Abstract
Satellite cells (SCs) sustain muscle growth and empower adult skeletal muscle with vigorous regenerative abilities. Here, we report that EZH2, the enzymatic subunit of the Polycomb-repressive complex 2 (PRC2), is expressed in both Pax7+/Myf5− stem cells and Pax7+/Myf5+ committed myogenic precursors and is required for homeostasis of the adult SC pool. Mice with conditional ablation of Ezh2 in SCs have fewer muscle postnatal Pax7+ cells and reduced muscle mass and fail to appropriately regenerate. These defects are associated with impaired SC proliferation and derepression of genes expressed in nonmuscle cell lineages. Thus, EZH2 controls self-renewal and proliferation, and maintains an appropriate transcriptional program in SCs.
Keywords: Polycomb, Ezh2, satellite cells, muscle growth, muscle regeneration
Adult skeletal muscle regenerates in response to traumatic injuries or degenerative conditions. This property is afforded mainly by satellite cells (SCs), a heterogeneous population of resident committed myogenic progenitors and noncommitted stem cells (Sherwood et al. 2004; Collins et al. 2005; Montarras et al. 2005; Kuang et al. 2007). In the mouse, postnatal SCs are mitotically active for the initial 2 wk after birth. After this period, they enter quiescence and their number declines. However, following muscle injury or degeneration, adult SCs undergo intense proliferation and efficiently differentiate. To replenish the reservoir, a subset of dividing SCs returns to the niche following a process of asymmetric and symmetric cell division (Shinin et al. 2006; Conboy et al. 2007; Kuang et al. 2007; Shea et al. 2010). Approximately 10% of noncommitted Pax7+/Myf5− SCs can asymmetrically generate a self-renewing, noncommitted Pax7+/Myf5− cell and a committed Pax7+/Myf5+ daughter cell in vivo. The noncommitted Pax7+/Myf5− cell returns to the niche to maintain the SC reservoir, while the committed Pax7+/Myf5+ SC undergoes several rounds of cell division and the ensuing cells eventually differentiate into pre-existing or newly formed myofibers (Kuang et al. 2008).
Polycomb group (PcG) proteins regulate differentiation of totipotent embryonic stem (ES) cells and maintenance of multipotent and progenitor stem cell populations (Sauvageau and Sauvageau 2010). The Polycomb-repressive complex 2 (PRC2) subunit EZH2 methylates histone H3 Lys 27 (H3K27me3), establishing an epigenetic mark that identifies repressed chromatin regions. Ablation of PRC2 members in ES cells impairs their differentiation (Pasini et al. 2007; Chamberlain et al. 2008; Shen et al. 2008) and results in unscheduled expression of mixed cell lineage genes (Boyer et al. 2006; Lee et al. 2006). While PcG establishes and maintains positional patterning of the body axis through regulation of Hox genes in both Drosophila and mammals, its role in conferring cell identity by repressing inappropriate cell lineage-specific transcription in animal development has not been demonstrated. Indeed, derepression of mixed cell lineage genes does not occur in epidermal, neuronal, or pancreatic cells of Ezh2 conditional null mice (Chen et al. 2009; Ezhkova et al. 2009; Hirabayashi et al. 2009).
We generated mice in which Ezh2 was conditionally ablated in SCs (Ezh2 muscle knockout, Ezh2mKO). While EZH2 was dispensable for fetal muscle development, it was required for postnatal muscle growth and adult muscle regeneration, ensuring appropriate homeostasis of the SC pool. Unlike other progenitor cells, reduced H3K27me3 in Ezh2mKO SCs was accompanied by RNA polymerase II (PolII) recruitment and transcriptional activation of genes normally repressed in SCs and expressed in other cell lineages, including cardiac progenitors, retinal cones, neurons, and chondrocytes. Thus, our findings indicate that EZH2, which regulates proliferation and maintains transcriptional identity of adult muscle stem cells, is an important molecular component of adult skeletal myogenesis.
Results and Discussion
Pax7 and Ezh2 are coexpressed in postnatal SCs
Pax7 is essential for SC specification and survival (Buckingham and Relaix 2007). While Pax7+ cells were present at embryonic day 15.5 (E15.5) and E17.5, EZH2 could not be detected in skeletal muscle but was expressed in the epidermis (Supplemental Fig. S1A; Ezhkova et al. 2009). At postnatal day 1 (P1), EZH2 was expressed in limb muscles, where it remained detectable, along with Pax7, for ∼14 d (P14) (Fig. 1A). Indeed, the vast majority of Pax7+ cells coexpressed EZH2 in muscle cross-sections of P8 animals (Fig. 1B). Both EZH2 and Pax7 decreased and became undetectable by immunoblot after P21 (Fig. 1A), consistent with cessation of myonuclei accretion.
Figure 1.
Ezh2 expression in limb muscles and SCs. (A) Limb muscle extracts from mice at P1–P60 probed with the indicated antibodies. (B) In P8 hindlimb muscles, EZH2 is expressed in Pax7+ cells. Arrowheads indicate cells positive for Pax7 and EZH2. Quantification of Pax7+/EZH2+ cells in P8 EDL muscles. (C, top panel) Myofiber-associated quiescent (0 h) Pax7+ SCs do not express a detectable level of EZH2. (Bottom panel) After 42 h, EZH2 is expressed in couplets of dividing Pax7+ SCs. (D) In single myofibers of Myf5-Cre/ROSA-YFP mice cultured for 42 h, EZH2 is expressed in both Pax7+/YFP− (top panel) and Pax7+/YFP+ (bottom panel) cells. Arrows indicate YFP+ cells, and arrowheads indicate YFP− cells. Bars, 50 μm.
Ezh2 is expressed in both noncommitted and myogenically committed proliferating SCs
To evaluate EZH2 expression in quiescent and activated (i.e., cells that have entered the cell cycle) SCs, individual myofibers and their associated SCs were isolated from the extensor digitorus longus (EDL) muscle of 2-mo-old mice (P60) and either fixed immediately or cultured for 42 h to stimulate SC proliferation. While Pax7 was expressed in both conditions, EZH2 was detected only in Pax7+ dividing SCs at 42 h (Fig. 1C). Neither Pax7 nor EZH2 was detected in myofiber nuclei (Fig. 1C). The low abundance of SCs at P21 prevented Pax7 detection (Fig. 1A).
The SC pool is composed of at least two hierarchical subpopulations (Kuang et al. 2007). Pax7+/Myf5− cells are noncommitted self-renewing stem cells maintaining the SC compartment and giving rise to Pax7+/Myf5+ myogenic committed precursors fated to differentiate into myofibers. To investigate EZH2 expression in these two subpopulations, we used a lineage-tracing approach based on the Myf5-Cre/ROSA-YFP mouse, where cells that express or have ever expressed Myf5-Cre are YFP+, while cells that have never expressed Myf5-Cre are YFP− (Kuang et al. 2007). Immunostaining with Pax7 and EZH2 antibodies of Myf5-Cre/ROSA-YFP myofibers cultured for 42 h in growth medium revealed that Pax7 and EZH2 were coexpressed in both YFP− (Fig. 1D, top panel) and YFP+ (Fig. 1D, bottom panel) cells (Fig. 1D). Thus, EZH2 is expressed in both dividing Pax7+/Myf5− noncommitted stem cells and Pax7+/Myf5+ muscle progenitors.
Mice with conditional ablation of Ezh2 in Pax7-derived muscle precursor cells have reduced muscle mass with smaller myofibers
Ezh2 was selectively ablated in Pax7-derived cells by crossing knock-in mice expressing Cre recombinase from the Pax7 locus (Pax7-Cre) (Keller et al. 2004) with mice bearing floxed Ezh2 alleles (Su et al. 2003). In Pax7-Cre+/−;Ezh2fl/fl mice (Ezh2mKO), both EZH2 RNA and protein were largely and specifically reduced in muscle tissue (Fig. 2A). Ezh2 deletion did not affect the expression of other members of the PRC2 complex, Suz12 and Eed, or the PRC1 protein Bmi1 (Supplemental Fig. S2A). Ezh2 floxed alleles were not deleted in the kidney, heart, and white fat (data not shown). Ezh2mKO mice had no obvious feeding difficulties and were apparently healthy. At E17.5, when EZH2 is not expressed in muscle tissues (Supplemental Fig. S1A), we did not observe gross morphological changes or appreciable abnormalities in Ezh2mKO fetal myofibers (Supplemental Fig. S1B). In addition, a comparable number of Pax7+ cells in the EDL of Ezh2mKO and littermate controls further suggested that EZH2 is not required for fetal muscle development (Supplemental Fig. S1B). However, at P8, the muscle size of Ezh2mKO animals was obviously smaller than that of littermate controls, and myofibers with a reduced cross-sectional area (CSA) were prevalent (Supplemental Fig. S2B,C). The myofiber number was comparable with that of littermate controls. These postnatal muscle defects persisted through adulthood, and both the body mass and the total muscle mass of P60 Ezh2mKO animals were significantly reduced (Fig. 2B,C), with a prevalence of smaller myofibers (Fig. 2D). Thus, Ezh2 ablation in Pax7-derived skeletal muscle cells results in defects of postnatal muscle growth characterized by reduced muscle mass with smaller muscle fibers.
Figure 2.
Conditional Ezh2 ablation results in postnatal skeletal muscle defects and an impoverished SC pool. (A) RNA (left) and protein (right) analysis of Ezh2 in wild-type (WT) (Pax7-Cre−/−; Ezh2fl/fl) and mKO (Pax7-Cre+/−; Ezh2fl/fl) littermates. EZH2fl and EZH2Δ correspond to floxed and SET-deleted Ezh2 RNAs, respectively. (B,C) Both body and muscle mass are significantly reduced in P60 mKO compared with control (wild-type) littermates. Data are presented as mean ± SD (n = 6); (***) P < 0.0005; (**) P < 0.005. (D) H&E staining of transverse sections of P60 TA reveals a reduced myofiber CSA in Ezh2mKO animals (n = 3). Ninety-five percent confidence intervals do not overlap. (E) EDL single myofibers from adult (P60) Ezh2mKO animals have a reduced number of SCs. Arrows indicate Pax7+ cells. Data are presented as mean ± SD (n = 6); (*) P < 0.05. (F,G) Pax7 immunostaining of FACS-isolated wild-type and Ezh2mKO SCs. Arrows indicate cells stained positive for Pax7 and DAPI. Data are presented as mean ± SD (n = 3); (**) P < 0.005.
Ezh2 is required for maintenance of the adult muscle SC pool
We examined the quiescent SC population of P60 mice. Pax7+ cells on freshly isolated individual myofibers were reduced by ∼40% in Ezh2mKO animals (Fig. 2E). To further substantiate these findings, we isolated the quiescent SC population from both wild-type and Ezh2mKO animals by FACS by gating on integrin α-7+ (positive selection) and Lin− (CD31−,CD45−,CD11−, Sca1−) (negative selection) cells. Although the percentage of the integrin-α7+Lin− population was similar in wild type and Ezh2mKO (Supplemental Fig. 3A), only ∼50% of the purified cells were Pax7+ in Ezh2mKO compared with ∼85% in wild type (Fig. 2F,G). In agreement with the results reported above, muscle cross-sections of Ezh2mKO animals contained only ∼50% of the Pax7+ cells of littermate controls (Supplemental Fig. S3B). Reduced Pax7 expression in Ezh2mKO is not the consequence of direct regulation by EZH2, since adenoviral Cre-mediated Ezh2 excision did not affect the Pax7 level (Supplemental Fig. S3C). Together, these data suggest that EZH2 regulates establishment and/or maintenance of the adult SC pool.
Ezh2 affects SC proliferation and differentiation
We evaluated the SC population by quantifying Pax7+ cells in P8 mice, when SCs are highly proliferative. Pax7+ cells, located under the basal lamina, were decreased by ∼40% in Ezh2mKO animals and, consistent with Ezh2 ablation, H3K27me3+ cells were hardly detected (Fig. 3A,B; Supplemental Fig. S3D). The reduced number of Pax7+ cells in Ezh2mKO animals suggested that SC proliferation may be impaired. To test this hypothesis, individual myofibers derived from either wild-type or Ezh2mKO EDL muscle were isolated and cultured for 3 d to allow for SC delamination and proliferation. The number of Pax7+ cells derived from 3-d cultured Ezh2mKO myofibers was significantly curtailed, when compared with that of wild-type myofibers (Fig. 3C). An equivalent number of FACS-isolated cells obtained from either wild-type or Ezh2mKO animals was plated and cultured for 14, 48, and 96 h, respectively. Fewer Ezh2mKO cells were present after 48 and 96 h in culture (Fig. 3D). 5-bromo-2′-deoxyuridine (BrdU) cell incorporation and immunostaining with an antibody detecting histone H3Ser10 phosphorylation—a marker of the G2/M mitotic transition—indicated that Ezh2mKO-derived cells had reduced proliferative capacity (Fig. 3E). Apoptosis was not increased in Ezh2mKO SCs (Supplemental Fig. S4A). The proliferation defects of Ezh2mKO SCs are likely the consequence of derepression of the cell cycle inhibitor p16 (Cdkn2a) (Fig. 3F; Supplemental Table S1; Bracken et al. 2007; Kotake et al. 2007; Chen et al. 2009; Ezhkova et al. 2009; Pereira et al. 2010). To directly test this hypothesis, we reduced p16 in FACS-isolated and cultured SCs by siRNA (Supplemental Fig. S4B). A higher percentage of Ezh2mKO SCs with reduced p16 scored positive for H3Ser10 phosphorylation compared with control siRNA transfected Ezh2mKO SCs. Reducing p16 did not affect H3Ser10 phosphorylation of wild-type SCs. (Supplemental Fig. S4B). Thus, p16 up-regulation is partly responsible for the proliferative defects of Ezh2mKO SCs. Despite the reduced number of Pax7+ cells per clone (Fig. 3C), the majority of Ezh2mKO SCs cultured for 72 h expressed myogenin (Fig. 3G), suggesting earlier differentiation potential (Juan et al. 2009). Overall, these results indicate that EZH2 plays an important role in the SC's proliferation and differentiation.
Figure 3.
Skeletal muscle progenitors derived from Ezh2mKO animals have proliferative defects and precociously activate myogenin. (A,B) Immunostaining of H3K27me3 and Pax7 of P8 EDL. Arrowheads indicate cells stained positive for both Pax7 and H3K27me3. Arrows indicate Pax7+/H3K27me3− cells. Data are presented as mean ± SD (n = 3); (***) P < 0.0005; (**) P < 0.005. (C) Pax7 immunostaining of EDL single myofibers isolated from wild-type (WT) and Ezh2mKO mice cultured for 3 d. (*) P < 0.05. (D) FACS-isolated wild-type and Ezh2mKO SCs after 1 or 4 d in culture. Cells (1.5 × 104) of wild-type and Ezh2 mKO SCs were seeded, and the total number obtained from counting cells present in 10 independent microscopic fields after 14, 48, and 96 h in culture was plotted in the curve graph (n = 4; P < 0.001 and P < 0.003 for 42-h and 96-h counts, respectively). (E) Percentage of FACS-isolated wild-type and Ezh2mKO SCs that have incorporated BrdU or that stained positive for histone-H3 (H3-P). Data are presented as mean ± SD for BrdU (n = 3; [**] P < 0.005) and H3-P (n = 4; [**] P < 0.005). (F) Increased p16 protein in Ezh2mKO muscles. GAPDH serves as a loading control. (G) Myogenin immunostaining of SC clones from wild-type and Ezh2mKO myofibers. Data are presented as mean ± SD; (*) P < 0.05.
Muscle regeneration is compromised in Ezh2mKO animals
The proliferative defects of SCs and the depletion of the quiescent SC pool observed in Ezh2mKO mice suggested that muscle regeneration may be compromised in these animals. To test this hypothesis, muscle damage was induced by injecting the tibialis anterior (TA) muscle of adult (P60) wild-type and Ezh2mKO animals with cardiotoxin (CTX). Three days after CTX injection, Pax7 and myogenin, which are normally undetectable by immunoblot in uninjured adult muscles (Fig. 1A), were readily seen in wild type but were reduced in Ezh2mKO animals (Fig. 4A). Reduced myogenin expression in Ezh2mKO animals is likely the consequence of defective SC proliferation rather than impaired differentiation, as Ezh2mKO SCs display anticipated myogenin expression (Fig. 3G). Seven days after CTX injection, proliferation of Pax7+ cells in the Ezh2mKO regenerating muscle was severely impaired, as indicated by an almost 10-fold reduction of double-positive Pax7+/phospho-H3+ cells (Fig. 4B). At this stage, while regenerating myofibers of littermate controls had almost extinguished expression of embryonic myosin heavy chain (eMyHC), a marker of muscle regeneration, Ezh2mKO myofibers exhibited intense eMyHC staining and were smaller, indicating that muscle repair in Ezh2mKO lagged behind that of control mice (Fig. 4C). Indeed, the majority of newly formed myofibers, identified by the presence of nuclei located in a central position, had a smaller CSA in Ezh2mKO animals (Fig. 4D). Overall, these findings indicate that EZH2 is required for appropriate SC proliferation during muscle regeneration.
Figure 4.
Impaired muscle regeneration in Ezh2mKO animals. (A) Immunoblots of limb muscle extracts from wild-type (WT) or Ezh2mKO animals (n = 2) 3 d after CTX, probed with myogenin, Pax7, and GAPDH antibodies. (B) Pax7 and H3-P immunostaining of TA muscles from wild-type and Ezh2mKO animals 7 d after CTX injection. Data are presented as mean ± SD (n = 3); (***) P < 0.0005; (*) P < 0.05. (C) Embryonic MyHC [MyHC(emb)] immunostaining in wild-type and Ezh2mKO mice in 7-d regenerating myofibers. (D) H&E staining of transverse sections of wild-type or Ezh2mKO regenerating TA muscles 7 d after CTX injection indicates that the CSA of centrally nucleated myofibers is reduced in Ezh2mKO animals (n = 3). (Right panel) Ninety-five percent confidence intervals do not overlap.
Ezh2 maintains the transcriptional identity of SCs
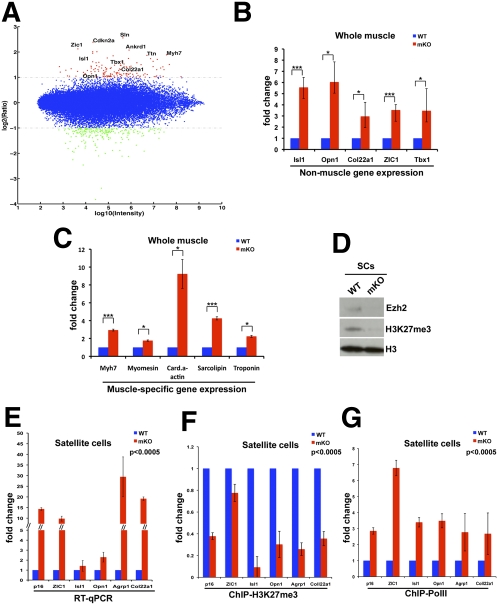
We compared transcriptional profiles of wild-type and Ezh2mKO muscles by microarray analysis (Fig. 5A; Supplemental Table S1). Among the down-regulated transcripts in Ezh2mKO muscle were several corresponding to developmental regulators and structural proteins (Supplemental Table S1). It is likely that reduced transcription is an indirect effect of Ezh2 ablation mediated by intermediate regulators, as H3K27me3 is not known to be directly associated with transcriptional activation. Gene ontology (GO) analysis for up-regulated genes found significant enrichment for functional classes, including those corresponding to muscle functions (Supplemental Table S2). Indeed, transcripts for muscle-expressed titin, sarcolipin, myosin Myh7, myomesin3, cardiac actin, Ankrd1, and troponin Tnni1 were increased in Ezh2mKO (Fig. 5A; Supplemental Table S1). In addition, several developmental regulators expressed in multiple lineages that are PRC targets in ES cells were also up-regulated (Fig. 5A; Supplemental Table S1). Among those were ZIC-1 (expressed in early somites and the cerebellum), Isl1 (expressed in cardiac progenitors and the pancreas), and Tbx1 (expressed in cardiac progenitors). Transcripts of structural genes expressed in other cell lineages were also up-regulated in Ezh2mKO. For instance, expression of the agouti-related protein homolog 1 (Agrp1), expressed in neurons of the arcuate nucleus of the hypothalamus (Hahn et al. 1998); the collagen Col22a1, specifically expressed in chondrocytes (Koch et al. 2004); and opsin1 (Opn1), encoding a retinal photopigment expressed in retinal cones (Shichida and Matsuyama 2009) was increased (Fig. 5A; Supplemental Table S1). Expression of some of the transcripts identified as up-regulated by microarray analysis was evaluated independently by quantitative PCR (qPCR) and confirmed to be increased in the limb muscles of Ezh2mKO animals. (Fig. 5B,C). Importantly, expression of mixed cell lineage genes and p16 was increased in FACS-isolated quiescent Ezh2mKO SCs (Fig. 5E). However, muscle-specific transcripts increased in Ezh2mKO muscles were not augmented in Ezh2mKO SCs (data not shown), suggesting that their misregulation occurs in myofibers.
Figure 5.
Ezh2 maintains transcriptional identity of SCs. (A) Log intensity versus ratio scatter plot of gene expression obtained from microarray data derived from muscles of wild-type (n = 2) and Ezh2mKO (n = 2) animals. The X-axes represents the log10 of the average intensity of the data, and the Y-axes represents the log2 of their ratio. Each dot represents one gene. Up-regulated and down-regulated genes are color-coded red and green, respectively (fold change >2). (B,C) Expression of selected up-regulated genes identified in A by qPCR in muscles of wild-type or Ezh2mKO animals. Data are presented as mean ± SD (n = 3); (***) P < 0.0005; (*) P < 0.05. (D) EZH2 and H3K27me3 immunoblots of FACS-isolated Ezh2mKO SCs. Total histone H3 serves as loading control. (E) qPCR on selected up-regulated genes from FACS-isolated wild-type or Ezh2mKO SCs. Data are presented as mean ± SD (n = 3); P < 0.0005 for all the values. (F,G) ChIP-qPCR for H3K27me3 and PolII at selected genes up-regulated in Ezh2mKO SCs. Data are presented as mean ± SD (n = 3); P < 0.0005 for all the values.
PolII recruitment to mixed cell lineage genes and the Ink4a/Arf locus in Ezh2mKO SCs
To mechanistically investigate how Ezh2 ablation causes gene derepression, we conducted chromatin immunoprecipitation (ChIP) assays with antibodies against H3K27me3 and PolII. ZIC-1, Ink4a/Arf (p16), Isl1, Opn1, and Coll22a1 genes were marked by H3K27me3, and PolII was undetectable in wild-type SCs. In contrast, H3K27me3 was significantly reduced and PolII was recruited at these genes in Ezh2mKO SCs (Fig. 5D,F,G; Supplemental Fig. S4C). Overall, these results indicate that SCs are the source of mixed cell lineage and p16 gene misexpression, and that reduced H3K27me3 in Ezh2mKO SCs allows PolII chromatin access, resulting in unwarranted gene transcription.
Conditional ablation of Ezh2 in MyoD-expressing cells recapitulates the muscle phenotypes observed in Ezh2mKO mice
Pax7 expression is not limited to SCs but extends to cells located in the CNS and craniofacial tissues (Jostes et al. 1990). Indeed, some Pax7-Cre- Ezh2mKO mice with CNS defects were retrieved at E15.5 and E17.5 (data not shown). To ablate Ezh2 in committed skeletal myogenic cells (MyoD+ cells), we bred MyoD-Cre (Chen et al. 2005) and floxed Ezh2 mice (Supplemental Fig. S5A). Body and muscle mass were greatly reduced in MyoD-Cre-Ezh2mKO mice (Supplemental Fig. S5B–E). Moreover, the number of Pax7+ cells was also reduced in muscle cross-sections (Supplemental Fig. S5F). Recent studies have documented an activation of the MyoD locus in prenatal SCs (Kanisicak et al. 2009). Thus, MyoD-Cre-Ezh2mKO mice may have deleted the Ezh2fl/fl alleles in SCs capable of self-renewal. While p16 derepression in MyoD-Cre-Ezh2mKO was comparable with that observed in Pax7-Cre-Ezh2mKO mice, expression of mixed-lineage genes ZIC-1, Agrp1, and Coll22a1 was much less pronounced (Supplemental Fig. S5G). Overall, the phenotypic similarities of Pax7-Cre-Ezh2mKO and MyoD-Cre-Ezh2mKO mice suggest that the observed muscle defects can be imputed to Ezh2 ablation in skeletal myogenic cells.
Conclusions
The present study revealed that EZH2 influences several aspects of SC biology, including self-renewal, proliferation, and cell identity.
Unlike other studies in which Ezh2 deletion was conditionally obtained in committed progenitors or differentiated cells (Chen et al. 2009; Ezhkova et al. 2009; Hirabayashi et al. 2009), we observed that Pax7-induced Ezh2 deletion resulted in derepression of developmental regulators and structural genes physiologically not expressed in SCs. We speculate that, in committed or differentiated cells, the chromatin structure at selected genomic regions may be insensitive to epigenetic modifications caused by Ezh2 ablation. Indeed, when Ezh2 was deleted in committed myogenic precursors (MyoD+), gene misexpression was barely observed (Supplemental Fig. S5G). A more naive and plastic chromatin environment, such as that of ES cells or noncommitted Pax7+/Myf5− stem cells, may react to PcG gene ablation by dysregulating gene expression (Boyer et al. 2006; Lee et al. 2006). In contrast to mixed-lineage genes, the Ink4a/Arf locus is derepressed in committed and differentiated Ezh2-ablated cells (MyoD-Cre-Ezh2mKO) (Chen et al. 2009; Ezhkova et al. 2009; Hirabayashi et al. 2009; this study). Thus, different genomic chromatin structures may be susceptible to PcG regulation only at defined developmental windows. Interestingly, derepression of some of the mixed-lineage genes reported here has been observed in the skin of Ezh1/Ezh2 double-knockout mice (Ezhkova et al. 2011).
EZH2 has been reported to repress Pax7 expression (Palacios et al. 2010). Specifically, EZH2 knockdown in cultured SCs results in Pax7 activation only if induced when the Pax7 level starts declining (Palacios et al. 2010). Since Cre expression in Pax7-Cre mice is coincident with Pax7 transcription (Keller et al. 2004), it is likely that Ezh2 ablation in Ezh2mKO mice occurred at the onset of Pax7 expression. In addition to the different temporal Ezh2 inactivation strategies, cultured SCs may not experience the same physiological regulation attained in the animal, and may thus explain the different experimental outcomes.
In conclusion, the processes regulated by EZH2 are pivotal for SC homeostasis. As SCs hold therapeutic potential to treat muscle-wasting diseases (Kuang et al. 2008), PRC2 may be exploited as a target to promote ex vivo expansion and in vivo SC self-renewal.
Materials and methods
Mice and animal care
Mice were housed in a pathogen-free facility, and all experiments were performed according to the National Institutes of Health's (NIH) Animal Care and Use regulations. For a detailed animal description, see the Supplemental Material.
Myotoxic injury
The right TA muscle of 8-wk-old wild-type and Ezh2mKO mice was surgically exposed and injected to holding capacity (∼50 μL) with 10 μM CTX, as described previously (Schertzer et al. 2007). The left TA muscle was not injected and served as an uninjured control.
Histology
For a detailed histology protocol, see the Supplemental Material.
FACS sorting, myofiber isolation, BrdU labeling, immunofluorescence, and image acquisition
SC FACS sorting was performed as described previously (Joe et al. 2010). BrdU staining and immunofluorescence protocols are described in the Supplemental Material.
Transferase dUTP nick end-labeling (TUNEL) assay
FACS-sorted SCs were examined by terminal deoxynucleotidyl TUNEL assay (Roche Applied Science) according to the manufacturer's instructions.
Gene expression analyses
Total RNA from either whole-muscle or FACS-isolated SCs was reverse-transcribed using a cDNA synthesis kit (Applied Biosystems) and subjected to qPCR analysis. Gene expression analyses are detailed in the Supplemental Material.
Immunoblots
Muscles or FACS-isolated SCs were lysed, and their extracts were resolved by SDS-PAGE and transferred onto nitrocellulose filters. Specific antibodies used to perform immunoblots are described in Supplemental Table 4.
ChIP assay
Cross-linking and ChIP were performed as described (Caretti et al. 2004) with minor modifications (see the Supplemental Material).
Acknowledgments
We thank Mario Capecchi, David Goldhamer, and Alexander Tarakhovsky for sharing the Pax7-Cre, MyoD-Cre, and Ezh2fl/fl animals, respectively. Fabio Rossi provided advice with the FACS experiments, Jeffrey Lay helped with cell sorting, and Gustavo Gutierrez-Cruz assisted with animal genotyping. Members of the NIAMS Laboratory Animal Care and Use Section and NIAMS Light Imaging Section are kindly acknowledged. J.G.R. was supported by an Overseas Biomedical Research Fellowship from the NH and MRC (Australia). This work was supported in part by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases of the National Institutes of Health.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2027911.
Supplemental material is available for this article.
References
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. 2007. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Relaix F 2007. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol 23: 645–673 [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T 2008. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Mortimer J, Marley J, Goldhamer DJ 2005. MyoD-cre transgenic mice: a model for conditional mutagenesis and lineage tracing of skeletal muscle. Genesis 41: 116–121 [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK 2009. Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 23: 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE 2005. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122: 289–301 [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Karasov AO, Rando TA 2007. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol 5: e102 doi: 10.1371/journal.pbio.0050102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn TM, Breininger JF, Baskin DG, Schwartz MW 1998. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci 1: 271–272 [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y 2009. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63: 600–613 [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM 2010. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol 12: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostes B, Walther C, Gruss P 1990. The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech Dev 33: 27–37 [DOI] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V 2009. mir-214-dependent regulation of the Polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 36: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ 2009. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev Biol 332: 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR 2004. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev 18: 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Schulze J, Hansen U, Ashwodt T, Keene DR, Brunken WJ, Burgeson RE, Bruckner P, Bruckner-Tuderman L 2004. A novel marker of tissue junctions, collagen XXII. J Biol Chem 279: 22514–22521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes Dev 21: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Gillespie MA, Rudnicki MA 2008. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell 2: 22–31 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M 2005. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067 [DOI] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, et al. 2010. TNF/p38α/Polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ 2010. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci 107: 15957–15962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G 2010. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer JD, Gehrig SM, Ryall JG, Lynch GS 2007. Modulation of insulin-like growth factor (IGF)-I and IGF-binding protein interactions enhances skeletal muscle regeneration and ameliorates the dystrophic pathology in mdx mice. Am J Pathol 171: 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, Brack AS 2010. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell 6: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ 2004. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119: 543–554 [DOI] [PubMed] [Google Scholar]
- Shichida Y, Matsuyama T 2009. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci 364: 2881–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S 2006. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol 8: 677–687 [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4: 124–131 [DOI] [PubMed] [Google Scholar]