Abstract
pRB-mediated inhibition of cell proliferation is a complex process that depends on the action of many proteins. However, little is known about the specific pathways that cooperate with the Retinoblastoma protein (pRB) and the variables that influence pRB's ability to arrest tumor cells. Here we describe two shRNA screens that identify kinases that are important for pRB to suppress cell proliferation and pRB-mediated induction of senescence markers. The results reveal an unexpected effect of LATS2, a component of the Hippo pathway, on pRB-induced phenotypes. Partial knockdown of LATS2 strongly suppresses some pRB-induced senescence markers. Further analysis shows that LATS2 cooperates with pRB to promote the silencing of E2F target genes, and that reduced levels of LATS2 lead to defects in the assembly of DREAM (DP, RB [retinoblastoma], E2F, and MuvB) repressor complexes at E2F-regulated promoters. Kinase assays show that LATS2 can phosphorylate DYRK1A, and that it enhances the ability of DYRK1A to phosphorylate the DREAM subunit LIN52. Intriguingly, the LATS2 locus is physically linked with RB1 on 13q, and this region frequently displays loss of heterozygosity in human cancers. Our results reveal a functional connection between the pRB and Hippo tumor suppressor pathways, and suggest that low levels of LATS2 may undermine the ability of pRB to induce a permanent cell cycle arrest in tumor cells.
Keywords: retinoblastoma tumor suppressor, LATS2, HIPPO pathway, DREAM complex, senescence, G1 arrest
The ability of the Retinoblastoma protein (pRB) to block cell proliferation is thought to be central to its functions as a tumor suppressor. There is no single binding partner that mediates these effects, but pRB has multiple activities that contribute to the initiation and maintenance of a permanent cell cycle arrest. This is illustrated by the sequence of changes that is seen following the reintroduction of pRB into pRB-deficient human osteosarcoma SaOS2 cells. Within 12 h of pRB expression, cells cease to enter S phase and the population starts to accumulate in the G1 phase of the cell cycle (Hinds et al. 1992; Ji et al. 2004). An early effect of pRB expression is the up-regulation of p27CIP/KIP, a change mediated by pRB's interaction with Skp2 and APC/C (Ji et al. 2004; Binne et al. 2007). While elevated p27CIP/KIP initiates cell cycle arrest in SaOS2 cells (Ji et al. 2004), pRB-initiated repression of E2F targets helps to promote cell cycle exit by preventing the expression of proliferation-promoting genes. pRB cooperates with a variety of repressor complexes, including the DREAM (DP, RB [retinoblastoma], E2F, and MuvB) complex, to establish a repressive chromatin structure and silence the expression of E2F targets (Blais and Dynlacht 2007; Burkhart and Sage 2008). In some cell types, pRB-arrested cells undergo differentiation, and pRB's ability to physically interact with differentiation-promoting factors such as Runx2 (Thomas et al. 2001) or RBBP2/KDM5A (Benevolenskaya et al. 2005) can be important for this activity. The sustained expression of pRB in SaOS2 cells leads to an irreversible arrest, and many cells begin to express senescence-associated β-galactosidase (SA-β-gal), a marker often found in senescent cells (Alexander and Hinds 2001).
Stress- or oncogene-induced senescence is a tumor suppressor mechanism induced by the activated oncogenes that drive cellular transformation and mediated by proliferative stress signals (Serrano et al. 1997; Ben-Porath and Weinberg 2005; Courtois-Cox et al. 2008). pRB has a unique role in the repression of E2F target genes during senescence (Chicas et al. 2010) and is needed for the formation of senescence-associated heterochromatin foci (SAHF), compact heterochromatin structures that are not essential for senescence (Kosar et al. 2011) but have been proposed to promote efficient silencing of E2F-regulated genes and permanent cell cycle exit (Narita et al. 2003; Ye et al. 2007). pRB's role in senescence is distinct from its role in differentiation and requires an intact LXCXE motif (Talluri et al. 2010) that mediates interaction with corepressor complexes like BRG1, BRM, HDAC's, HP1, and Suv3-9H1/2 (Nielsen et al. 2001; Blais et al. 2007; Burkhart and Sage 2008). In response to senescence signals, pRB and E2F localize to PML bodies (Ferbeyre et al. 2000; Zhang et al. 2005; Vernier et al. 2011) and repress E2F-responsive promoters, perhaps through SAHF. In the formation of SAHF, pRB acts in parallel with histone chaperones HIRA/ASF1a. Phosphorylation of HIRA and HP1γ promotes the formation of SAHFs, containing macroH2A, HP1γ, and HMGA (Narita et al. 2003; Zhang et al. 2005, 2007; Adams 2007, 2009). More recently, SAHF have been shown to dampen the DNA damage response, thereby suppressing apoptosis and promoting senescence (Di Micco et al. 2011).
The evidence that pRB-mediated inhibition of cell proliferation is a complex, multistep process that depends on the combined activity of many different components raises the possibility that its ability to stop cell proliferation is influenced by cooperating pathways. Currently, little is known about the pathways that modify pRB-induced phenotypes. Here, we took advantage of SaOS2-TR-pRB cells that allow pRB to be re-expressed in a tightly controlled manner. Like many tumor cells, SaOS2 cells contain multiple mutations. For example, these cells are mutant for both pRB and p53, and it is likely that the effects of re-expressing pRB in SaOS2 cells will differ somewhat from cell cycle control or senescence responses that occur in primary cells. Nevertheless SaOS2-TR-pRB cells provide a system to study the effects of functional pRB in cancer cells and are amenable to screening.
Using two separate lentiviral shRNA knockdown screens, we identified kinases that are necessary for pRB to suppress cell proliferation in SaOS2-TR-pRB cells and kinases that impact pRB-induced increase of SA-β-gal activity. The results show that these readouts of pRB action require overlapping but different sets of proteins. Unexpectedly, we found that pRB induction of some markers of senescence is strongly suppressed by reduced levels of LATS2. Subsequent experiments showed that pRB-induced repression of E2F targets is compromised when LATS2 levels are low, and that reduced LATS2 leads to defects in the recruitment of DREAM complexes to E2F-regulated promoters. This functional interaction is especially intriguing because LATS2 is physically linked to RB1 on human chromosome 13 and the copy number of LATS2 is reduced in many tumor cell lines. We suggest that low levels of LATS2 in cells that retain an intact copy of RB1 may compromise pRB's ability to silence E2F targets, potentially creating an environment that helps cells to evade oncogene-induced senescence.
Results
Identification of kinases that are important for pRB-induced inhibition of cell proliferation
To identify kinases that impact the ability of pRB to arrest cell proliferation, duplicate sets of SaOS2-TR-pRB cells, grown in 96-well plates, were infected with lentiviral vectors carrying a shRNA library (one vector per well). The screening library consisted of 2112 lentiviral shRNA vectors targeting 415 human kinases, with an average of 4.5 shRNAs targeting each kinase (Moffat et al. 2006; Grueneberg et al. 2008). Four days after lentiviral infection and puromycin selection, pRB expression was induced in one set of the infected plates by changing to tetracycline-containing medium. The second set of plates was kept in non-tetracycline-containing medium. Changes in cell proliferation and viability were monitored over a 4-d time period following pRB expression by staining cells with Alamar blue, a redox-sensitive dye that gives an absorbance that is directly proportional to the number of viable cells (Grueneberg et al. 2008).
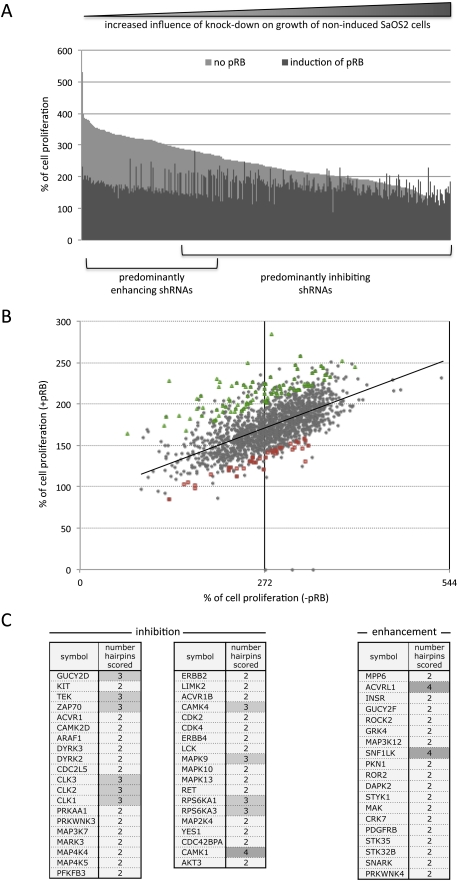
The re-expression of pRB in SaOS2 cells causes a robust G1 arrest. Over time, the inhibition of cell proliferation results in a measurable decrease in cell number when compared with uninduced cells. Using Alamar blue staining data gathered at the start of tetracycline treatment (time 0) and after 4 d of pRB expression, we calculated a factor of repression (FR). The 4-d time point was chosen so that even minor changes in proliferation would give a clear difference in cell number. The FR represents the ratio between the proliferation of the cell population seen after 4 d in cultures that lack pRB versus pRB-expressing cells. For example, control cells infected with scramble shRNA grew to 335% of the starting cell number in the absence of pRB, but this was reduced to 206% when pRB expression was induced, and is represented by a FR of 1.63 (P < 0.001 × 10−74) (Supplemental Fig. S1A).
FR values were calculated for each of the shRNA constructs, attributing a numerical value to the change in cell proliferation caused by pRB in the presence of each kinase-targeting shRNA. These were converted to a Z-score to evaluate the impact of each individual shRNA on pRB-mediated inhibition of cell proliferation, in comparison with the whole-screen data set. We assumed that shRNA treatments that modify the degree of pRB-induced arrest would alter the FR from that of scramble shRNA controls (FR = 1.63, confidence interval of Z0.99 between 1.6 and 1.66) (Supplemental Fig. S1A). Figure 1A shows all shRNAs that changed the FR with a Z-score of >|2|. Supplemental Table S1 lists all of the kinases that gave a Z-score of >|3| and gave similar effects when targeted by at least two independent hairpins.
Figure 1.
shRNAs that modify pRB-induced growth arrest. After infection of SaOS2 TR-pRB cells with shRNAs targeting different kinases and selection with puromycin for 3 d, cells were induced to express pRB or kept without tetracycline to serve as controls. Cell viability was analyzed using the Alamar blue assay. The percentage of growth was calculated relative to the 0-h time point. The FR after 96 h was calculated as the ratio between percent proliferation − pRB/percent proliferation + pRB and was used to score for shRNAs that influenced pRB-mediated cell cycle arrest. (A) Percentage of growth of uninduced (light) or induced (dark) cells for all hairpins that either reduce or increase the FR with a Z-score of >|2|. The shRNAs in the graph are ranked according to their effect on the growth of the uninduced cells. shRNAs that enhanced proliferation arrest (increased FR) are clustered mainly to the left, while those that reduce the effect of pRB (decreased FR) are to the right. Hits are evident due to their aberrant pattern. (B) Two-dimensional scatter plot of the pRB-induced proliferation arrest screen results. Each dot represents percent growth values for the same shRNA from uninduced (−pRB) cells (X-axis) and from cells with induction of pRB (Y-axis). The majority of shRNAs shows similar ratios of percent proliferation between uninduced (−pRB) and induced (+pRB) cells, and therefore appears along the trend line. The shRNAs that change this ratio appear either above (inhibitors of the pRB-mediated arrest) or below (enhancers of the pRb-mediated arrest) this line. The distance from the trend line corresponds to the strength of the effect. Shown in green (inhibitors) or red (enhancers) are the genes that significantly alter pRB-induced cell cycle arrest. (C) List of kinases whose knockdown inhibited or enhanced G1 arrest. Shown are the genes that differed at least three standard deviations from the trend line and scored with two or more shRNAs.
Many shRNAs have an effect on cell proliferation themselves, and these effects obscure changes in pRB-induced arrest. To adjust for this, the proliferation data were analyzed using a two-dimensional scatter plot (Fig. 1B) that compares the influence of shRNAs on cell proliferation induced by pRB with their effects on non-pRB-expressing cells. Using the two-dimensional analysis, and minimizing off-target effects by considering only kinases that were targeted by at least two independent shRNAs, we identified 39 genes whose knockdown significantly reduced pRB's ability to suppress proliferation (Fig. 1B, green rectangles) and 19 genes that enhanced pRB's effects (Fig. 1B, red squares).
As expected, kinases identified by this screening approach included several that have been linked previously to tumorigenesis or that have known effects on cell proliferation. Kinases whose knockdown reduced the effects of pRB include MAP2K4 (mutated or deleted in 5% of tumors) (Su et al. 2002), ACVR1 (knockdown of ACVR1 prevents TGFβ-SMAD signaling [Craft et al. 2007], and inhibition of ACVR1B leads to pancreatic carcinomas [Su et al. 2001]), and ZAP70 (involved in the inhibition of ERK/JNK signaling) (Alonso et al. 2003; Rahmouni et al. 2006). On the other hand, the list of kinases whose knockdown enhanced the effect of pRB included genes like ROCK2, CRK7, and GUCY2F that, conversely, are overexpressed or show gain-of-function mutations in cancer cells (Kamai et al. 2003; Capra et al. 2006; Wood et al. 2006).
Kinases that impact pRB-induced increase of SA-β-gal activity
Cell cycle arrest is a relatively immediate consequence of pRB expression and occurs within 24 h after pRB induction. Approximately 72 h after pRB expression, SA-β-gal, a marker that is often seen in senescent cells, begins to appear in the population of SaOS2 TR-pRB cells. Senescent cells are thought to be irreversibly arrested, and there is accumulating evidence that premature cellular senescence has an important role in tumor suppression (Kuilman et al. 2010).
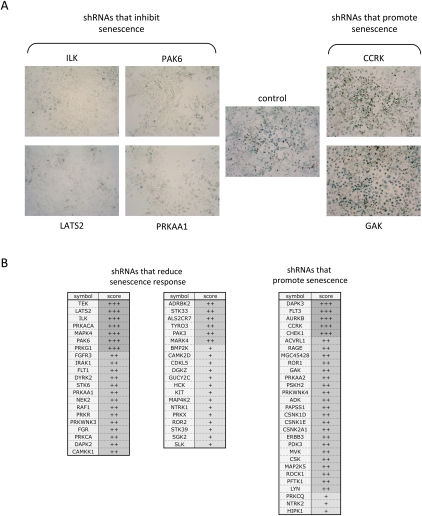
In parallel with the screen that scored the effects on cell number, we screened the library of shRNAs for constructs that affected a marker of pRB-induced senescence. The shRNA library was transduced into SaOS2-TR-pRB cells, pRB expression was induced with tetracycline, and, after 84 h of pRB expression, cells were fixed and analyzed for SA-β-gal (Supplemental Fig. S1B). At this time point, not all cells were SA-β-gal positive, making it possible to score enhancement or suppression. Because SA-β-gal is difficult to quantify in a high-throughput format, we examined the staining under the microscope and compared the intensity and percentage of stained cells with controls on a scale between 1 and 5, with 3 representing the control level, 1 being a strong decrease, and 5 being a strong increase.
Overall, fewer shRNA constructs scored in the SA-β-gal screen than the proliferation screen. This may in part be due to the lower sensitivity of the assay compared with the measurements of cell proliferation. shRNAs that either increased or decreased the extent of pRB-induced SA-β-gal staining were identified, and, as described above, we focused only on kinases that were targeted by two or more independent hairpins (examples are shown in Fig. 2A, with the complete lists of shRNA targets in B). As expected, several of the shRNAs that scored in this screen target kinases that are known to regulate senescence pathways, including PAK6, RAF1, or IRAK (Courtois-Cox et al. 2006; Ranuncolo et al. 2008; Orjalo et al. 2009). In addition, some of the shRNAs that enhanced the SA-β-gal activity had targets that have been shown to be up-regulated in tumors or cancer cell lines, including HIPK1 (up-regulated in breast cancer) (Kondo et al. 2003), MAP2K5 (increased expression associated with metastatic prostate cancer) (Mehta et al. 2003) and RAGE (activated in cancer and it promotes chronic inflammation, a favorable microenvironment for tumor formation) (Gebhardt et al. 2008).
Figure 2.
Knockdown of different kinases alter the pRB-induced increase in SA-β-gal activity. SaOS2 TR-pRB cells were infected, selected, and induced as described in Figure 1. After 84 h of pRB expression, cells were fixed and stained for SA-β-gal (blue), a classical marker of senescence. The intensity of the staining was analyzed by light microscopy (10× objective) and scored using a scale from 1 (reduced) to 5 (enhanced). (A) Examples of SA-β-gal staining of SAOS2 cells following shRNA knockdown of indicated genes and 84 h of induction of pRB from analyzed 96-well plates. (B) List of shRNAs that inhibited (green) or enhanced (red) the senescence phenotype induced by pRB. The hairpins are shown ranked in decreasing order, depending on number of hairpins that scored per gene and intensity of staining. (+++) Strong change with two or more hairpins; (++) strong and medium change with two or more hairpins; (+) medium change with two hairpins.
Pathway analysis
Ingenuity Pathway Analysis (IPA) software was used to find networks and functional themes in the kinases identified by two screens. Each data set gave multiple networks, ranked by scores between 13 and 48 (representing P-values of P ≤ 10−13 to 10−48 for the probability that these genes were included in the network by chance alone) (summarized in Supplemental Tables S3–S5). The highest-scoring networks from both screens contained subgroups of stress-activated protein kinases (SAPKs), including MAPK9, MAPK10 (JNK1/2), MAPK13 (p38MAPK), and MAP2K4 (cell proliferation screen), and MAP2K5, MAP4K2, and MAP2K1 (senescence screen). Stress-activated kinases (p38MAPK) are known to contribute to p16INK4A up-regulation, a key activator of pRB in senescent cells (Deng et al. 2004). Diverse cellular stressors—such as proinflammatory cytokines, irradiation, reactive oxygen species (ROS), osmotic shock, DNA damage, and activated oncogenes—trigger activation of these kinases. For instance, chronic Ras/ERK signaling or ROS are shown to converge on p38MAPKs mediated by MKK3/6 or MINK (Deng et al. 2004; Nicke et al. 2005). The IPA analysis suggests that both pRB-induced cell cycle arrest and a pRB-induced senescence-like state are stimulated via stress-activated pathways. Additional networks assemble around central molecules like the ERK1/2, NFkB, Hsp90, TGFβ, PI3K/AKT, p38MAPK, and Ras signaling pathways (Supplemental Tables S3–S5) that have all been connected previously to pRB (Wang et al. 1999; Shim et al. 2000; Takebayashi et al. 2003).
All three major MAPK cascades (ERKs, JNKs, and p38MAPKs) have been linked to stress-induced premature senescence. Likewise, Hsp90, Ras, NFkB, TGFβ, and PI3K/AKT have been connected to oncogene-induced senescence (Courtois-Cox et al. 2006; Maruyama et al. 2009; Ren et al. 2009; Restall and Lorimer 2010). The finding that components of these pathways scored strongly in our screens is consistent with the idea that SaOS2 cells have evolved to escape oncogene-induced senescence, and that this process can be reactivated by re-expression of pRB.
Our dual-screening strategy allowed us to compare the effects of shRNAs on two different readouts of pRB function that have been proposed to be associated with its role as a tumor suppressor. Cell cycle arrest and exit is a prerequisite for induction of senescence, and we expected to find overlap between the results of the two screens. shRNAs that scored in both assays are listed in Supplemental Tables S1 and S2. Since proliferation arrest is necessary for senescence, it is unlikely that shRNAs would bypass arrest without affecting senescence. The automated cell proliferation assay was more accurate and sensitive than the manually scored SA-β-gal assays, and it is possible that many of the shRNAs that scored solely in the proliferation screen may have had effects on SA-β-gal activity levels that we failed to detect. However, we were interested to note that 24 of the 41 kinases identified by the SA-β-gal screen did not score in the cell proliferation assays (including all Z-scores >|2|). This suggests that there may be specific pathways that are not required for pRB to arrest the cell cycle, but that may be important for pRB-arrested cells to enter a senescence-like state (as reflected by SA-β-gal activity). This distinction is potentially significant because pRB's activity is regulated by cdk phosphorylation, and cells arrested in G1 by pRB can potentially be driven back into cycle, whereas senescent cells are permanently arrested. IPA analysis placed 18 out of these 24 kinases together in a highly scoring network that focuses around ERK1/2, JNK, PI3K/AKT, and p38MAPK as central components (Supplemental Fig. S2).
Potentially, changes in the activity of any number of these kinases may allow tumor cells to evade some of the effects of pRB. In this regard, one of the more intriguing hits is LATS2. LATS2 is part of a well-studied tumor suppressor pathway (the Hippo pathway) that was first discovered in Drosophila (Edgar 2006; Harvey and Tapon 2007). The Hippo pathway plays a central role in growth regulation by inhibiting proproliferative and anti-apoptotic functions of YAP1 and YAP1-related proteins (Zeng and Hong 2008). Although the Hippo pathway is conserved from flies to humans, and the homozygous inactivation of hpo or wts causes tissue overgrowth in flies (Justice et al. 1995; Xu et al. 1995; Harvey et al. 2003; Huang et al. 2005), currently, there are few examples of homozygous mutation of LATS2 in human tumors. LATS2 depletion enhances H-RasV12-dependent cell migration, providing evidence that loss of LATS2 can enhance some aspects of cell transformation (Aylon et al. 2009).
shRNAs targeting LATS2 were among the strongest and most consistent suppressors of pRB-induced SA-β-gal. This is intriguing because LATS2 is located near RB1 on chromosome 13 (LATS2 maps to 13q11-q12, while RB1 maps to 13q14.2), and genomic studies have shown that a large number of cancer cell lines are heterozygous for both RB1 and LATS2 (see the Discussion). Our results suggest that LATS2 functionally cooperates with pRB, and we sought to understand why low levels of LATS2 impacts some pRB-induced phenotypes.
LATS2 is important for pRB induction of a senescence-like state and promotes the complete silencing of E2F target genes
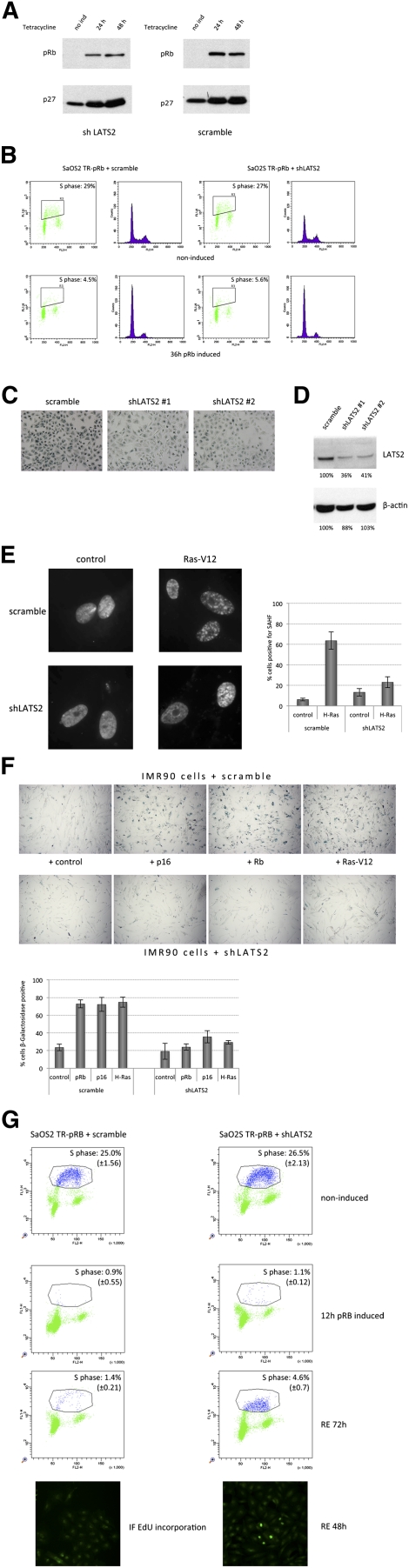
First, we examined the effects of LATS2 depletion on pRB-induced phenotypes. Depletion of LATS2 did not change the amount of pRB induced in the SaOS2-TR-pRB cells or pRB's ability to induce the expression of p27CIP/KIP (Fig. 3A). Consistent with this, two-dimensional FACS analysis showed that pRB suppressed S-phase entry and drove the accumulation of cells in G1 in LATS2-depleted cells (Fig. 3B), similar to control-treated cells.
Figure 3.
LATS2 is required for pRB-dependent senescence. The effect of shRNA-mediated LATS2 knockdown on pRB-dependent G1 arrest and senescence. (A) Knockdown of LATS2 does not interfere with p27CIP/KIP induction following pRB expression. Protein extracts were prepared, and equal amounts of protein were subjected to SDS-PAGE and Western analysis for the indicated proteins. (B) Two-dimensional FACS analysis of SaOS2 cells after pRB induction. Control or LATS2 shRNAs were transduced into SaOS2 cells. After 36 h of tetracycline induction, cells were pulsed with BrdU, fixed, and subjected to flow cytometric analysis. Knockdown of LATS2 does not interfere with pRB-induced G1 arrest. The box highlights S-phase cells marked by BrdU. The cell cycle profile is represented in a FL2-H histogram (DNA content analyzed by propidium iodide staining). (C) shRNAs against LATS2 decrease SA-β-gal staining in SaOS2 cells after 84 h of induction of pRB expression. Cells were transduced with either a scrambled control shRNA or two different shRNAs against LATS2. LATS2 shRNAs show a reduced level of SA-β-gal staining (blue) compared with control shRNA-containing cells (scramble). (D) Western blot analysis of LATS2 expression after knockdown using two independent shRNAs. Protein abundance was determined by digital quantification (Adobe Photoshop) from Western blot scans. Each value was converted to percentage of protein level in the scramble shRNA control sample. (E) Reduced LATS2 expression diminishes SAHF formation after H-RasV12 expression. IMR90 cells with either LATS2 shRNA or control shRNA (scramble) knockdown were infected with lentiviruses containing either empty vector control or H-RasV12. After 9 d post-infection of incubation, cells were fixed and stained with DAPI. Enlarged images of nuclei are shown at 100× magnification. (Bar graph) Quantification shows the percentage of cells positive for SAHF. (F) LATS2 knockdown reduces SA-β-gal staining in IMR90 cells that were induced to senesce by expressing p16, pRB, or H-rasV12. Cells were fixed and stained for the senescence marker SA-β-gal 9 d after introduction of the indicated constructs or a control vector. (Bar graph) Quantification shows the percentage of cells positive for β-gal. (G) Depletion of LATS2 enables cells to escape pRB-induced G1 arrest. FACS analysis is shown for SaOS-TR-pRB cells containing shRNA against LATS2 or scramble control. The top panel shows uninduced cells. The second panel shows cells after 12 h of pRB induction. The third panel shows cells after 12 h of pRB induction and a 72-h chase (in the absence of induction). Although control cells remain arrested, a subset of shLATS2 cells starts incorporation of EdU after 72 h of release. The bottom panel shows immunofluorescence images of the EdU-incorporating cells (green nuclei) in addition to S-phase percentages shown by flow cytometry dot plots.
Depletion of LATS2 strongly reduced both the number of cells that stained positive for SA-β-gal following pRB induction and the intensity of the staining (Fig. 3C). shRNAs often provide only a partial knockdown of the target. Two independent shRNAs targeting LATS2 strongly suppressed pRB-induced SA-β-gal activity even though LATS2 levels were only reduced to slightly less than half of their normal levels.
Expression of pRB in SaOS2 cells has been established as a model for pRB induction of a senescence-like state (Alexander and Hinds 2001). Several additional experimental systems have been described in which pRB is required for senescence (Serrano et al. 1997; Lin et al. 1998; Dimri et al. 2000; Steiner et al. 2000; Alexander et al. 2003; Campisi 2005). Depletion of LATS2 reduced the number of SAHF induced by H-RasV12 in IMR90 fibroblasts (Fig. 3E) and suppressed SA-β-gal staining induced by pRB, p16INK4A, and H-RasV12 in both IMR90 and RPE cells (Fig. 3F; Supplemental Fig. S4). Thus, in multiple assay systems in which pRB is activated, reducing the level of LATS2 strongly suppressed the appearance of markers of senescence.
Formation of SAHF is thought to be a multistep process. PML bodies are proposed to play a mediatory role in SAHF formation by acting as sites for the assembly of macromolecular regulatory complexes and protein modification (Zhang et al. 2005, 2007). Depletion of LATS2 had only a slight effect on the appearance of PML bodies (Supplemental Fig. S3A) and a minor effect on the changes in cell morphology (increased cell size and increased granularity) associated with pRB-induced senescence in SaOS2 cells (Supplemental Fig. S3B). This suggests that reduced LATS2 influences some aspects of senescence but does not affect pRB-induced changes in morphology.
The strong defects in SAHF formation and reduction in SA-β-gal staining suggested that LATS2-depleted cells have difficulty establishing a fully senescent state. Consistent with this, SaOS2 cells that were arrested with a pulse of pRB expression were more easily able to re-enter the cell cycle, as revealed by EdU incorporation, if first depleted of LATS2 (Fig. 3G).
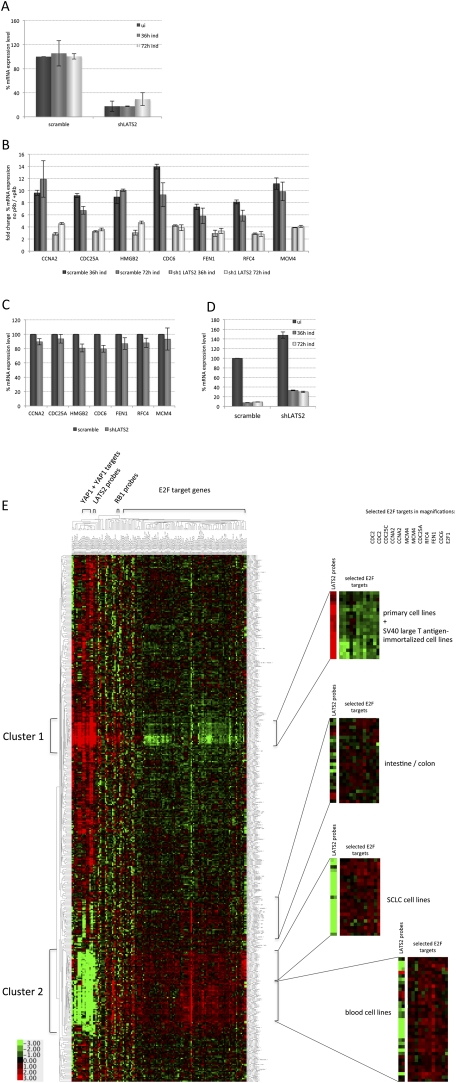
The formation of SAHF has been linked to the silencing of E2F target genes and requires an intact pRB pathway (Narita et al. 2003). Quantitative PCR (qPCR) was used to measure the repression of E2F target genes by pRB in control SaOS2-TR-pRB cells or cells expressing a LATS2-targeting shRNA. In control cells, E2F target genes are strongly down-regulated after 24 h of pRB expression. Although knockdown of LATS2 does not interfere with induction of p27CIP/KIP or with pRB-induced cell cycle arrest, we observed a clear defect in pRB-initiated repression of E2F target genes in cells with reduced LATS2 (Fig. 4A,B). In most cases, the knockdown of LATS2 had no effect on gene expression in the absence of pRB, suggesting that reduced LATS2 impairs the repression of E2F targets in response to pRB (Fig. 4C). The failure to completely silence E2F transcription provides a simple explanation for the defect in some aspects of senescence: In previous studies, inhibition of E2F-mediated repression has been shown to interfere with pRB-mediated induction of senescence markers (Rowland et al. 2002; Talluri et al. 2010). The expression of E2F1, itself an E2F-regulated target, was slightly increased when LATS2 is depleted in the absence of pRB (Fig. 4D). Even though this did not result in a simultaneous increase in expression of E2F targets, it is possible that a small change in the levels of activator E2Fs could also contribute to the failure to silence E2F targets. In summary, reducing the level of LATS2 impairs the repression of E2F target genes, and this likely contributes to a partial bypass of senescence.
Figure 4.
LATS2 is required for complete repression of E2F target genes by pRB. E2F target gene expression was analyzed after knockdown of LATS2 and puromycin selection. Total RNA of SaOS2 cells was extracted and reverse-transcribed to prepare cDNA. qPCR was carried out and the mRNA expression levels of indicated genes were analyzed using the level of GAPDH as a reference. The Y-axis represents the percentage of mRNA expression of the indicated genes relative to the expression level of noninduced scrambled shRNA-containing control cells normalized to GAPDH (set to 100%). Error bars represent the standard deviation of three data points. (A) qPCR analysis confirms the reduction of LATS2 expression level in the shRNA-treated cells that were analyzed for E2F target gene expression (shown in B) without and with induction of pRB expression. (B) Comparison of E2F target gene expression in control cells and LATS2 shRNA-containing cells before and after induction of pRB expression for 36 h or 72 h. The bar graph represents the fold change of E2F target gene expression in −pRB versus +pRB cells. (C) The effect of LATS2 knockdown on E2F target gene expression in uninduced SaOS2-TR-pRB cells analyzed by qPCR. (D) qPCR analysis of E2F1 mRNA levels in control and LATS2 knockdown cells. (E) Microarray expression data derived from tumor cell lines support the idea of cooperation between the Hippo pathway kinase LATS2 and the pRB–E2F pathway. The heat map represents the hierarchical clustering analysis of the gene expression data for E2F target genes, RB1, LATS2, and other Hippo pathway members such as YAP and YAP target genes. Horizontal columns represent individual genes and vertical rows represent the separate cell lines from a panel of 428 cancer and normal cell lines (Supplemental Table S6). Red and green correspond to the high and low expression of the gene transcripts, respectively. The bar indicates relative expression of transcripts using a log2 transformed scale. The statistical analysis of the correlation between expression of LATS2 and E2F target genes resulted in a P-value of 0.014.
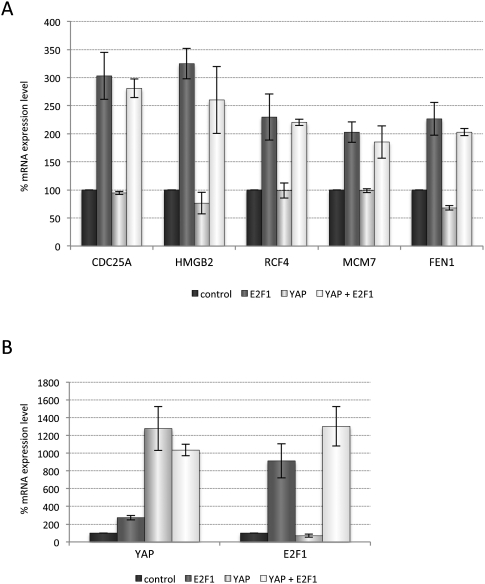
These results suggest that LATS2 facilitates the repression of E2F targets. To test the feasibility of such a model, we compared the levels of expression of LATS2 and E2F-regulated genes across a panel of 428 cancer and normal cell lines. A heat map of expression data for LATS2 and E2F target genes, as well as RB1 and the transcriptional coactivators of the Hippo pathway (TEAD, YAP1, and WWTR1), is shown in Figure 4E. Several prominent clusters are clearly evident within the data, including a cluster of nontransformed cells that show high expression of LATS2 and low expression of E2F targets and a cluster of blood cell lines and small cell lung cancer cells that have the converse pattern. Overall, the data show an inverse relationship between LATS2 expression and the expression of E2F target genes. The statistical correlation of LATS2 expression with E2F target genes compared with a random gene set resulted in a significant two-sided P-value of 0.014. These observations are consistent with the idea that LATS2 promotes E2F repression, while low levels of LATS2 correlate with increased expression of E2F targets.
LATS2 promotes DREAM-mediated silencing of E2F targets
The best-known target of the LATS kinases is the transcriptional coactivator YAP1. The mammalian LATS1 and LATS2 proteins regulate the nuclear localization of YAP1 and repress its transcriptional activity. Chromatin immunoprecipitation (ChIP) and microarray (ChIP–chip) experiments for the TEAD transcription factor, one of the main binding partners of YAP1, show that the mammalian genome contains many potential YAP1-regulated binding sites (Zhao et al. 2008). Therefore, one model to explain the effect of LATS2 on the expression of E2F targets was the possibility that YAP1 and E2F might converge on many of the same promoters. To test whether E2F1 and YAP1 synergistically activate E2F-dependent promoters, we asked whether E2F1 and an activated variant of YAP1 (YAP1S127A) can increase transcription of E2F target genes when they are coexpressed. We examined several E2F targets that showed increased expression in the presence of pRB in the LATS2-depleted cells and performed qPCR analysis of their mRNA after exogenous expression of E2F1, YAP1, or the combination of proteins. Although YAP1S127A activated expression from its targets (Supplemental Fig. S4), and E2F1 expression induced expression of multiple E2F-dependent genes, no additive effect was seen when E2F1 and YAP1 were coexpressed, even when both transcription factors were strongly overexpressed (Fig. 5A,B). Consistent with this, we note that, although LATS2 and E2F-regulated genes show an inverse pattern of gene expression across the panel of cancer cell lines, low levels of LATS2 do not strongly correlate with the elevated expression of previously described YAP1 targets such as CTGF and SERPINE1 (Fig. 4E), making it unlikely that changes in YAP1 activity could explain a connection between LATS2 and E2F.
Figure 5.
Analysis of YAP1- or E2F1-dependent activation of E2F target genes. YAP1, the transcriptional coactivator that is negatively regulated by the Hippo pathway, does not increase transcriptional activation of E2F target genes. (A) Analysis of the indicated E2F target gene mRNA expression levels in SaOS2 cells expressing a constitutively active YAP1 variant (S127A), tetracycline-induced E2F1, or the combination of both. (B) The YAP1 and E2F1 expression levels were analyzed in parallel to the E2F targets in A to confirm the increase in expression levels.
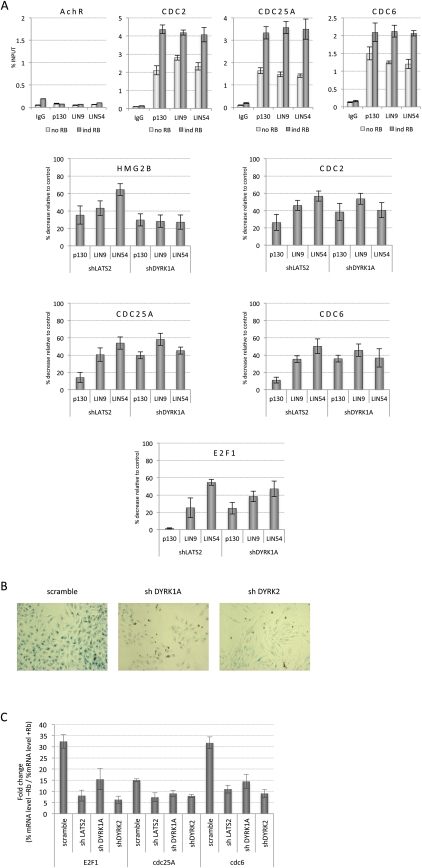
An alternative possibility is that LATS2 has additional substrates. Previous studies have shown that p130/DREAM repressor complexes are major repressors of E2F-regulated promoters in quiescent and senescent cells, and that depletion of the DREAM protein LIN9 from SaOS2 cells strongly suppresses pRB-induced SA-β-gal staining (Gagrica et al. 2004). ChIP experiments with antibodies to the DREAM complex components p130, LIN54, and LIN9 showed that the binding of these proteins to E2F target promoters increased when pRB was expressed (Fig. 6A), suggesting that DREAM repressor complexes mediate the pRB-initiated silencing of E2F-regulated promoters in SaOS2 cells (Fig. 6A, top panel). Strikingly, the binding of LIN54 and LIN9 to E2F-regulated promoters was reduced when LATS2 was depleted (Fig. 6A, bottom panel), indicating that the depletion of LATS2 reduces either the formation of DREAM complexes or the ability of DREAM complexes to bind stably to E2F-regulated promoters. Consistent with this result, shRNAs targeting LIN9 or LIN54 strongly suppressed pRB-induced SA-β-gal staining in SaOS2-TR-pRB cells, confirming that DREAM contributes to pRB-mediated induction of senescence markers. Indeed, the effect of targeting LIN9 or LIN54 was similar to the effects caused by the knockdown of LATS2 (Supplemental Fig. S6).
Figure 6.
LATS2 promotes DREAM complex-mediated silencing of E2F target genes. shRNA-mediated knockdown of LATS2 and the DYRK kinases causes similar defects in pRB-mediated phenotypes. (A) Analysis of mammalian DREAM complex components bound to E2F target gene promoters. SaOS2 cells with or without induced pRB expression and containing shRNAs targeting LATS2, DYRK1A, or scramble control were analyzed by ChIP using antibodies against p130, LIN9, and LIN54. (Top panel) Binding to the indicated E2F target gene promoters or control AchR promoter was measured by qPCR and is presented as percentage of input. The bottom panel shows the percent decrease of binding in either shLATS2- or shDYRK1A-containing cells compared with scramble control cells after 36 h of pRB expression. (B) Decrease in pRB-induced appearance of the SA-β-gal senescence marker after knockdown of DYRK1A or DYRK2. shRNAs against the DYRK1A and DYRK2 decrease SA-β-gal staining in SaOS2 cells after induction of pRB expression. (C) DYRK1A or DYRK2 knockdown impairs the pRB-mediated repression of E2F target genes. qPCR analysis of E2F target gene mRNA levels after knockdown of DYRK1A, DYRK2, or LATS2 and induction of pRB for 36 h. The mRNA level of GAPDH is used as a reference. The expression level of noninduced scramble control shRNA-containing cells normalized to GAPDH is set to 100%. The graph shows the fold change of gene expression after pRB induction. Error bars represent the standard deviation of three data points.
Recent work has identified the DYRK family of kinases as important regulators of DREAM complex assembly, with DYRK1A phosphorylation of LIN52 promoting the formation of the repressor complex (Litovchick et al. 2011). Interestingly, shRNAs targeting the DYRK family of kinases scored multiple times in the screens (Figs. 1B, 2B; Supplemental Tables S1, S2), providing additional support for the idea that DREAM repressor complexes are important for pRB-induced arrest. Knockdown of DYRK1A and DYRK2 also suppressed pRB-induced SA-β-gal activity in SaOS2-TR-pRB cells (Fig. 6B) and reduced pRB-initiated repression of E2F targets (Fig. 6C). Indeed, depletion of DYRK1A caused a reduction in the binding of LIN54 and LIN9 to E2F-regulated promoters (Fig. 6A) that was comparable with the change seen in LATS2-depleted cells.
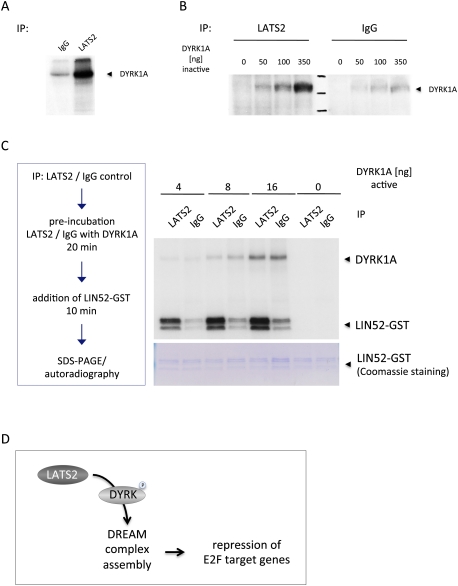
The similar phenotypes resulting from the depletion of LATS2 and DYRK suggest that these kinases may act in the same pathway. The amino acid sequences of both DYRK1A and DYRK2 contain putative consensus phosphorylation sites for the LATS2 kinase (HxH/R/KxxS/T) (Hao et al. 2008). The LATS2 kinase phosphorylates DYRK1A in a dose-dependent manner when the proteins are incubated in vitro (Fig. 7A,B). To test whether LATS2 modulates DYRK1A activity, we took advantage of the recent discovery that DYRK1A phosphorylates LIN52 (Litovchick et al. 2011). We immunoprecipitated LATS2 from 293 cells, incubated it with purified DYRK1A and kinase buffer containing ATP, and then examined the ability of DYRK1A to phosphorylate LIN52-GST in vitro. Preincubation with LATS2 strongly enhanced DYRK1A-dependent phosphorylation of LIN52-GST, compared with controls (Fig. 7C). LATS2 complexes showed no activity toward LIN52-GST, suggesting that the effects of LATS2 on LIN52-GST phosphorylation were mediated via DYRK1A. Taken together, these results show that LATS2 can phosphorylate DYRK1A and enhances DYRK1A's ability to modify LIN52. Since DYRK1A-mediated phosphorylation of LIN52 promotes the assembly of DREAM complexes (Litovchick et al. 2011), this provides a simple model to explain how LATS2 levels influence DREAM-mediated repression. LATS2 cooperates with DYRK kinases to promote the assembly of DREAM repressor complexes at E2F-regulated promoters (Fig. 7D). This silencing of E2F target genes is not essential for pRB to arrest the cell cycle, perhaps because pRB also induces p27. However, it is important for pRB induction of some senescence markers and does seem to affect the ability of pRB-arrested cells to re-enter the cell cycle.
Figure 7.
LATS2 promotes DYRK1A activity. LATS2 phosphorylates DYRK1A and enhances its kinase activity in vitro. (A) LATS2 kinase modifies DYRK1A. LATS2-V5 was expressed in 293T cells, immunoprecipitated, and subjected to a 32P-γATP in vitro kinase assay with 1 μg of inactivated DYRK1A as a substrate. Phosphorylation of DYRK1A is visualized by autoradiography. (B) LATS2 in vitro kinase assay as shown in A, with increasing amounts of inactivated DYRK1A provided as substrate. (C) LATS2 increases the ability of DYRK1A to phosphorylate LIN52. The DYRK1A kinase was preincubated with LATS2-V5 in kinase buffer containing unlabeled ATP and was subsequently assayed in an in vitro kinase reaction as shown in A. Phosphorylation of purified LIN52-GST is used to assess DYRK1A activity. Potentially, the increase in phosphorylated LIN52-GST could be due to an increase in DYRK1A enzymatic activity or enhanced substrate specificity toward LIN52. Autophosphorylation of DYRK1A results in the indicated band. (D) Model illustrating the relationship between LATS2 and E2F target gene repression. See the text for details.
Discussion
In this study, we identify sets of kinases that modify the cellular response of tumor cells to pRB. pRB is a key component of the tumor-suppressive mechanisms that cells use to respond to insults and oncogenic signals. pRB's ability to stop cell proliferation and drive cells into a permanently arrested state is an important barrier against cell transformation. Although pRB is famously mutated in some cancers, most human cancer cells express an intact pRB protein, and the extent to which the function of this protein can be reactivated and/or enhanced is unknown. The results described here show that there are many kinases that impact the ability of pRB to stop cell proliferation, and the lists of kinases reported here represent an important starting point for the identification of new ways to enhance pRB function.
Currently, it is uncertain which of pRB's many molecular functions are most critical for its role as a tumor suppressor. In our screens, we assessed two related properties (pRB's ability to suppress cell proliferation, and its ability to induce the appearance of a senescence marker) that are different readouts of pRB action. Irreversible proliferation arrest is a hallmark of senescence, and expression of SA-β-gal frequently characterizes this state. However, cells can also enter a reversibly arrested state that is distinct from senescence, and these cells typically do not express SA-β-gal. Both senescence and reversible proliferation arrest are likely relevant to tumor suppression. While some kinases were identified with both assays, many scored preferentially in just one. Several of the shRNAs that scored in these screens target stress-activated kinases and components of well-known signaling pathways (ERB2 and KIT) (Tang et al. 2008). These hits are consistent with the idea that the SaOS2 cells used for screening are addicted to specific signaling pathways, and thus are primed to undergo oncogene-induced senescence following the reintroduction of pRB. However, other hits from the screens were completely unexpected, and reveal levels of functional cooperation between pathways that had not been appreciated previously.
The functional interaction between LATS2 and pRB is particularly intriguing. The knockdown of LATS2 compromises pRB-induced repression of E2F-regulated genes and suppresses the increase in SA-β-gal activity and formation of SAHF. These findings are consistent with previous studies showing that the transcriptional silencing of E2F target genes is important for the establishment of a stable arrest and is implemented by changes in chromatin organization that are characteristic marks of senescent cells (Narita et al. 2003; Zhang et al. 2007). It also is in accord with experiments demonstrating that the failure to fully repress E2F targets interferes with aspects of pRB-dependent senescence (Rowland et al. 2002; Talluri et al. 2010). Our finding that LATS2 depletion reduces the recruitment of the DREAM complex to E2F targets is also consistent with evidence that the DREAM components LIN9 and LIN54 are needed for pRB-induced SA-β-gal staining in SaOS2 cells (Supplemental Fig. S6; Gagrica et al. 2004), and that pRB and p130 are present at the promoters of many proliferation-related genes in senescent cells (Chicas et al. 2010). In keeping with studies showing that the initial cell cycle arrest induced by pRB in SaOS2 cells is primarily dependent on the up-regulation of p27CIP/KIP, the defect in E2F repression in LATS2-depleted cells was not sufficient to prevent pRB from arresting cells in G1, but these cells more easily re-entered the cell cycle.
The discovery that LATS2 cooperates with pRB to establish a senescence-like arrest reveals an unexpected link between the pRB and Hippo tumor suppressor pathways. We suggest that the level of LATS2 expression may have an impact on pRB's tumor suppressor activity by reducing the ability of some pRB-arrested cells to permanently exit the cell cycle. Remarkably, LATS2 and RB1 fall within a region on 13q that frequently exhibits loss of heterozygosity (LOH) in primary human cancers (Yabuta et al. 2000). An examination of array comparative genomic hybridization (CGH) data available at the Sanger database revealed that 43% (336 of 772) of human tumor cell lines analyzed show LOH for either RB1, LATS2, or both genes. Of these, 82% (276 of 336) show LOH for both RB1 and LATS2. The evidence that a modest reduction in the levels of LATS2 compromises the activity of pRB raises the possibility that these large deletions may reduce the functionality of protein expressed from the remaining RB1 allele. Intriguingly, the specific mutation of the remaining RB1 allele has been found in only a small percentage of cell lines carrying 13q deletions, suggesting that, in most cases, the remaining RB1 allele is intact. Using the Catalogue of Somatic Mutations in Cancer (COSMIC) obtained from the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/CGP/cosmic), we examined the frequency of RB1 mutation, p16INK4 deletions/mutations, and CCND1 amplifications in tumor cell lines with 13q deletions. We found that 76% of the cell lines that showed LOH for RB1 but not LATS2 had either a specific mutation in the remaining RB1 allele, loss of p16INK4, or gains in CCND1 that would compromise the activity of any wild-type pRB. However, only 56% of cells that showed LOH for both LATS2 and RB1 had these additional changes (P < 0.02, significance tested by a two-tailed Z-test for two proportions). Such a difference is consistent with the idea that LOH for LATS2 may reduce the selective pressure for further mutations within the “pRB pathway.” Clearly, additional experiments will be necessary to test this hypothesis.
Several studies have noted that the expression of LATS2 is low in tumor cells (Jiang et al. 2006; Steinmann et al. 2009; Strazisar et al. 2009), and the results described here help to explain why this change might be advantageous. These findings are also consistent with reports showing that increased levels of LATS2 inhibit cell proliferation and suppress tumorigenicity (Li et al. 2003), and with evidence that LATS2 cooperates with p53 to promote an oncogenic stress checkpoint that opposes H-Ras-dependent oncogenic transformation (Aylon et al. 2009).
Functional cooperation between coamplified genes has been documented in several studies, including analysis of the 17q12 amplicon in breast cancer (Kao and Pollack 2006) and gastrointestinal adenocarcinomas (Maqani et al. 2006). The interactions between pRB and LATS2 suggest that 13q deletions may have the converse effect of targeting the cooperative effects of synergistic tumor suppressor pathways. It is noteworthy that the BRCA2 tumor suppressor is also frequently codeleted with RB1 and LATS2, and it is tempting to speculate that reduced levels of BRCA2 may have additional effects on either pRB or LATS2.
These results suggest that some Hippo pathway signals may converge with the pRB pathway at the level of E2F regulation. Interestingly, functional interactions between the pRB and Hippo tumor suppressor pathways appear to be present in both flies and humans. Genetic studies show that inactivation of the Drosophila rbf1 and wts genes have strongly synergistic effects on cell proliferation. Cells that are mutant for both suppressors have increased proliferation and continue to cycle even when they have initiated differentiation (Nicolay et al. 2010). Curiously, the mechanisms underlying the interactions between these pathways are related, but distinct, in the two species. In both flies and humans, the two pathways converge on E2F regulation. In Drosophila, the mutation of wts elevates E2F-dependent transcription, causing ectopic cell proliferation that is de2f1-dependent. The functional interaction between rbf1 and wts mutants occurs because dE2F1 and Scalloped/Yorkie (the orthologs of mammalian YAP1/TEAD proteins) share common targets and cooperate in transcriptional activation (Nicolay et al. 2011). In contrast, in mammalian cells, LATS2 depletion has a more general effect on E2F targets and increases E2F-dependent transcription primarily by interfering with the assembly of E2F-mediated repressor complexes. Nevertheless, the conservation of this genetic interaction suggests that the functional cooperation between the pRB and Hippo tumor suppressor pathways is an important component of cell proliferation control.
These results also add to the emerging view that the mammalian Hippo pathway is neither linear nor simple. YAP1 and its homolog, TAZ, are both substrates of the Hippo pathway in human cells (Huang et al. 2005; Lei et al. 2008). Recently, LATS1 has also been shown to phosphorylate FOXL2 and enhance its activity as a transcriptional repressor. LATS2 interacts with Omi, which enhances its protease activity (Kuninaka et al. 2007), and, by cooperating with the Androgen receptor (AR), LATS2 can act as a corepressor of AR-dependent transactivation (Powzaniuk et al. 2004). In Drosophila, WWC1/KIBRA complexes with Wts, leading to reduced Yki phosphorylation (Genevet et al. 2010), but neither protein has yet been shown to be a direct phosphorylation target of the LATS2 kinase. Our results indicate that DYRK1A may be an additional target of LATS2, and that LATS2 cooperates with DYRK family kinases to regulate the DREAM complex (Litovchick et al. 2011), thus promoting the stable repression of E2F targets. Potentially, this connection may enable the central kinase cascade of the mammalian HIPPO pathway (Fig. 7D) to influence the expression of many proliferation-related genes.
Materials and methods
Cell lines and DNA constructs
IMR-90, hTERT-RPE-1, 293T, and SAOS-TR-pRB cells were grown in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Stable SAOS-TR-pRB cells (Binne et al. 2007) and stable SAOS-TR-E2F1 cells (Morris et al. 2008) were described before. pRB or E2F1 expression was induced by addition of tetracycline (0.5 μg mL−1 or 0.25 μg mL−1, respectively).
DNA fragments encoding LATS2 were amplified by PCR, and were used with the pENTR Directional TOPO Cloning Kit (Invitrogen) according to manufacturer's instructions to create a pENTR-LATS2. LATS2 cDNA was subsequently transferred into pLenti6/V5-DEST (Invitrogen) with LR clonase (Invitrogen) to obtain the pLenti6-LATS2-V5 construct. Lentiviral constructs containing p16INK4A and pRB (pWPI-p16INK4A and pWPI-pRB) were created using pDONR221 plasmids obtained from PlasmidID (DF/HCC DNA Resource Core) and transferring the p16INK4A and pRB cDNA fragments into pWPI-DEST (Addgene, Didier Trono) using the gateway cloning procedure (LR clonase, Invitrogen). The DNA preparation, transfection, and virus preparation methods have been published elsewhere (Pearlberg et al. 2005). The pWPI-H-RasV12 and pWPI-YAP1-S127A plasmid constructs were kindly provided by Anurag Singh and Jianmin Zhang, respectively. LKO.1 shRNA vectors targeting LATS2 (NM_014572; sh#1, TRCN0000000880; sh#2, TRCN0000000883), DYRK1A (NM_001396; TRCN0000000525), DYRK2 (NM_003583; TRCN0000000651), LIN9 (NM_173083; TRCN0000115874), and LIN54 (NM_194282; TRCN0000107669) were obtained from the RNAi Consortium.
shRNA library, virus production, and infections
The kinase shRNA library used for screens is a subset of the lentiviral shRNA library in the LKO.1 vector from the RNAi Consortium. Kinase shRNAs were arrayed onto 96-well plates along with scrambled shRNAs and were described before (Conery et al. 2010). DNA preparation, transfection, and virus preparation methods have been published elsewhere (Pearlberg et al. 2005). The volume of virus to be used for infections was determined in preliminary experiments in which virus amount was titrated using resistance to puromycin and the effectiveness of known lethal shRNAs. For both screens in SaOS2-TR-pRB cells, 2200 cells were plated in duplicate 96-well plates. The next day, 12 μL of lentivirus was added to each well along with 8 μg/mL polybrene (Sigma). Plates were centrifuged at 2250 rpm for 30 min and incubated overnight at 37°C. After ∼16 h, the virus was removed and fresh medium with 1 μg/mL puromycin (Sigma) was added. After cells were incubated for 3 d, pRB expression was induced in one of the plates infected in parallel by changing to tetracycline-containing medium. The second plate was kept in non-tetracycline-containing medium.
Cell proliferation and viability were analyzed over a 4-d time period (0 h [before induction], and 48 h and 96 h of induction) by using Alamar blue (Resazurin, Sigma) staining. Alamar blue is a redox-sensitive dye that provides a readout for cell proliferation and viability and where absorbance was directly proportional to cell number (Grueneberg et al. 2008).
For the SA-β-gal screen, cells were grown, infected, and induced as described above. pRB expression was induced for 84 h, and cells were fixed in PBS containing 2% formaldehyde/0.2% glutaraldehyde and stained for SA-β-gal (as described below) to assess the level of senescence after pRB induction. After staining, cells were analyzed using a light microscope (10× objective) and scored for intensity and relative amount of stained cells on a scale between 1 and 5, with 3 representing the control level, 1 representing inhibition, and 5 representing enhancement. Each screen was carried out twice with an average of 4.5 independent hairpins per gene, and SA-β-gal assays were also scored twice independently.
Primary screen data analysis and hit determination
The Z-score for each shRNA was determined by comparing the FR (ratio between percent of growth after 4 d of non-pRB-expressing vs. pRB-expressing cells) for each shRNA with the mean FR of the scramble shRNA controls for the data set and calculating the number of standard deviations by which they differed. Hits were determined by comparing the Z-scores for each kinase, and a hit was declared if the Z-score was >|3| (Supplemental Table S1).
Senescent assay, immunofluorescence, and flow cytometry
Cells were fixed and stained for the senescence marker β-Galactosidase (β-Gal) as described before (Dimri et al. 1995). Briefly, cells were washed in PBS, fixed for 5–10 min in PBS containing 2% formaldehyde/0.2% glutaraldehyde, washed, and incubated for 12–16 h (without CO2) in staining solution (1 mg/mL X-Gal [Sigma] in 40 mM citric acid/sodium phosphate buffer at pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, 2 mM MgCl2) at 37°C. Cells were washed briefly with ddH2O, and staining was analyzed by light microscopy. The number of senescent cells and the total number of cells were counted in multiple fields (10× objective) to determine the percentage of senescent cells.
PML bodies were detected by immunofluorescence staining, as described before, using an anti-PML antibody (PGM-3, 1:200; Santa Cruz Biotechnology) (Ferbeyre et al. 2000).
For visualization of SAHF, cells were fixed in 4% PFA and stained with 0.2 μg/mL DAPI in PBS/0.1% Tween20/2.5% BSA. Images of fixed nuclei were captured with a Hamamatsu Orca AG cooled CCD camera mounted on a Nikon TI/Yokagawa CSU-10 spinning-disk confocal microscope with a 100×, 1.4 NA objective. The number of SAHF-containing cells and the total number of cells were counted in multiple fields (40× objective) to determine the percentage of SAHF-positive cells.
Flow cytometry was performed as described earlier (Binne et al. 2007). Briefly, cells were labeled for 1 h with BrdU (GE), fixed in 75% ethanol, denatured in 2 M HCl and 0.5% Triton X-100, and neutralized in 0.1 M borate (pH 8.5). Cells were incubated with anti-BrdU antibody (BD; 1:80 in PBS, 0.5% Tween-20, 1% BSA) and secondary anti-mouse FITC-conjugated antibody (1:400; BD). Cells were stained with 5 μg/mL propidium iodide in PBS, 1% BSA, and 250 μg/mL RNase A. Cells labeled with EdU were detected using the Click-iT EdU Flow Cytometry Assay kit (Invitrogen) with modification of the protocol by fixation of cells in 75% ethanol. Cell morphology (cell size and granularity) was determined using flow cytometry on cells fixed in 1% paraformaldehyde/PBS. All samples were analyzed using a BD FACS Calibur flow cytometer and CellQuest software.
Pathway and network analysis
Kinases identified in our screens were imported into IPA (Ingenuity Systems, http://www.ingenuity.com) for canonical pathway analyses. The statistical significance for each network was determined by the IPA core analysis using a Fisher Exact test. The networks were ranked by scores that represent the probability that these genes from our input list were included in the network by chance alone. For example, networks with scores of >6 have a P-value of <1 × 10−6, and therefore a 99.999999% confidence of not being generated by random chance (Calvano et al. 2005).
Gene expression microarray analyses
Comparative whole-genome expression profiling was performed on Affymetrix U133 X3P microarrays. Gene expression microarray analysis was done as described before (Singh et al. 2009). Expression data were normalized using the MAS5 algorithm. Normalized expression values are provided in Supplemental Table S6. To generate heat maps of microarray expression data, average linkage hierarchical clustering was performed using Cluster 3.0 and Treeview Software (Eisen et al. 1998). By using a comparable set of random genes, the statistical significance of the correlation between LATS2 and E2F target genes was analyzed. The correlation between expression of LATS2 and the average expression of E2F targets was computed. In addition, the average expression of a random set of genes (10,000 times) was used to calculate the correlation to LATS2 expression. By comparison of the calculated correlations, a two-sided P-value of 0.014 was obtained. Repeating the same calculations with correlation between expression of RB1 and E2F targets yielded a two-sided P-value of 0.013.
Immunoblotting and antibodies
Western blot analysis was performed using standard procedures. Protein extracts were prepared with lysis buffer (50 mM HEPES at pH 7.8, 500 mM NaCl, 1% NP40, 5 mM EDTA, 1 mM DTT, protease inhibitor cocktail, phosphatase inhibitor cocktail; Roche). Lysates were subjected to SDS-PAGE and transferred to PVDF membrane. The primary antibodies used were anti-LATS2 (Bethyl Laboratories), anti-p27CIP/KIP (cl57, BD), anti-pRB (G3-245, BD), anti-p130 (Cell Signaling), anti-LIN9 and anti-LIN54 (Litovchick et al. 2007), β-actin (mouse; Sigma), and anti-V5 tag (Invitrogen). Primary antibodies were used at 1:1000 in 5% milk in TBS containing 0.1% Tween20. Secondary antibodies were obtained from GE and were used at 1:10,000 dilutions.
RNA extraction and real-time qPCR
Total RNA extraction was carried out using the RNeasy kit (Qiagen) as recommended by the manufacturer's instructions and was quantified using an absorption of 260 nm. Reverse transcription of RNA samples (RT–PCR) was performed using TaqMan Reverse Transcription reagents (PE Applied Biosystems). Relative levels of specific mRNA were determined using the LightCycler 480 SYBR Green I Master reagent (Roche). Quantification by the comparative CT method was carried out as described in the procedures manual provided by the manufacturer. Amplification with GAPDH-specific primers was used for normalization. All sequences of gene-specific primers are available upon request.
ChIP
ChIP was performed essentially as described before (Rayman et al. 2002) with minor modifications in cell fixation. Cells were prefixed for 15 min in 1.5 mM disuccinimidyl glutarate (EGS), followed by fixing in 1% formaldehyde for 15 min at room temperature. The cross-linking reaction was quenched with 0.125 M glycine follow by two washes with cold PBS. Precleared chromatin was incubated with 2 μg of primary antibodies for 14 h at 4°C. Washes and recovery of precipitated DNA were carried out as described before. DNA was analyzed using the real-time qPCR reaction. PCR primer sequences are available upon request.
Kinase assay
The LATS2 kinase assays was carried out as described before (Zhao et al. 2007). 293T cells were transfected with LATS2-V5. Forty-eight hours post-transfection, cells were lysed (50 mM HEPES at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM DTT, 1× Complete protease inhibitor cocktail, 1× PhosSTOP [Roche]) and immunoprecipitated with 2 μg of anti-V5 antibody. The immunoprecipitates were washed with lysis buffer, followed by wash buffer (40 mM HEPES, 200 mM NaCl) and kinase assay buffer (30 mM HEPES, 50 mM potassium acetate, 5 mM MgCl2, 5 mM MnCl2). The immunoprecipitated LATS2 was used in a kinase assay reaction in the presence of 1 μM cold ATP and 5 μCi of [γ-32P]-ATP for 30 min at 30°C, with 1 μg (or as indicated) of GST-DYRK1A (Invitrogen) as substrate. For analysis of DYRK1A-mediated LIN52 phosphorylation, 0.5 μg of purified LIN52-GST (Litovchick et al. 2011) was used in each sample. The reactions were terminated with SDS sample buffer and subjected to SDS-PAGE and autoradiography.
Acknowledgments
We thank Jianmin Zhang for the pWPI-YAP(S127A) expression vector, and Anurag Singh for providing the pWPI-H-Ras12V expression vector. We also thank Endre Anderssen for advice and help with statistical data analysis, and Patricia Greninger for assistance with the microarray gene expression analysis. We are also very grateful to W. Endege and members of the Dyson and Harlow laboratories for helpful discussions and technical assistance and reading of the manuscript. We especially thank Peter Adams and Phil Hinds for valuable advice and critical reading of the manuscript. This work was supported by NIH grants GM81607 and CA64402 (to N.D). K.T. was supported by the German Research Foundation (DFG) post-doctoral research fellowship TS 235/1-1. A.R.C. was supported by ACS post-doctoral fellowship number PF-07-030-01-CCG. N.D. is the MGH Cancer Center Saltonstall Foundation Scholar.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2000211.
Supplemental material is available for this article.
References
- Adams PD 2007. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene 397: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 36: 2–14 [DOI] [PubMed] [Google Scholar]
- Alexander K, Hinds PW 2001. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol 21: 3616–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander K, Yang HS, Hinds PW 2003. pRb inactivation in senescent cells leads to an E2F-dependent apoptosis requiring p73. Mol Cancer Res 1: 716–728 [PubMed] [Google Scholar]
- Alonso A, Rahmouni S, Williams S, van Stipdonk M, Jaroszewski L, Godzik A, Abraham RT, Schoenberger SP, Mustelin T 2003. Tyrosine phosphorylation of VHR phosphatase by ZAP-70. Nat Immunol 4: 44–48 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Yabuta N, Besserglick H, Buganim Y, Rotter V, Nojima H, Oren M 2009. Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene 28: 4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG Jr 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 18: 623–635 [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Weinberg RA 2005. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 37: 961–976 [DOI] [PubMed] [Google Scholar]
- Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG Jr, Naar AM, Dyson NJ 2007. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol 9: 225–232 [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD 2007. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol 19: 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD 2007. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol 179: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. 2005. A network-based analysis of systemic inflammation in humans. Nature 437: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Campisi J 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, Boldorini R, Pallotti F, Viale G, Gishizky ML, et al. 2006. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res 66: 8147–8154 [DOI] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conery AR, Sever S, Harlow E 2010. Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal stability by activating the GTPase dynamin during cytokinesis. Proc Natl Acad Sci 107: 15461–15466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K 2006. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell 10: 459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cichowski K 2008. Many roads lead to oncogene-induced senescence. Oncogene 27: 2801–2809 [DOI] [PubMed] [Google Scholar]
- Craft CS, Romero D, Vary CP, Bergan RC 2007. Endoglin inhibits prostate cancer motility via activation of the ALK2–Smad1 pathway. Oncogene 26: 7240–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Liao R, Wu BL, Sun P 2004. High intensity ras signaling induces premature senescence by activating p38 pathway in primary human fibroblasts. J Biol Chem 279: 1050–1059 [DOI] [PubMed] [Google Scholar]
- Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, Dal Zuffo R, Matti V, d'Ario G, Montani E et al. 2011. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol 13: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Itahana K, Acosta M, Campisi J 2000. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol 20: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA 2006. From cell structure to transcription: Hippo forges a new path. Cell 124: 267–273 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev 14: 2015–2027 [PMC free article] [PubMed] [Google Scholar]
- Gagrica S, Hauser S, Kolfschoten I, Osterloh L, Agami R, Gaubatz S 2004. Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J 23: 4627–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Riehl A, Durchdewald M, Nemeth J, Furstenberger G, Muller-Decker K, Enk A, Arnold B, Bierhaus A, Nawroth PP, et al. 2008. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med 205: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N 2010. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 18: 300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueneberg DA, Degot S, Pearlberg J, Li W, Davies JE, Baldwin A, Endege W, Doench J, Sawyer J, Hu Y, et al. 2008. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc Natl Acad Sci 105: 16472–16477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X 2008. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496–5509 [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N 2007. The Salvador–Warts–Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer 7: 182–191 [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467 [DOI] [PubMed] [Google Scholar]
- Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70: 993–1006 [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434 [DOI] [PubMed] [Google Scholar]
- Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, Zhu L 2004. An Rb–Skp2–p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell 16: 47–58 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Li X, Hu J, Zhou W, Jiang Y, Li G, Lu D 2006. Promoter hypermethylation-mediated down-regulation of LATS1 and LATS2 in human astrocytoma. Neurosci Res 56: 450–458 [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534–546 [DOI] [PubMed] [Google Scholar]
- Kamai T, Tsujii T, Arai K, Takagi K, Asami H, Ito Y, Oshima H 2003. Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res 9: 2632–2641 [PubMed] [Google Scholar]
- Kao J, Pollack JR 2006. RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer 45: 761–769 [DOI] [PubMed] [Google Scholar]
- Kondo S, Lu Y, Debbas M, Lin AW, Sarosi I, Itie A, Wakeham A, Tuan J, Saris C, Elliott G, et al. 2003. Characterization of cells and gene-targeted mice deficient for the p53-binding kinase homeodomain-interacting protein kinase 1 (HIPK1). Proc Natl Acad Sci 100: 5431–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J 2011. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16 (ink4a). Cell Cycle 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS 2010. The essence of senescence. Genes Dev 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninaka S, Iida SI, Hara T, Nomura M, Naoe H, Morisaki T, Nitta M, Arima Y, Mimori T, Yonehara S, et al. 2007. Serine protease Omi/HtrA2 targets WARTS kinase to control cell proliferation. Oncogene 26: 2395–2406 [DOI] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL 2008. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pei J, Xia H, Ke H, Wang H, Tao W 2003. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene 22: 4398–4405 [DOI] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA 2011. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev (this issue). doi: 10.1101/gad.2034211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqani N, Belkhiri A, Moskaluk C, Knuutila S, Dar AA, El-Rifai W 2006. Molecular dissection of 17q12 amplicon in upper gastrointestinal adenocarcinomas. Mol Cancer Res 4: 449–455 [DOI] [PubMed] [Google Scholar]
- Maruyama J, Naguro I, Takeda K, Ichijo H 2009. Stress-activated MAP kinase cascades in cellular senescence. Curr Med Chem 16: 1229–1235 [DOI] [PubMed] [Google Scholar]
- Mehta PB, Jenkins BL, McCarthy L, Thilak L, Robson CN, Neal DE, Leung HY 2003. MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene 22: 1381–1389 [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ 2008. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature 455: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Nicke B, Bastien J, Khanna SJ, Warne PH, Cowling V, Cook SJ, Peters G, Delpuech O, Schulze A, Berns K, et al. 2005. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol Cell 20: 673–685 [DOI] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV 2010. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet 6: e1000918 doi: 10.1371/journal.pgen.1000918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV 2011. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev 25: 323–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J 2009. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci 106: 17031–17036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlberg J, Degot S, Endege W, Park J, Davies J, Gelfand E, Sawyer J, Conery A, Doench J, Li W, et al. 2005. Screens using RNAi and cDNA expression as surrogates for genetics in mammalian tissue culture cells. Cold Spring Harb Symp Quant Biol 70: 449–459 [DOI] [PubMed] [Google Scholar]
- Powzaniuk M, McElwee-Witmer S, Vogel RL, Hayami T, Rutledge SJ, Chen F, Harada S, Schmidt A, Rodan GA, Freedman LP, et al. 2004. The LATS2/KPM tumor suppressor is a negative regulator of the androgen receptor. Mol Endocrinol 18: 2011–2023 [DOI] [PubMed] [Google Scholar]
- Rahmouni S, Cerignoli F, Alonso A, Tsutji T, Henkens R, Zhu C, Louis-dit-Sully C, Moutschen M, Jiang W, Mustelin T 2006. Loss of the VHR dual-specific phosphatase causes cell-cycle arrest and senescence. Nat Cell Biol 8: 524–531 [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Wang L, Polo JM, Dell'Oso T, Dierov J, Gaymes TJ, Rassool F, Carroll M, Melnick A 2008. BCL6-mediated attenuation of DNA damage sensing triggers growth arrest and senescence through a p53-dependent pathway in a cell context-dependent manner. J Biol Chem 283: 22565–22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev 16: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren JL, Pan JS, Lu YP, Sun P, Han J 2009. Inflammatory signaling and cellular senescence. Cell Signal 21: 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restall IJ, Lorimer IA 2010. Induction of premature senescence by hsp90 inhibition in small cell lung cancer. PLoS ONE 5: e11076 doi: 10.1371/journal.pone.0011076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, Denissov SG, Douma S, Stunnenberg HG, Bernards R, Peeper DS 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2: 55–65 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Shim J, Park HS, Kim MJ, Park J, Park E, Cho SG, Eom SJ, Lee HW, Joe CO, Choi EJ 2000. Rb protein down-regulates the stress-activated signals through inhibiting c-Jun N-terminal kinase/stress-activated protein kinase. J Biol Chem 275: 14107–14111 [DOI] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J 2009. A gene expression signature associated with ‘K-Ras addiction’ reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner MS, Wang Y, Zhang Y, Zhang X, Lu Y 2000. p16/MTS1/INK4A suppresses prostate cancer by both pRb dependent and independent pathways. Oncogene 19: 1297–1306 [DOI] [PubMed] [Google Scholar]
- Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH 2009. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep 22: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Strazisar M, Mlakar V, Glavac D 2009. LATS2 tumour specific mutations and down-regulation of the gene in non-small cell carcinoma. Lung Cancer 64: 257–262 [DOI] [PubMed] [Google Scholar]
- Su GH, Bansal R, Murphy KM, Montgomery E, Yeo CJ, Hruban RH, Kern SE 2001. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci 98: 3254–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su GH, Song JJ, Repasky EA, Schutte M, Kern SE 2002. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat 19: 81. [DOI] [PubMed] [Google Scholar]
- Takebayashi T, Higashi H, Sudo H, Ozawa H, Suzuki E, Shirado O, Katoh H, Hatakeyama M 2003. NF-kappa B-dependent induction of cyclin D1 by retinoblastoma protein (pRB) family proteins and tumor-derived pRB mutants. J Biol Chem 278: 14897–14905 [DOI] [PubMed] [Google Scholar]
- Talluri S, Isaac CE, Ahmad M, Henley SA, Francis SM, Martens AL, Bremner R, Dick FA 2010. A G1 checkpoint mediated by the retinoblastoma protein that is dispensable in terminal differentiation but essential for senescence. Mol Cell Biol 30: 948–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Song WX, Luo J, Haydon RC, He TC 2008. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res 466: 2114–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell 8: 303–316 [DOI] [PubMed] [Google Scholar]
- Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G 2011. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev 25: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Nath N, Minden A, Chellappan S 1999. Regulation of Rb and E2F by signal transduction cascades: divergent effects of JNK1 and p38 kinases. EMBO J 18: 1559–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Calhoun ES, Silliman N, Ptak J, Szabo S, Powell SM, Riggins GJ, Wang TL, Yan H, Gazdar A, et al. 2006. Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum Mutat 27: 1060–1061 [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Yabuta N, Fujii T, Copeland NG, Gilbert DJ, Jenkins NA, Nishiguchi H, Endo Y, Toji S, Tanaka H, Nishimune Y, et al. 2000. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics 63: 263–270 [DOI] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD 2007. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Hong W 2008. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13: 188–192 [DOI] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30 [DOI] [PubMed] [Google Scholar]
- Zhang R, Chen W, Adams PD 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2343–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]