RING-associated (SRA) domain of the HKMT KRYPTONITE (KYP) binds to methylated DNA. In effect, KYP targets nucleosomes on methylated DNA and methylates H3K9 to form H3K9me2. Reciprocally, H3K9me2 recruits the DNA methyltransferase CHROMOMETHYLASE 3 to induce DNA methylation.
There are functional links between H3K9 methylation and DNA methylation in mammals as well. DNA methylation at major satellite repeats is reduced in Suv39h1 and Suv39h2 double-knockout embryonic stem (ES) cells (Lehnertz et al. 2003). Since the pericentromeric heterochromatin recruitment of HP1α and HP1β depends on Suv39h-mediated H3K9 methylation, and since HP1α and HP1β interact with DNA methyltransferase 3b (Dnmt3b), functional roles for Suv39h-mediated H3K9 methylation and HP1 recruitment in the regulation of DNA methylation have been proposed (Lehnertz et al. 2003).
DNA methylation is also affected in G9a or GLP knockout mouse ES cells (Dong et al. 2008; Epsztejn-Litman et al. 2008; Tachibana et al. 2008). However, in this case, this G9a/GLP-dependent DNA methylation is independent of the in vivo histone methyltransferase activity; i.e., catalytically inactive G9a partially restored the aberrant DNA methylation pattern in G9a−/− cells. There are some reports that DNMT1 regulates H3K9 methylation, although this matter is still open for debate. In HeLa cells, DNMT1 interacts with G9a, regulating chromatin loading of G9a, and knockdown of DNMT1 induces a reduction in H3K9me2 levels (Estève et al. 2006). Furthermore, DNMT1 knockout HCT116 human colon cancer cells also show a reduction in H3K9me2 and H3K9me3 levels (Espada et al. 2004). However, there is no reduction of H3K9me2 in Dnmt1 knockout or Dnmt1, Dnmt3a, and Dnmt3b triple-knockout mouse ES cells (Tsumura et al. 2006).
The hemimethylated DNA-binding molecule UHRF1/ICBT90/NP95 forms a complex with Dnmt1 and mediates Dnmt1-mediated maintenance DNA methylation during replication (Bostick et al. 2007; Sharif et al. 2007). Furthermore, the tandem Tudor domain and the plant homeodomain (PHD) of UHRF1 bind H3K9me2 and H3K9me3 (Hashimoto et al. 2009; Karagianni et al. 2008; Rottach et al. 2010). Although the phenotypes in UHRF1 knockout or knockdown studies indicate that the localization of UHRF1 to replicating heterochromatin is dependent on DNA methylation (Bostick et al. 2007; Sharif et al. 2007), the ability of UHRF1 to bind methylated H3K9 may facilitate the recruitment of Dnmt1 to chromatin containing both H3K9 methylation and hemimethylated DNA, and, hence, Dnmt1-mediated full methylation.
Nonhistone substrates of G9a and GLP
As mentioned previously, most of the SET domain lysine methyltransferases have been described as HKMTs. However, the SET domain molecule SETD6 methylates RelA (K310), a subunit of NF-κB, but not histones (Levy et al. 2011). In addition to self-methylation, G9a and GLP also methylate nonhistone substrates, including p53 (K372), Wiz (K305), CDYL1 (K135), ACINUS (K654), and Reptin (K67) (Sampath et al. 2007; Rathert et al. 2008; Huang et al. 2010; Lee et al. 2010). Currently, the biological roles of these nonhistone methylations are not well understood.
How are target specificities of G9a and GLP regulated?
As described above, G9a and GLP are the main HKMTs for H3K9me1 and H3K9me2 in euchromatin. Genome-wide analysis of different histone modifications suggests that the H3K9me2 mark is linked with transcriptional repression (Barski et al. 2007). In comparison with other histone methyl marks, H3K9me2 shows much longer spatial and continuous distribution on megabase-long stretches of genomic DNA (Wen et al. 2009). Thus, these data suggest that G9a/GLP-mediated H3K9me2 is integral to the establishment of these facultative heterochromatin domains, which are contained within larger expanses of euchromatin. Furthermore, in mouse ES cells, most of the G9a-repressed genes that are marked with H3K9me2 by G9a and derepressed by G9a inactivation are localized to the nuclear periphery (Yokochi et al. 2009). In human cells, nuclear lamina-associated large chromatin domains are transcriptionally repressed and relatively highly methylated with H3K9me2 (Guelen et al. 2008). Whether this nuclear lamina association is important for target specificities of H3K9me2 deposited by the G9a–GLP complex or is simply linked with transcriptional silencing is not clear. In any case, the H3K9me2 mark is not essential for the nuclear peripheral localization of specific chromatin loci because the G9a-repressed genes remain in the nuclear periphery in ES cells after G9a inactivation and a reduction in H3K9me2 levels (Yokochi et al. 2009).
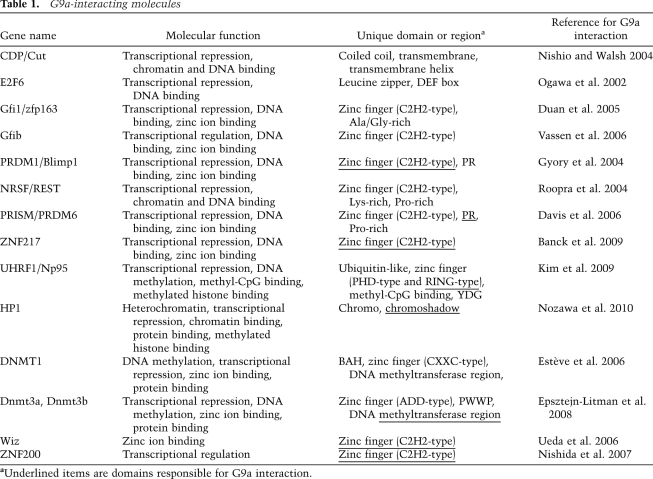
Many G9a-interacting molecules have been reported (Table 1). Most of these are characterized as repressive chromatin proteins and also multi-zinc finger molecules. Biochemically, G9a and GLP efficiently methylate histone octamers in vitro, but exhibit an extremely low rate of catalysis for nucleosomal histones (Tachibana et al. 2005; Shinkai 2007). Based on these molecular properties of G9a and GLP, we propose the following model for the G9a–GLP-mediated H3K9 methylation reaction in vivo. The G9a–GLP complex localizes to specific chromatin loci due to multiple interactions of the complex with chromatin or sequence-specific DNA-binding molecules. During replication, the G9a–GLP complex deposits K9me2 marks on newly synthesized H3 just before its incorporation into chromatin. Consistent with this notion, G9a colocalizes with replication foci during S phase (Estève et al. 2006).
Table 1.
G9a-interacting molecules
H3K9me2 spreading and H3K9me binding by G9a and GLP ankyrin repeats (ANKs)
Because the chromodomain of HP1 binds to H3K9me2 and H3K9me3, and because H3K9 HKMT Suv39h1 itself interacts with HP1, a self-enforcing spreading mechanism has been proposed (Bannister et al. 2001; Lachner et al. 2001). The prevalence of H3K9me2 in megabase-long chromatin domains suggests that the contribution of the spreading mechanism is much greater for H3K9me2 than for H3K9me3. HP1 also exists in G9a- and GLP-containing complexes (Ogawa et al. 2002; Nishio and Walsh 2004; Nozawa et al. 2010), and self-methylation of G9a is involved in the G9a–HP1 interaction (Sampath et al. 2007). Furthermore, the ANKs of G9a and GLP also bind to H3K9me1 and H3K9me2 (Collins et al. 2008). Therefore, once the G9a–GLP complex initiates an H3K9 methylation reaction at a specific target locus, these multi-H3K9me-binding modules (HP1 chromodomain plus G9a and GLP ANKs) may enhance the spreading of H3K9me2 marks, again via G9a/GLP-mediated methylation reactions (Collins and Cheng 2010).
Development of G9a and GLP inhibitors
Lysine methyltransferases, including SET domain-containing molecules, transfer one to three methyl groups from S-adenosyl-L-methionine (SAM) to the ɛ-amino group of the target lysine residue. Chaetocin, a competitive inhibitor of SAM, was the first HKMT inhibitor to be discovered, and it specifically inhibits the enzymatic activities of HKMTs belonging to members of the SUV39 family, including SUV39H1, dSU(VAR)3-9, G9a, DIM-5, GLP, and ESET (Greiner et al. 2005). Another more specific H3K9 HKMT inhibitor, BIX01294 (BIX), has also been described (Kubicek et al. 2007). BIX was originally reported as a G9a-specific inhibitor (half maximal inhibitory concentration [IC50] = 1.7 μM), but has been shown lately to exhibit a more potent inhibitory activity against GLP (G9a IC50 = 1.9 μM vs. GLP IC50 = 0.7 μM [Chang et al. 2009]; G9a IC50 = 250 nM vs. GLP IC50 = 27 nM, [Quinn et al. 2010]). More potent G9a/GLP inhibitors have been developed by further modification of BIX (E72: GLP IC50 = 100 nM [Chang et al. 2010]; UNC0321: G9a IC50 = 9.0 nM and GLP IC50 = 15 nM, [Liu et al. 2010]). Structural comparison of GLP bound to BIX with that of GLP bound to a substrate peptide suggests that the BIX family of small molecules acts as competitive inhibitors of the N-terminal peptides of H3 (K4 to R8 or K9) (Chang et al. 2010). It is worth noting that the G9a/GLP inhibitor activity of BIX is robust if an H3 N-terminal oligopeptide is used as a substrate for the in vitro methyltransferase assay, but is not significant (no inhibition at 10 μM) if full-length H3 is used (Y Shinkai, unpubl.).
Roles of G9a and GLP: relevance to various biological processes in mammals and human diseases
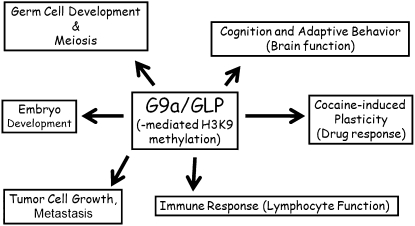
Since G9a and GLP are expressed ubiquitously and function as major euchromatic H3K9me1 and H3K9me2 HKMTs, multiple biological roles have been proposed (Fig. 2). Both G9a and GLP play an important role in mouse development, as shown by the fact that G9a and GLP knockout lead to embryonic lethality around embryonic day 9.5 (E9.5) due to severe growth defects (Tachibana et al. 2002, 2005). Importantly, mutant mice with catalytically inactive G9a HKMT also have an embryonic lethal phenotype similar to that of G9a knockout mice (M Tachibana and Y Shinkai, unpubl.), suggesting that G9a-mediated H3K9 methylation is important for mouse development. Furthermore, G9a may regulate the expression of some imprinted genes (Nagano et al. 2008; Wagschal et al. 2008). Germ lineage-specific G9a knockout also induces developmental defects of germ cells, and completion of meiosis is not observed in either sex (Tachibana et al. 2007). In the case of lymphocyte-specific G9a knockout, lymphocyte development is mostly intact; however, some immune responses are affected or impaired. B-cell-specific G9a knockout mice have a reduced usage of Igλ L chains, and a reduction in Igλ gene assembly in bone marrow precursor cells is also seen. G9a-deficient B cells do not respond well to LPS + IL-4 in vitro (Thomas et al. 2008). T-cell development in the thymus is intact in T-cell-specific G9a knockout mice; however the expression of lineage-specific Th2-associated cytokine genes such as IL-4, IL-5, and IL-13 are impaired, and the mice fail to defend themselves against parasitic infections (Lehnertz et al. 2010).
Figure 2.
Multiple biological roles of G9a/GLP. Multiple biological roles of G9a/GLP are proposed, based mainly on the results of studies using G9a or GLP knockout mice.
Phenotypes of the postnatal and forebrain neuron-specific G9a or GLP knockout mice are interesting (Shaefer et al. 2009). In such mice, multiple nonadult neuronal genes are derepressed in the forebrain, but there are no obvious neuronal developmental or architectural defects. However, these mice show various abnormal phenotypes, especially defects in cognition and adaptive behaviors. Similar roles of G9a in neuronal function are conserved in Drosophila, and EHMT/G9a-deficient flies show defects in learning and memory (Kramer et al. 2011). One copy of the EHMT1/GLP gene is deleted or disrupted in 9q34.3 subtelomeric deletion syndrome patients with severe mental retardation (Kleefstra et al. 2005, 2006). Indeed, not only forebrain-specific GLP homozygous knockout mice but also the heterozygous knockout mice show abnormal behaviors, including reduced locomotor activity and exploration (Shaefer et al. 2009). In addition, complete GLP knockout heterozygous mice also show reduced exploration, increased anxiety, and altered social behavior (Balemans et al. 2010). These results suggest that reduced function of G9a, GLP, or the G9a–GLP complex in the forebrain may, in part, lead to the phenotypes of 9q34.3 subtelomeric deletion syndrome patients.
Interesting findings have further been reported regarding a critical role of G9a-mediated H3K9me2 in cocaine-induced neuronal responses (Maze et al. 2010). Repeated cocaine exposure induces alterations in gene expression in the nucleus accumbens (NAc) neurons in mice, and this change is critical for cocaine-induced changes in neural morphology and behavior that may underlie cocaine addiction. Of the known H3K9 and H3K27 HKMT and HKDM genes, the expression levels of G9a and GLP are specifically diminished in the NAc neurons by repeated cocaine exposure in mice. Consistent with this finding, levels of H3K9me2 are also significantly reduced. If this cocaine-induced G9a reduction is complemented by exogenous G9a expression, the cocaine-induced neuronal morphological and behavioral changes are suppressed. On the other hand, if G9a function is inactivated specifically in NAc neurons, the neuronal morphological changes are enhanced even without cocaine exposure, and the behavioral preference for cocaine is further enhanced. Repeated cocaine exposure induces the accumulation of the transcription factor ΔFosB in NAc neurons. ΔFosB represses G9a expression, and the consequent reduction of H3K9me2 induces up-regulation of G9a-repressed genes. If these molecular mechanisms in mice can be extrapolated to cocaine addiction in humans, G9a (G9a–GLP complex) or the HKMT activity of G9a would be a novel target for controlling cocaine addiction.
In addition, G9a and GLP have also both been found to be up-regulated in various types of human cancer cells (Huang et al. 2010). Consistent with this finding, knockdown of G9a suppresses tumor cell growth in vitro and the invasion of cancer cells in nude mice (Kondo et al. 2008; Chen et al. 2010).
Finally, G9a is involved in provirus silencing. A provirus is a viral genome that is integrated into the host genome, and it is generally in the cell's best interest to silence proviruses. Such silencing in mouse ES cells is effected by another H3K9 HKMT, ESET/SETDB1/KMT1E (Matsui et al. 2010). G9a and GLP control H3K9me2 and DNA methylation of endogenous retroviruses (ERVs) in mouse ES cells (Dong et al. 2008). However, ERVs are not reactivated in G9a or GLP knockout ES cells, since ESET and ESET-mediated H3K9me3 are still maintained on the ERVs (Dong et al. 2008; Matsui et al. 2010). Further characterization indicates that newly integrated active proviruses are not silenced efficiently in G9a knockout ES cells, suggesting that G9a is dispensable for maintenance but is critical for establishment of proviral silencing in ES cells (Leung et al. 2011).
Concluding remarks
H3K9 methylation is a highly conserved histone post-translational modification and is commonly linked with both facultative and constitutive heterochromatin formation and transcriptional repression. Furthermore, the proteins that bind to methylated H3K9 (HP1/Swi6) are also conserved. Studies using G9a or GLP knockout/knockdown mice and cells also reported that G9a/GLP-mediated H3K9me2 is highly linked with transcriptional silencing. However, it is still unclear whether HP1 recruitment and its downstream signaling pathway are the primary mechanisms for H3K9me2- or H3K9me3-mediated gene silencing, or whether other unknown mechanisms are more important. Recently, a subset of G9a/GLP has been shown to form a complex with Suv39h1 and ESET (Fritsch et al. 2010). As discussed for the G9a/GLP complex, it is interesting to address the functional significance of a complex possessing multiple (four in this case) HKMTs with the same substrate (H3K9) specificity. Finally, in plants and the yeast Schizosaccharomyces pombe, the RNAi machinery is integrated with the regulation of H3K9 methylation (Grewal 2010; Tamaru 2010). While the RNAi machinery is also conserved in mammals, it is not clear whether this system is involved in G9a/GLP-mediated H3K9 methylation.
Dysregulation of histone methylation may have a role in various human diseases. Recent reports strongly support this notion, and it is likely that more evidence will be reported in the future. It is obvious that efficient animal cloning by somatic nuclear transfer and the establishment of induced pluripotent stem (iPS) cells depends on the efficiency of epigenetic reprogramming. If dysregulation of histone modification is the underlying cause of some human diseases and this epigenetic dysregulation can be reprogrammed, then it may be possible for such diseases or abnormalities to be reset or cured. From this point of view, the abnormal behavioral phenotypes of the forebrain-specific G9a/GLP knockout mice (Schaefer et al. 2009) and the obese phenotype of the H3K9 HKDM Jmjd1a/JHDM2A/KDM3a knockout mice (Inagaki et al. 2009; Tateishi et al. 2009) are quite interesting. If genetic or pharmacological approaches can improve these knockout mice after the onset of phenotypes, then an epigenetic reprogramming strategy may be applicable to a variety of biological aims.
Acknowledgments
We thank Matthew Lorincz for providing unpublished information, and James Hejna for his assistance with the English editing of the manuscript. Y.S. and M.T. are supported by a Grant-in-Aid from the Ministry of Education, Science, Technology, and Culture of Japan.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2027411.
References
- Balemans MC, Huibers MM, Eikelenboom NW, Kuipers AJ, van Summeren RC, Pijpers MM, Tachibana M, Shinkai Y, van Bokhoven H, Van der Zee CE 2010. Reduced exploration, increased anxiety, and altered social behavior: autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behav Brain Res 208: 47–55 [DOI] [PubMed] [Google Scholar]
- Banck MS, Li S, Nishio H, Wang C, Beutler AS, Walsh MJ 2009. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics 4: 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X 2009. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol 16: 312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, Bedford MT, Shinkai Y, Snyder JP, et al. 2010. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J Mol Biol 400: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, et al. 2010. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res 70: 7830–7840 [DOI] [PubMed] [Google Scholar]
- Collins R, Cheng X 2010. A case study in cross-talk: the histone lysine methyltransferases G9a and GLP. Nucleic Acids Res 38: 3503–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X 2005. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem 280: 5563–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X 2008. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 15: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S, Owens GK, Olson EN 2006. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol 26: 2626–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol 25: 10338–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 15: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M 2004. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem 279: 37175–37184 [DOI] [PubMed] [Google Scholar]
- Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S 2010. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37: 46–56 [DOI] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A 2005. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol 1: 143–145 [DOI] [PubMed] [Google Scholar]
- Grewal SI 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5: 299–308 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Cheng X 2009. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4: 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Berger SL 2008. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158 [DOI] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL 2010. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 285: 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Tachibana M, Magoori K, Kudo H, Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y, Sakai J 2009. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 14: 991–1001 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G 1998. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 54: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagianni P, Amazit L, Qin J, Wong J 2008. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol 28: 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Estève PO, Jacobsen SE, Pradhan S 2009. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 37: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Smidt M, Banning MJ, Oudakker AR, Van Esch H, de Brouwer AP, Nillesen W, Sistermans EA, Hamel BC, de Bruijn D, et al. 2005. Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet 42: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakke AR, Nillesen WM, Magee A, Geneviève D, Cormier-Daire V, van Esch H, Fryns JP, et al. 2006. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet 79: 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y 2007. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa JP 2008. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS ONE 3: e2037 doi: 10.1371/journal.pone.0002037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Kochinke K, Oortveld MA, Marks H, Kramer D, de Jong EK, Asztalos Z, Westwood JT, Stunnenberg HG, Sokolowski MB, et al. 2011. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol 9: e1000569 doi: 10.1371/journal.pbio.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, et al. 2010. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell 39: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Northrop JP, Antignano F, Burrows K, Hadidi S, Mullaly SC, Rossi FM, Zaph C 2010. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J Exp Med 207: 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. 2011. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci doi: 10.1073/pnas.1014660108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, et al. 2011. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat Immunol 12: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, Senisterra G, Chau I, Siarheyeva A, et al. 2010. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem 53: 5844–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J 2002. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. 2010. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79: 155–179 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Nicholson TB, Chen T 2009. LSD1 demethylates histone and non-histone proteins. Epigenetics 4: 129–132 [DOI] [PubMed] [Google Scholar]
- Nishida M, Kato M, Kato Y, Sasai N, Ueda J, Tachibana M, Shinkai Y, Yamaguchi M 2007. Identification of ZNF200 as a novel binding partner of histone H3 methyltransferase G9a. Genes Cells 12: 877–888 [DOI] [PubMed] [Google Scholar]
- Nishio H, Walsh MJ 2004. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci 101: 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, Kimura H, Obuse C 2010. Human POGZ modulates dissociation of HP1α from mitotic chromosome arms through Aurora B activation. Nat Cell Biol 12: 719–727 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Pikaard CS 2010. Arabidopsis histone lysine methyltransferases. Adv Bot Res 53: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Zhou MM 2006. SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci 63: 2755–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A 2010. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors. Mol Biosyst 6: 782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A 2008. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4: 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF 2004. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell 14: 727–738 [DOI] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H 2010. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res 38: 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Selker EU 2010. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity 105: 38–44 [DOI] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbräuker I, Viale A, Rudensky E, Zhou MM, Chait BT, et al. 2007. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell 27: 596–608 [DOI] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P 2009. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Shinkai Y 2007. Regulation and function of H3K9 methylation. Subcell Biochem 41: 337–350 [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276: 25309–25317 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N, Shinkai Y 2007. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J 26: 3346–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y 2008. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H 2010. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Dev 24: 1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Tateishi K, Okada Y, Kallin EM, Zhang Y 2009. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LR, Miyashita H, Cobb RM, Pierce S, Tachibana M, Hobeika E, Reth M, Shinkai Y, Oltz EM 2008. Functional analysis of histone methyltransferase G9a in B and T lymphocytes. J Immunol 181: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D 2009. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. J Biol Chem 284: 8395–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 43: 811–816 [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. 2006. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11: 805–814 [DOI] [PubMed] [Google Scholar]
- Ueda J, Tachibana M, Ikura T, Shinkai Y 2006. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 281: 20120–20128 [DOI] [PubMed] [Google Scholar]
- Vassen L, Fiolka K, Möröy T 2006. Gfi1b alters histone methylation at target gene promoters and sites of γ-satellite containing heterochromatin. EMBO J 25: 2409–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Sutherland HG, Woodfine K, Henckel A, Chebli K, Schulz R, Oakey RJ, Bickmore WA, Feil R 2008. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol Cell Biol 28: 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM, Dundr M, Garcia BA, Daujat S, Schneider R 2010. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin 3: 7 doi: 10.1186/1756-8935-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP 2009. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 41: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Chen X, Xiong J, Li Y, Li H, Ding X, Liu S, Chen S, Gao S, Zhu B 2011. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell Res 21: 365–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci 106: 19363–19368 [DOI] [PMC free article] [PubMed] [Google Scholar]