Abstract
Autophagy is a major pathway for degradation of cytoplasmic proteins and organelles, and has been implicated in tumor suppression. Here, we report that mice with systemic mosaic deletion of Atg5 and liver-specific Atg7−/− mice develop benign liver adenomas. These tumor cells originate autophagy-deficient hepatocytes and show mitochondrial swelling, p62 accumulation, and oxidative stress and genomic damage responses. The size of the Atg7−/− liver tumors is reduced by simultaneous deletion of p62. These results suggest that autophagy is important for the suppression of spontaneous tumorigenesis through a cell-intrinsic mechanism, particularly in the liver, and that p62 accumulation contributes to tumor progression.
Keywords: autophagy, tumorigenesis, model mouse, oxidative stress
Macroautophagy (hereafter referred to as autophagy) is one of the major degradation pathways in the cell, along with the ubiquitin–proteasome system. In autophagy, intracellular components are sequestered by autophagosomes and then degraded upon fusion with lysosomes (Mizushima 2007). Genetic studies of yeast have identified >30 autophagy-related (ATG) genes that are essential for various types of autophagy (not only macroautophagy). Among them, ATG genes that are required for macroautophagy are highly conserved in higher eukaryotes (Yang and Klionsky 2010). Using reverse genetic approaches, many important roles of autophagy in mammals have been demonstrated, including adaptive response to starvation, quality control of intracellular proteins and organelles, preimplantation embryonic development, elimination of intracellular microbes, and presentation of cytoplasmic antigens (Rubinsztein 2006; Mizushima 2007; Cecconi and Levine 2008; Mizushima et al. 2008; Deretic and Levine 2009).
Evidence has suggested that autophagy is also involved in tumor suppression (Levine and Kroemer 2008; Chen and Debnath 2010; White et al. 2010). Possible tumor-suppressive mechanisms thus far proposed in cell culture and allografted tumor models include suppression of tumorigenic inflammation (Degenhardt et al. 2006), mitigation of metabolic stress and genomic damage (Karantza-Wadsworth et al. 2007; Mathew et al. 2007), and p62 (also called SQSTM1) degradation (Mathew et al. 2009). However, results from currently available in vivo models have been limited. Mice with heterozygous disruption of Beclin 1, the mammalian homolog of yeast Atg6, induce an increased frequency of spontaneous cancers, lung cancers, hepatocellular carcinomas, and lymphomas (Qu et al. 2003; Yue et al. 2003). Several Beclin 1-interacting proteins—including Ambra 1 (Fimia et al. 2007), Bif-1 (Takahashi et al. 2007), and UVRAG (Liang et al. 2006)—were also shown to have tumor-suppressive or anti-proliferation effects. However, these factors are not specific for autophagy and are considered to have multiple functions, such as involvement in the endocytic pathway (Funderburk et al. 2010; He and Levine 2010; Thoresen et al. 2010), which has many roles in tumorigenesis (Mosesson et al. 2008). Therefore, the suppressive role of autophagy in tumorigenesis in vivo should be further evaluated using a different model deficient for a factor more specific to autophagy. However, because systemic deletion of Atg3, Atg5, Atg7, Atg9, and Atg16L1 causes neonatal lethality (Kuma et al. 2004; Komatsu et al. 2005; Saitoh et al. 2008, 2009; Sou et al. 2008), the long-term effects of defects in autophagy have not been analyzed.
In this study, we analyzed a new mouse model in which the ATG5 genes were mosaically deleted in various tissues. Our results showed that multiple benign tumors developed only in the liver, but not in other tissues. Swollen mitochondria and oxidative stress and genomic damage responses were detected in the hepatic tumor cells. Liver-specific ATG7-deficient mice also developed liver tumors, but their size was reduced by concomitant knockout of the p62 gene. Our study suggests that continuous autophagy is important for suppression of tumorigenesis in the liver, and accumulation of p62 caused by autophagy deficiency contributes to tumor progression. Generation of only benign tumors, not cancers, also suggests that autophagy may be required for progression beyond the benign state.
Results and Discussion
Mosaic deletion of Atg5 causes multiple liver tumors
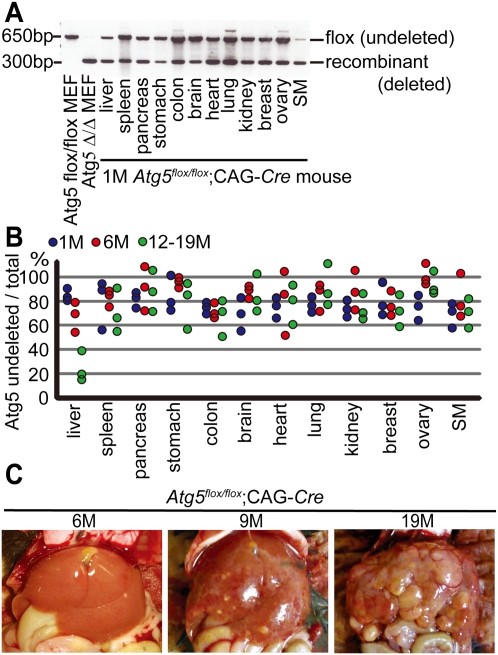
To overcome the lethal phenotype of Atg5−/− mice (Kuma et al. 2004), we analyzed mice with mosaic deletion of Atg5 (Hara et al. 2006). Originally, we intended to generate systemically deleted mice by crossing Atg5flox/flox mice with CAG-Cre transgenic mice in which Cre recombinase is ubiquitously expressed. However, the resultant Atg5flox/flox;CAG-Cre mice showed only partial deletion of the Atg5 gene in the liver, heart, skeletal muscle, and brain (Hara et al. 2006). This incomplete deletion was observed in all tissues as far as we examined; PCR analyses of genomic DNA extracted from various organs showed both recombinant (deleted) and flox (undeleted) alleles (Fig. 1A). In addition, real-time quantitative PCR analysis showed that, in most cases, ∼60%–90% of cells in various organs of Atg5flox/flox;CAG-Cre mice possessed the undeleted Atg5 flox allele (Fig. 1B). Although the exact cause of the incomplete deletion is unknown, mosaic deletion occurred reproducibly. Atg5flox/flox;CAG-Cre mice were viable and could survive >19 mo. With this mouse model, we were able to follow the fate of autophagy-deficient cells systemically and in a nonbiased manner for a long period.
Figure 1.
Mosaic deletion of Atg5 causes multiple liver tumors. (A) Genotyping of Atg5flox/flox;CAG-Cre mouse organs by PCR at 1 mo. Positions of the Atg5 flox (undeleted) and recombinant (deleted) alleles are indicated. Atg5Δ/Δ mouse embryonic fibroblasts (MEFs) indicates Atg5flox/flox MEFs infected with the adenovirus vector coding Cre recombinase. (SM) Skeletal muscle. (B) Real-time quantitative PCR with the Atg5 flox allele-specific primers. Genomic DNA is extracted from various tissues of Atg5flox/flox;CAG-Cre mice and Atg5flox/flox mice at 1 mo (blue circles), 6 mo (red circles), and 12–19 mo (green circles) (n = 3). The relative DNA quantity of the Atg5 flox allele is normalized to that of Atg14. Ratio (percentage) of flox allele in Atg5flox/flox;CAG-Cre organs to that in Atg5flox/flox organs is shown. (C) Representative gross anatomy of Atg5flox/flox;CAG-Cre mice analyzed at 6 mo (n = 3), 9 mo (n = 6), and 18–19 mo (n = 9).
Gross anatomy revealed severe hepatomegaly in Atg5flox/flox;CAG-Cre mice within 6 mo after birth. By 6–9 mo, multiple small tumors were detected in the liver (Fig. 1C). These tumors increased in both size and number, and the liver was finally occupied by multiple tumors at 19 mo of age. Tumors were detected in all Atg5flox/flox;CAG-Cre mice after 9 mo (n = 17), but not in Atg5flox/+;CAG-Cre mice (n = 14) and Atg5flox/flox mice (n = 4). Tumor formation was so far observed only in the liver; there was no evidence of tumors or any other macroscopic abnormalities in other organs (Supplemental Fig. 1).
Histological findings of Atg5flox/flox;CAG-Cre mice
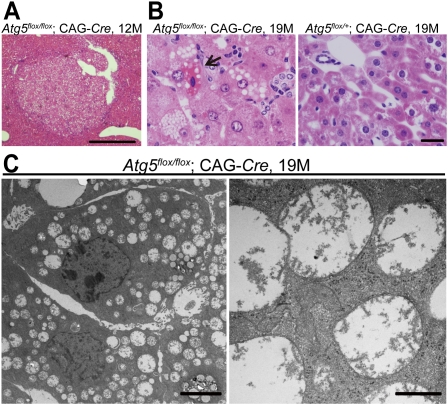
Hematoxylin and eosin staining and silver impregnation staining revealed the focal expansive growth of abnormal cells in the Atg5flox/flox;CAG-Cre mouse liver (Fig. 2A, Supplemental Fig. 2A). At high magnification, hepatocellular enlargement was observed in the Atg5flox/flox;CAG-Cre liver at 6 mo (Supplemental Fig. 2B). Only slight lymphocytic infiltration and intracellular vacuolation were observed at 9 mo. These findings became clearer and were observed almost throughout the liver at 12–19 mo. At 19 mo, many foamy hepatocytes and a small number of necrotic cells were observed (Fig. 2B). However, no typical signs of malignancy—such as abnormal nuclear morphology, pleomorphism, and local invasion—were detected even at 19 mo. Thus, the histological diagnosis of the tumor was benign tumor (adenoma), not malignant cancer. Oil-red O staining revealed that the cytoplasmic vacuoles were lipid droplets (Supplemental Fig. 2C). There was no histological abnormality in the Atg5flox/+;CAG-Cre liver (Fig. 2B). Abnormally swollen mitochondria were observed by electron microscopy in the cytoplasm of most of the Atg5flox/flox;CAG-Cre hepatocytes at 19 mo (Fig. 2C). On the other hand, no morphologic abnormalities were observed by light microscopy in tissues other than the liver in Atg5flox/flox;CAG-Cre mice (Supplemental Fig. 1).
Figure 2.
Histological findings of the liver in Atg5flox/flox;CAG-Cre mice. (A) Hematoxylin and eosin (H&E) staining of Atg5flox/flox;CAG-Cre (n = 2). Bar, 500 μm. (B) High-power-field images of H&E staining of the livers from Atg5flox/flox;CAG-Cre mice (n = 4) and an Atg5flox/+; CAG-Cre mouse (n = 4) at 19 mo. The arrow shows a necrotic hepatocyte observed in a Atg5flox/flox;CAG-Cre mouse liver at 19 mo. Bar, 20 μm. (C) Electron micrographs of Atg5flox/flox;CAG-Cre mouse hepatocytes (n = 2). (Right) Magnified image shows swollen mitochondria. Bars: left, 4 μm; right, 500 nm.
Liver tumors originate from autophagy-deficient hepatocytes
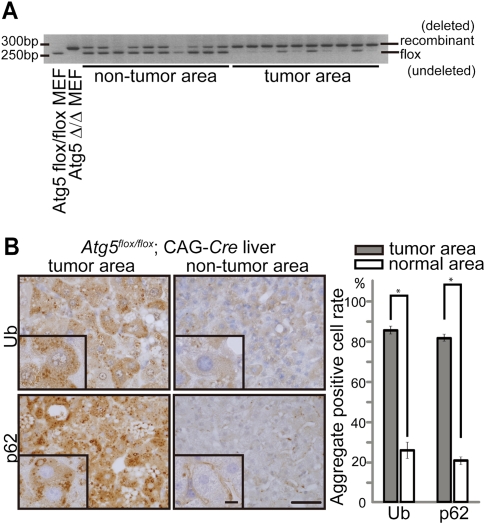
Next, we examined the origin of these liver tumors in the mosaically deleted mice. Although a defect in autophagy could cause tumors by an intrinsic mechanism, it is also possible that tumors developed as a secondary result of inflammation, as suggested previously (Degenhardt et al. 2006). If the latter is the case, tumors could develop from Atg5-undeleted cells. To this end, we analyzed genotypes of cells obtained by laser capture microdissection of liver section samples. PCR analysis revealed that the tumor areas mostly contained the recombinant (deleted) allele, whereas the nontumor areas contained both flox (undeleted) and recombinant alleles (Fig. 3A). Small amounts of the flox allele were occasionally detected in the tumor areas, which may be derived from infiltrating lymphocytes. These data suggest that the liver tumors in Atg5flox/flox;CAG-Cre mice originate from Atg5-deficient hepatocytes.
Figure 3.
Liver tumors are derived from Atg5-deleted hepatocytes. (A) Genotyping of liver samples microdissected from the tumor and nontumor areas of 9-mo-old Atg5flox/flox;CAG-Cre mice. Positions of the Atg5 flox (undeleted) and recombinant (deleted) alleles are indicated. Atg5Δ/Δ MEFs indicate Atg5flox/flox MEFs infected with the adenovirus vector coding Cre recombinase. (B) Immunohistochemical staining for ubiquitin and p62 of the tumor and nontumor areas of Atg5flox/flox;CAG-Cre mouse livers. The graph shows the percentage of cells with ubiquitin- or p62-positive aggregates in the tumor (gray) and nontumor (white) areas. One-hundred hepatocytes in 10 tumor and nontumor areas (total 1000 cells for each) were analyzed at 9 mo. Data are mean ± SE values from four mice. (*) P < 0.01. Bars: 40 μm; insets, 10 μm.
To confirm that the tumor cells are indeed autophagy-defective, we analyzed the formation of ubiquitin-positive aggregates because accumulation of ubiquitinated proteins/aggregates is one of the hallmarks of autophagy deficiency (Komatsu et al. 2005, 2006; Hara et al. 2006). In the tumor areas of the Atg5flox/flox;CAG-Cre mouse liver, a significantly higher proportion of hepatocytes (85.4% ± 2.0%) showed ubiquitin aggregates than that in nontumor areas (24.6% ± 4.1%) (Fig. 3B). We also analyzed p62, which is a selective substrate of autophagy (Bjørkøy et al. 2005), and found a higher accumulation rate of p62-positive dots in the tumor areas (80.8% ± 1.8%) than in the nontumor areas (22.5% ± 2.3%) (Fig. 3B). These data support the idea that the origin of these liver tumors is ATG5-deleted cells.
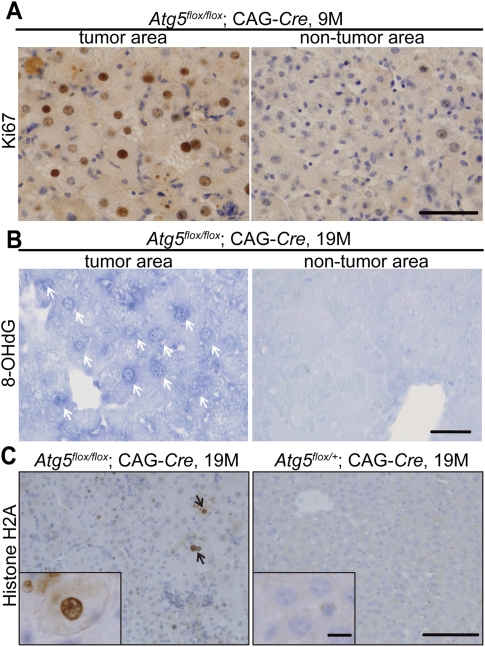
Time-course analysis revealed that liver DNA contents of the flox (undeleted) allele decreased and that of the recombinant (deleted) allele increased with age (Fig. 1B, Supplemental Fig. 3A). In addition, many hepatocytes in the tumor areas of the Atg5flox/flox;CAG-Cre liver at 19 mo were positive for the cell proliferation marker Ki-67 (49.6% ± 1.3% in the tumor area vs. 2.7% ± 0.5% in the nontumor area, P < 0.01), and this was mostly negative in the Atg5flox/+;CAG-Cre liver (Fig. 4A; Supplemental Fig. 3B). These data suggest that Atg5-deficient hepatocytes have a growth advantage over normal hepatocytes under long-term experimental conditions in vivo.
Figure 4.
Induction of the oxidative stress and DNA damage responses in Atg5flox/flox;CAG-Cre mouse livers. (A) Immunohistochemical analysis of the tumor and nontumor areas in Atg5flox/flox;CAG-Cre mouse livers at 19 mo for Ki-67 expression. n = 3. Bar, 50 μm. (B) 8-OHdG staining of the tumor and nontumor areas in the liver of Atg5flox/flox;CAG-Cre mice at 19 mo. White arrows indicate anti-8-OHdG antibody-positive nuclei (n = 3). Bar, 40 μm. (C) Phospho-histone H2A.X staining in the liver of Atg5flox/flox;CAG-Cre (n = 3) and Atg5flox/+;CAG-Cre (n = 3) mice at 19 mo. Black arrows indicate phospho-histone H2A.X-positive nuclei. Bars: 100 μm; insets, 10 μm.
In other organs of 19-mo-old Atg5flox/flox;CAG-Cre mice, a small population of cells showed high levels of diffuse and aggregated ubiquitin and p62 signals, which were not observed in Atg5flox/+;CAG-Cre mice (Supplemental Fig. 4A,B). The data suggest that these organs indeed possess Atg5-deficient cells, even though they do not develop spontaneous tumors, at least by 19 mo.
Induction of the oxidative stress and DNA damage responses in the Atg5flox/flox;CAG-Cre mouse liver
The presence of abnormally enlarged mitochondria in Atg5flox/flox;CAG-Cre hepatocytes suggests that these cells might be affected by reactive oxygen species (ROS), which have been implicated in tumorigenesis. We performed immunostaining of 8-hydroxydeoxyguanosine (8-OHdG), which is considered to be an indicator of the oxidative stress (Cadet et al. 2003). In Atg5flox/flox;CAG-Cre mice, most of the hepatocytes, especially in the tumor areas, were 8-OHdG-positive (41.8% ± 2.0% in the tumor area vs. 0.9% ± 0.1% in the nontumor area, P < 0.01), whereas those in Atg5flox/+;CAG-Cre mice were mostly negative (Fig. 4B; Supplemental Fig. 3C). An increase in 8-OHdG staining was observed as early as 6 mo (Supplemental Fig. 3C). In addition, an increase in the levels of the detoxifying enzyme glutathione-S-transferase (GST), which is a typical oxidative stress-inducible protein, was observed in liver tumor areas, as reported previously (Supplemental Fig. 5; Matsumoto et al. 2008; Mathew et al. 2009). Moreover, part of the hepatocytes in the same liver expressed phospho-histone H2A.X in the nuclei (2.1% ± 0.4% in Atg5flox/flox;CAG-Cre vs. 0.4% ± 0.1% in Atg5flox/+;CAG-Cre hepatocytes, P < 0.05) (Fig. 4C). Because phosphorylation of the histone variant H2A.X is a rapid response to DNA damage, this result suggests that DNA damage response is promoted in Atg5flox/flox;CAG-Cre hepatocytes. In contrast, 8-OHdG-positive and phospho-histone H2A.X-positive cells were mostly negative in other organs (Supplemental Fig. 6). These data are consistent with the recent in vitro and in vivo studies (Karantza-Wadsworth et al. 2007; Mathew et al. 2007).
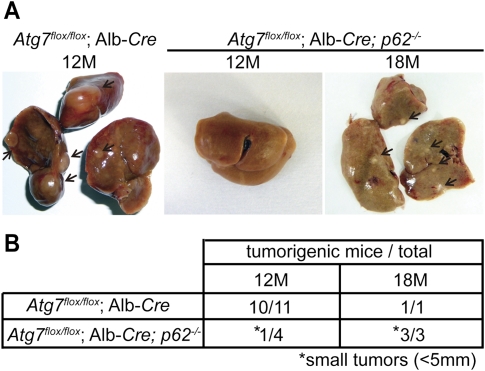
Tumor progression in liver-specific ATG7-deficient mice was partially suppressed by p62 deletion
To test whether the tumor-suppressive function is specific to Atg5, we analyzed liver-specific ATG7-deficient mice (Atg7flox/flox;Alb-Cre) (Komatsu et al. 2007). Although sporadically some of these mice died due to liver dysfunction 3 mo after ATG7 gene deletion (M. Komatsu, unpubl.), others survived for >1 yr and developed liver tumors, as observed in Atg5flox/flox;CAG-Cre mice (Fig. 5A,B). Thus, tumor formation is the shared phenotype between Atg5 and Atg7 deficiency, and is not specific to Atg5 deletion.
Figure 5.
Suppression of tumor progression in liver-specific ATG7-deficient mice by p62 deletion. (A) Representative gross anatomy of Atg7flox/flox;Alb-Cre mice analyzed at 12 mo (n = 11) and Atg7flox/flox;Alb-Cre; p62−/− mice analyzed at 12 mo (n = 4) and 18 mo (n = 3). Tumors are indicated by arrows. (B) Summary of tumor formation in Atg7flox/flox;Alb-Cre and Atg7flox/flox;Alb-Cre; p62−/− mice. (*) Small tumors (<5 mm).
Next, using this mouse model, we determined the relationship between tumor formation and p62 accumulation, as suggested previously (Mathew et al. 2009). The hepatic damage caused by Atg7 deficiency was ameliorated by simultaneous deletion of p62 (Atg7flox/flox;Alb-Cre, p62−/−) (Komatsu et al. 2007). In these double-knockout mice, tumors were still generated, but their size was much reduced (Fig. 5A,B). It has been shown that p62 accumulation traps and inactivates Keap1, a key regulator of the oxidative stress response, leading to activation of the transcription factor Nrf2 (Komatsu et al. 2010; Lau et al. 2010). Indeed, accumulation of Keap1 in a detergent-insoluble fraction and up-regulation of NAD(P)H dehydrogenase quinone 1 (Nqo1), an Nrf2-responsive factor were observed both in the tumor and nontumor areas in Atg7flox/flox;Alb-Cre mice (Supplemental Fig. 7). Keap1 sequestration and Nqo1 induction were restored in Atg7flox/flox;Alb-Cre, p62−/− mice (Supplemental Fig. 7). These data suggest that p62 accumulation due to autophagy suppression contributes to tumor formation, probably at the progression step.
In the present study, we determined the long-term effect of defects in autophagy in vivo for the first time and found that systemic mosaic deficiency of Atg5 leads to cell-type specific tumorigenesis; that is, benign tumor development only or at least preferentially in the liver. There were no apparent histological and physiological abnormalities in other organs except the accumulation of p62 and ubiquitinated proteins. Although neurodegeneration was observed in neural cell-specific Atg5- and Atg7-deficient mice (Hara et al. 2006; Komatsu et al. 2006), mosaically Atg5-deleted mice do not develop any neurological abnormality, which may be compensated by Atg5-undeleted neuronal cells. Liver tumors were observed also in liver-specific Atg7-deficient mice in the present study.
The pattern of tumor formation is different from that of Beclin 1-heterozygous mutant mice, in which malignant cancers develop in multiple organs, including the liver, lung, and lymphoid tissues (Qu et al. 2003; Yue et al. 2003). Beclin 1 is included in at least two different class III PI3-kinase complexes: the Beclin 1–Atg14–Vps34–Vps15 complex that is specifically required for autophagy, and the Beclin 1–UVRAG–Vps34–Vps15 complex that is involved in the endocytic pathway (Funderburk et al. 2010; He and Levine 2010; Thoresen et al. 2010). It is possible that the liver tumors that develop in Beclin 1 heterozygous mutant mice are caused by autophagy deficiency, but other tumors might be caused by loss of Beclin 1 functions other than in autophagy.
Regarding the underlying mechanism of tumor suppression by autophagy, several hypotheses have been proposed. Our observation that the hepatic tumors originated from Atg5-deficient cells even though intact cells (with undeleted Atg5flox allele) remained supports the hypothesis of an intrinsic mechanism rather than secondary tumorigenesis as a result of chronic inflammation. An interesting hypothesis is that p62 accumulation could cause tumorigenesis through enhancing oxidative stress and genomic instability (Mathew et al. 2009). Indeed, we observed a reduction in tumor size in Atg7flox/flox;Alb-Cre, p62−/− mice. However, small tumors were still detected in the Atg7flox/flox;Alb-Cre, p62−/− mouse liver, suggesting that p62 accumulation is important for the tumor progression step, rather than the initiation step. Activation of the Nrf2 pathway caused by p62 accumulation (Supplemental Fig. 7) can be important for progression of tumors and is consistent with reports that somatic mutations in either Keap1 or Nrf2 that cause constitutive Nrf2 activation have been found in human cancers (Padmanabhan et al. 2006; Shibata et al. 2008). Another possible mechanism is that p62 accumulation deregulates the canonical NF-κB pathway, which leads to activation of the noncanonical NF-κB pathway and tumorigenesis in the liver (Mathew et al. 2009). Further analysis of these downstream events may also answer the question of why tumors are generated only in the liver even though p62 accumulated in small populations of cells. Alternatively, lack of tumors in other organs may simply be because autophagy deficiency caused cell death or growth retardation.
Although we showed that autophagy is indeed a tumor suppressor, other studies have also suggested that autophagy could support tumor progression and survival (Levine and Kroemer 2008; Chen and Debnath 2010; White et al. 2010). These hypotheses are not mutually exclusive; generation of only benign tumors in our models suggests that autophagy may be important for progression beyond the benign state. This possibility needs to be further examined.
Materials and methods
Mice
Atg5flox/flox mice (Hara et al. 2006), CAG-Cre transgenic mice expressing Cre recombinase under the control of the CAG (CMV enhancer and chicken β-actin promoter) (Sakai and Miyazaki 1997), Atg7flox/flox mice (Komatsu et al. 2005), p62−/− mice (Okada et al. 2009), and Atg7flox/flox;Alb-Cre, p62−/− mice (Komatsu et al. 2007) were described previously. Wild-type C57BL/6 mice were obtained from Japan SLC, Inc. All animal experiments were approved by the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University and the Ethics Review Committee for Animal Experimentation of the Tokyo Metropolitan Institute of Medical Science.
Antibodies
A monoclonal antibody against ubiquitin (1B3) was purchased from Medical and Biological Laboratories (MBL) and used for immunohistochemistry. The following antibodies were also used: anti-p62 polyclonal antibody (PROGEN), anti-Ki67 monoclonal antibody (Thermo), anti-8-OHdG antibody (Japan Institute for the Control of Aging), phospho-histone H2A.X (Ser139) rabbit monoclonal antibody (Cell Signaling Technology), and anti-GST (B-14) monoclonal antibody (Santa Cruz Biotechnology). Anti-Atg7 (Komatsu et al. 2005), anti-Keap1 (Proteintech Group, Inc.), anti-Nqo1 (Abcam, Inc.), and anti-actin (MAB1501R, Chemicon International, Inc.) antibodies were used for immunoblotting.
Immunohistochemistry, electron microscopy, and laser capture microdissection
Detailed information of experimental procedures is provided in the Supplemental Material.
Statistics
Results are expressed as means ± SEM, and significance was determined by unpaired Student's t-test.
PCR
Tissue-genotyping PCR was performed with the following primers: check2 (5′-ACAACGTCGAGCACAGCTGCGCAAGG-3′) and short2 (5′-GTACTGCATAATGGTTTAACTCTTGC-3′) for the Atg5 flox allele, and 5Long2 (5′-CAGGGAATGGTGTCTCCCAC-3′) and short2 for the recombinant allele. PCR analysis with laser capture microdissection samples were performed with the following primers: NR1 (5′-CCTCTTGCAAAACCACACTGCTCGACATTG-3′) and short2 for the Atg5 flox allele, and 5Long2 and short2 for the recombinant allele. For real-time PCR analysis, genomic DNA was extracted from paraffin-embedded tissues with a DEX-PAT system (TAKARA) or from nonembedded tissues with proteinase K treatment. Real-time PCR analysis was performed with the SYBR PrimeScript RT–PCR system (TAKARA) and specific primers as follows: NR1 and short2 for the Atg5 flox allele, RecF (5′-GCCGCACGTCTAAGAAACC-3′) and short2 for the Atg5 recombinant allele, and mAtg14 ex5 F′ (5′-GAAGCTTTACAGCCGAGCAC-3′) and mAtg14 ex3 R′ (5′-TCACTTCGTCGATTGGGAAT-3′) for Atg14.
Acknowledgments
We thank N. Miyasaka for his mentorship and encouragement. We also thank P.N. Quy, K. Uchida, A. Furukawa, and Y. Suzuki for technical assistance; J. Miyazaki for the kind donation of CAG-Cre transgenic mice; and T. Ishii for p62 knockout mice. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (to M.K. and N.M.), Grants-in-Aid for Young Scientist (S) (to M. K.), and Specially Promoted Research (to K. T.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; Funding Program for Next Generation World-Leading Researchers (to M.K. and N.M.); Takeda Science Foundation; and Research Award Grant of Japanese Society of Anti-Aging Medicine (to N.M.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2016211.
Supplemental material is available for this article.
References
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Douki T, Gasparutto D, Ravanat JL 2003. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat Res 531: 5–23 [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B 2008. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Debnath J 2010. Autophagy and tumorigenesis. FEBS Lett 584: 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5: 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. 2007. Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z 2010. The Beclin 1–VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol 20: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- He C, Levine B 2010. The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E 2007. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21: 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata JI, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD 2010. A non-canonical mechanism of Nrf2 activation by autophagy deficiency: a direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU 2006. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8: 688–699 [DOI] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E 2007. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 21: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Ezaki J, Komatsu M, Takahashi K, Mineki R, Taka H, Kikkawa M, Fujimura T, Takeda-Ezaki M, Ueno T, et al. 2008. Comprehensive proteomics analysis of autophagy-deficient mouse liver. Biochem Biophys Res Commun 368: 643–649 [DOI] [PubMed] [Google Scholar]
- Mizushima N 2007. Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Mills GB, Yarden Y 2008. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer 8: 835–850 [DOI] [PubMed] [Google Scholar]
- Okada K, Yanagawa T, Warabi E, Yamastu K, Uwayama J, Takeda K, Utsunomiya H, Yoshida H, Shoda J, Ishii T 2009. The α-glucosidase inhibitor acarbose prevents obesity and simple steatosis in sequestosome 1/A170/p62 deficient mice. Hepatol Res 39: 490–500 [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21: 689–700 [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen E-L, Mizushima N, Ohsumi Y, et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112: 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443: 780–786 [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456: 264–268 [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci 106: 20842–20846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J 1997. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun 237: 318–324 [DOI] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S 2008. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci 105: 13568–13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19: 4762–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, et al. 2007. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 9: 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen SB, Pedersen NM, Liestol K, Stenmark H 2010. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 316: 3368–3378 [DOI] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R 2010. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 22: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]