SUMMARY
Poly-HAMP domains are widespread in bacterial chemoreceptors, but previous studies have focused on receptors with single HAMP domains. The Pseudomonas aeruginosa chemoreceptor, Aer-2, has an unusual domain architecture consisting of a PAS sensing domain sandwiched between three N-terminal and two C-terminal HAMP domains, followed by a conserved kinase control module. The structure of the N-terminal HAMP domains was recently solved, making Aer-2 the first protein with resolved poly-HAMP structure. The role of Aer-2 in P. aeruginosa is unclear, but here we show that Aer-2 can interact with the chemotaxis system of Escherichia coli to mediate repellent responses to oxygen, carbon monoxide and nitric oxide. Using this model system to investigate signaling and poly-HAMP function, we determined that the Aer-2 PAS domain binds penta-coordinated b-type heme and that reversible signaling requires four of the five HAMP domains. Deleting HAMP 2 and/or 3 resulted in a kinase-off phenotype, whereas deleting HAMP 4 and/or 5 resulted in a kinase-on phenotype. Overall, these data support a model in which ligand-bound Aer-2 PAS and HAMP 2 and 3 act together to relieve inhibition of the kinase control module by HAMP 4 and 5, resulting in the kinase-on state of the Aer-2 receptor.
Keywords: PAS, poly-HAMP, heme, chemoreceptor, signal transduction
INTRODUCTION
Pseudomonas aeruginosa is a ubiquitous bacterium whose complex lifestyle is facilitated by 26 chemoreceptors and four chemotaxis-like systems that include: the Pil-Chp system (gene Cluster IV), which is involved in pilus-mediated twitching (Darzins, 1994; Kearns et al., 2001); the Wsp system (Cluster III), which controls biofilm formation (D'Argenio et al., 2002; Hickman et al., 2005); the Che system (Clusters I and V), which is required for flagellar-mediated chemotaxis (Kato et al., 1999; Masduki et al., 1995); and the Che2 system (Cluster II), which has a largely unknown role (Ferrandez et al., 2002; Guvener et al., 2006; Hong et al., 2004). The Cluster II system (PA0173-PA0179) contains a set of genes (cheY2, A2, W2, R2, B2, D) that, with the exception of cheD, are homologous to the chemotaxis genes of E. coli (Guvener et al., 2006). In P. aeruginosa, these genes are expressed in stationary phase (Hong et al., 2005; Schuster et al., 2004), and their products form a cluster at the cell pole that is held together by a single Cluster II receptor called Aer-2 [(Guvener et al., 2006), Fig. 1, also known as McpB]. In 2004, Hong and colleagues reported that Aer-2, along with classical Aer (Aer-1), mediates aerotaxis by P. aeruginosa (Hong et al., 2004). However, other studies, including our own unpublished work, have found no role for the Cluster II proteins, including Aer-2, in aerotaxis or chemotaxis by P. aeruginosa (Ferrandez et al., 2002; Guvener et al., 2006). There is also little known about the function of Cluster II and Aer-2 orthologs [e.g., Vibrio cholerae VCA1092, or Shewanella oneidensis SO2123 (Baraquet et al., 2009)] in other systems.
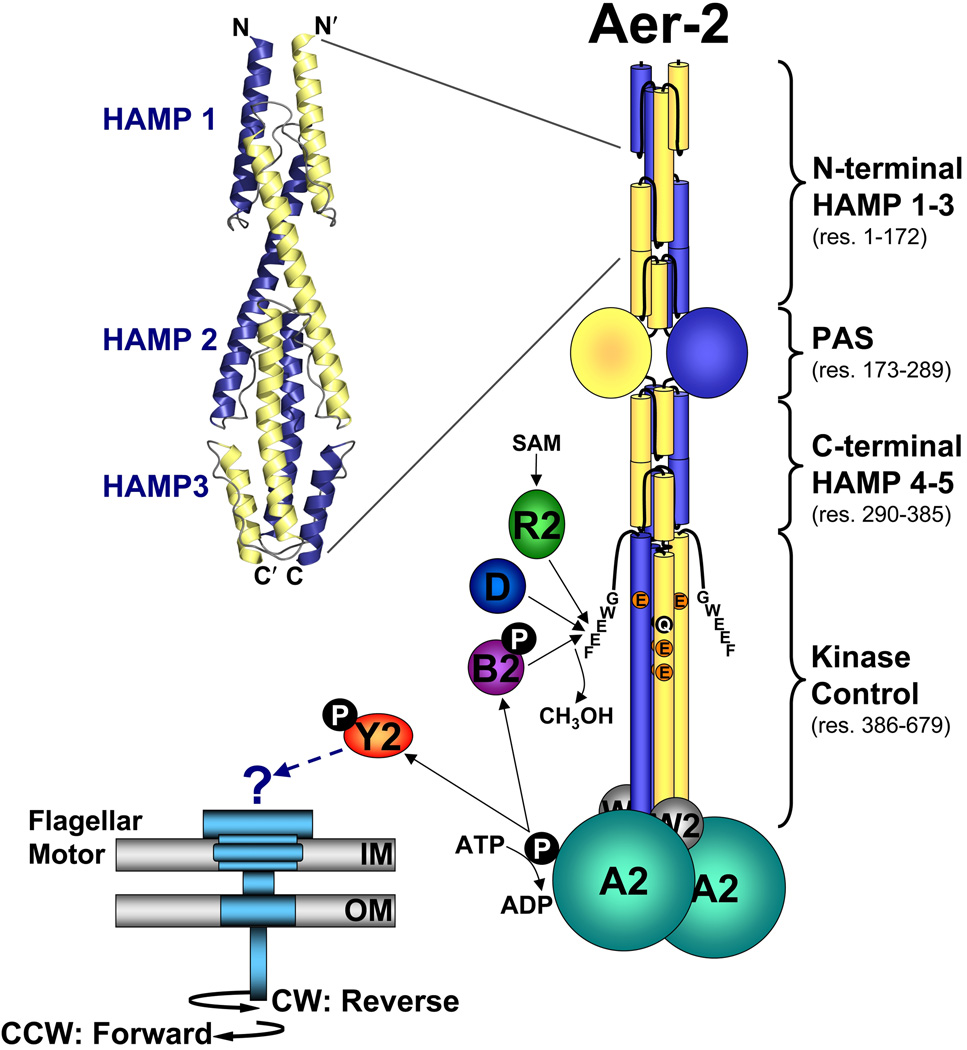
FIG. 1.
Cartoon showing the proposed domain organization of an Aer-2 dimer and the proteins associated with the Cluster II chemotaxis-like system. Each Aer-2 monomer contains three N-terminal HAMP domains, followed by a PAS sensing domain, two C-terminal HAMP domains, and a C-terminal kinase control domain. The structure of the three N-terminal HAMP domains was recently solved by x-ray crystallography [3LNR (Airola et al., 2010)]. By analogy to other methyl-accepting chemotaxis proteins, the kinase control domain is predicted to contain four methylation sites (QEEE, res. 414, 421, 428 and 610) and a C-terminal pentapeptide (GWEEF, res. 675–679) for binding the adaptation enzymes CheR2, CheB2 and CheD. Aer-2 is also predicted to form a ternary complex with the CheA2 and CheW2 proteins. The binding of an oxy-gas ligand to the Aer-2 PAS-heme domain is expected to alter the autophosphorylation rate of CheA2 and the subsequent transfer of phosphate to CheY2. It is currently unclear whether phospho-CheY2 binds to the flagellar motor or whether it interacts with a different response system. Abbreviations: A, W, Y, R, B, and D, Che proteins; SAM, S-adenosylmethionine; IM, inner membrane; OM, outer membrane; CW, clockwise; CCW, counterclockwise.
Aer-2 has an unusual architecture that includes three N-terminal HAMP domains, whose structure has been resolved (Airola et al., 2010), and two C-terminal HAMP domains (ExPASy: http://expasy.org/tools/scanprosite/, and S. Dunin-Horkawicz and A. Lupas, personal communication), separated by a PAS sensing domain (SMART: http://smart.embl-heidelberg.de/) (Fig. 1). These domains precede a kinase control module that is typical of chemosensory proteins (SMART: http://smart.embl-heidelberg.de/). Unlike many well-studied chemoreceptors (Hazelbauer et al., 2008), Aer-2 contains no predicted membrane-spanning segments (Hong et al., 2004).
PAS sensing domains comprise a superfamily of some 22,000 motifs that, in bacteria, sense light, oxygen, redox potential and energy [(Nambu et al., 1991; Taylor and Zhulin, 1999), SMART: http://smart.embl-heidelberg.de/]. The PAS domain of the E. coli aerotaxis receptor, Aer, senses changes in cellular redox potential via reduction of bound FAD (Bibikov et al., 1997; Rebbapragada et al., 1997). This is also the proposed role of the P. aeruginosa Aer-homolog, Aer-1 (Hong et al., 2004). In contrast, the Aer-2 PAS domain has only 18% sequence identity with Aer-1 PAS (Hong et al., 2004), and Aer-2 is missing key sequence features that are common to FAD-binding PAS domains, like that of Aer or the recently discovered AerC (Xie et al., 2010). The absence of these features suggests that the Aer-2 PAS domain does not bind FAD.
HAMP domains are important signal-conversion modules that communicate between signal-input and signal-output domains (Aravind and Ponting, 1999), and more than 12,500 have been identified by SMART. The first HAMP structure to be resolved was that of the Archaeoglobus fulgidus Af1503 protein (Hulko et al., 2006). This NMR-derived structure consists of a parallel four-helix bundle [made up of two helices from each monomer, called amphipathic sequence 1 and 2 (AS-1 and AS-2)] with an unusual knobs-to-knobs packing arrangement (Hulko et al., 2006). This arrangement has since been verified by biochemical and genetic studies of HAMP domains from several bacterial chemoreceptors (Swain and Falke, 2007; Watts et al., 2008; Zhou et al., 2009). Recently, the structures of the three N-terminal HAMP domains of Aer-2 were solved by crystallography (Airola et al., 2010). In this first poly-HAMP structure, the HAMP 1 domain is separated from HAMP 2 by a helical linker, whereas the HAMP 2 and 3 domains form an integrated, di-HAMP structure (Airola et al., 2010). HAMP domains 1 and 3 adopt conformations that are similar to the NMR structure of Af1503, although they lack knobs-to-knobs packing, whereas HAMP 2 forms a distinct trapezoidal four-helix bundle that may represent an alternative HAMP signaling state (Airola et al., 2010).
HAMP domains 4 and 5 of Aer-2 adjoin a kinase control domain, which, in chemoreceptors, interacts with a CheA histidine kinase and the CheW docking protein to form a stable ternary complex (Borkovich et al., 1989; Erbse and Falke, 2009; Gegner et al., 1992; McNally and Matsumura, 1991). This complex allows receptor-bound repellents and attractants to modulate the autophosphorylation rate of CheA (Borkovich and Simon, 1990; Falke and Hazelbauer, 2001), which in turn determines the level of transphosphorylated CheY (Swanson et al., 1993). Once phosphorylated, CheY binds to the flagellar motor and reverses the direction of flagella rotation from counterclockwise (CCW) to clockwise (CW) (Bren and Eisenbach, 1998; Welch et al., 1993). In P. aeruginosa, the Cluster I CheA, CheW and CheY proteins are involved in flagellar-mediated chemotaxis (Kato et al., 1999), but attempts to show similar involvement of the Cluster II orthologs has not been successful (Guvener et al., 2006). It is currently unclear whether the P. aeruginosa Cluster II system, and specifically CheY2, controls flagellar rotation or some other process (Fig. 1).
In this study we addressed the question of Aer-2 sensing and signaling. Although, we have not observed Aer-2-mediated swimming responses in P. aeruginosa, we found that Aer-2 can interact with and modulate the chemotaxis pathway of E. coli to mediate repellent responses to oxygen, carbon monoxide and nitric oxide. We show that the PAS sensing domain binds penta-coordinated b-type heme and is likely responsible for sensing these gases. In addition, we have begun to define the signaling roles of the N- and C-terminal poly-HAMP domains. Based on the data presented, we have developed a working model for Aer-2 signaling.
RESULTS
Historically, PAS/HAMP signaling has been studied in receptors that contain single PAS and HAMP domains (Campbell et al., 2010; Elliott and Dirita, 2008; Watts et al., 2004). The P. aeruginosa Aer-2 receptor provides an opportunity to study PAS/HAMP signaling in a receptor that contains one PAS and five HAMP domains (Fig. 1). Previously, Aer-2 was reported to be an aerotaxis receptor in P. aeruginosa (Hong et al., 2004), but we have observed no Aer-2-mediated behavioral responses when P. aeruginosa is assayed in a gas perfusion chamber (K. Watts and M. Gabra, unpublished data). Because attempts to identify Aer-2-mediated signals are complicated by the presence of four chemotaxis-like systems in P. aeruginosa, we decided to investigate Aer-2 signaling by its interaction with the single, well-characterized chemotaxis system of E. coli. This approach seemed reasonable because the kinase control domain of Aer-2 belongs to the same heptad-repeat class as that of the E. coli Tsr chemoreceptor [36 heptad repeats (Alexander and Zhulin, 2007)]. Moreover, the Aer-2 and Tsr kinase control domains share 61% sequence identity, have four putative methylation sites (QEEE in Aer-2 compared to QEQE for Tsr) and a C-terminal pentapeptide (GWEEF in Aer-2 compared to NWETF for Tsr) for binding adaptation enzymes (Fig. 1).
The Aer-2 receptor from P. aeruginosa activates the E. coli chemotaxis cascade
We cloned aer-2 (PAO176) from P. aeruginosa PAO1 DNA and expressed it from pProEX (renamed pLH1) in chemoreceptor-less E. coli BT3388 (tar, tsr, trg, tap, aer). In BT3388, Aer-2 was stable, and as previously predicted (Hong et al., 2004), soluble, because full-length Aer-2 (76.5 kDa, including the His-tag) partitioned primarily into the supernatant fraction from lysed cells that were centrifuged at low (10,000 × g) and then high (485,000 × g) speed.
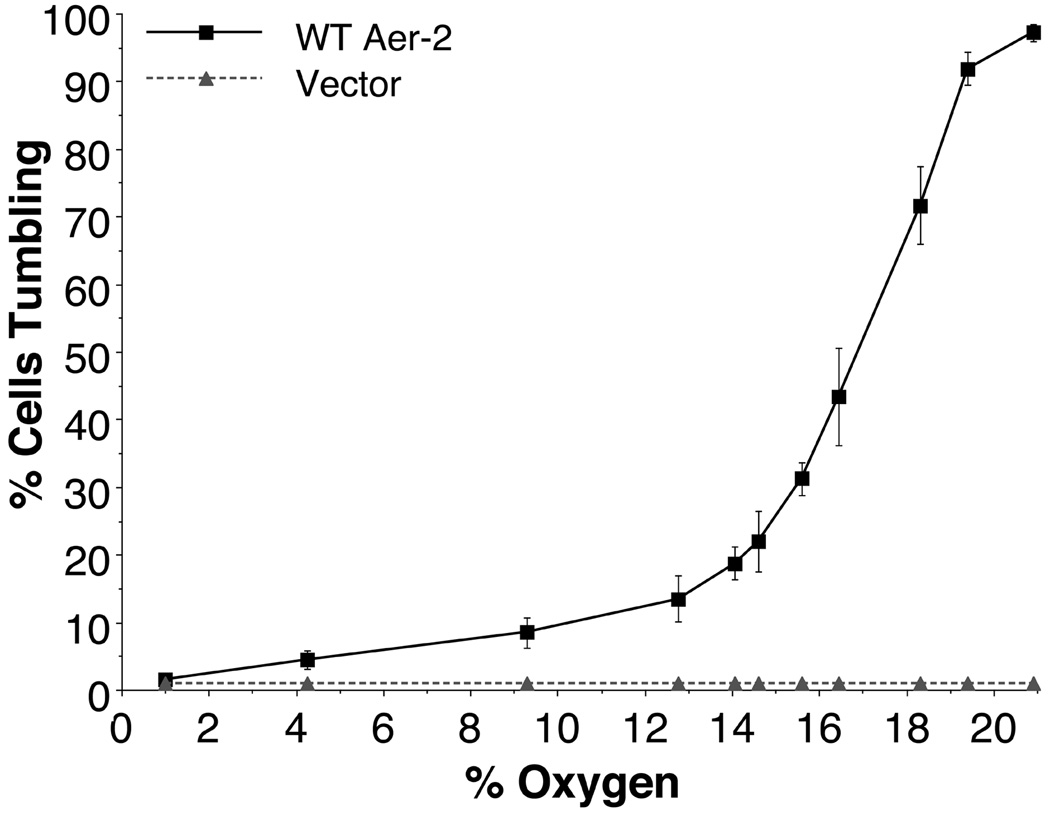
To test whether Aer-2 could mediate responses to oxygen (O2) in E. coli, we expressed Aer-2 from pLH1 (with 0 to 1000 µM IPTG) in BT3388 and monitored the swimming behavior in a gas perfusion chamber (Taylor et al., 2007). At induction levels of 15 µM IPTG and above, cells expressing Aer-2 tumbled constantly in air (20.9%, or 250 µM O2, Fig. 2) and swam smoothly when O2 was replaced by nitrogen (N2), unlike control cells, which contained the pProEX vector and did not respond to changes in O2 concentration (cells remained smooth-swimming, Fig. 2). The Aer-2-mediated phenotype was reversible when the gas mixture was toggled between O2 and N2, demonstrating that Aer-2 can reversibly activate the E. coli chemotaxis cascade. Notably, this response was opposite to the classic Aer-mediated aerotaxis response in which E. coli cells tumble in response to a drop in O2 concentration (Rebbapragada et al., 1997). To estimate the concentration of O2 required to maintain Aer-2 in an activated state, we expressed Aer-2 at levels approximately equal to the total number of chemoreceptors in wild-type (WT) E. coli (200 µM IPTG) and monitored the behavior of BT3388/Aer-2 cells as a function of O2 concentration. As the O2 concentration was decreased stepwise, there was a progressive drop in the fraction of tumbling cells, reaching 50% at 17% (203 µM) O2 (Fig. 2). At each O2 concentration, the cells were viewed for approximately 1 min, and during that time the percentage of tumbling cells remained constant, with no evidence of adaptation.
FIG. 2.
Steady-state behavior of receptor-less E. coli BT3388 cells expressing WT Aer-2 or vector alone, at decremental O2 concentrations between 20.9% and 1%. WT Aer-2 (pLH1) and the vector control (pProEX) were induced with 200 µM IPTG. Error bars represent the standard deviation from multiple titration experiments.
Aer-2 does not adapt to O2 signals in E. coli
As BT3388 cells expressing Aer-2 did not appear to adapt at any level of O2 (Fig. 2), temporal assays were repeated in BT3312, a strain that contains the E. coli high-abundance chemoreceptor Tar. Tar assists the adaptation of E. coli receptors lacking the canonical CheR/B binding pentapeptide by forming “assistance neighborhoods” with these receptors (Li and Hazelbauer, 2005). However, when Aer-2 was expressed in BT3312, the cells likewise did not adapt (data not shown). Aer-2 can therefore activate the E. coli chemotaxis system, but it may not form assistance neighborhoods with Tar nor interact effectively with E. coli CheR and/or CheB.
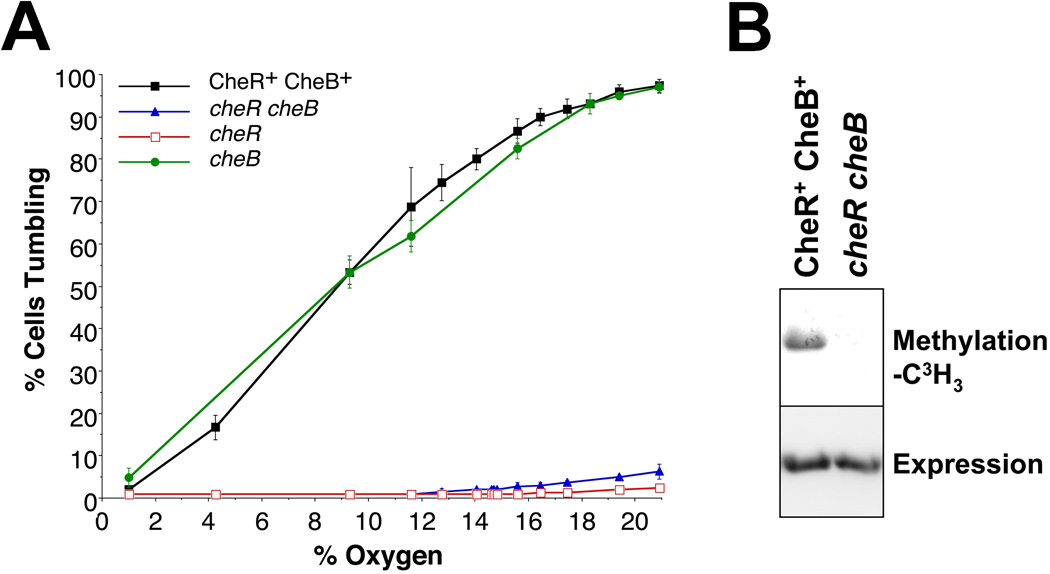
In E. coli, the adaptation enzymes CheR and CheB, respectively, methylate and deamidate/demethylate specific glutamyl residues on chemoreceptors and thereby reset the signal bias of the cell [reviewed by (Hazelbauer et al., 2008)]. Thus, Tsr mediates smooth swimming in a cheR strain because glutaminyl and glutamyl residues involved in adaptation are deamidated but not methylated, and it mediates constant tumbling in a cheB strain because all four glutamyl residues are amidated or methylated (Parkinson and Houts, 1982). In an adaptation minus cheR cheB strain, Tsr also generates a strong tumbling bias because the two adaptation glutamine residues are not deamidated by CheB, and these residues mimic methyl glutamates (Parkinson and Houts, 1982; Sherris and Parkinson, 1981). To examine Aer-2-mediated behavior in the presence and absence of CheR and CheB, we obtained an isogenic set of receptor-less E. coli adaptation mutants (kindly provided by J.S. Parkinson). The parental CheR+ CheB+ strain, UU2612 (tar, tsr, trg, tap, aer) has a similar chemotaxis genotype to BT3388 (tar, tsr, trg, tap, aer), but the Aer-2-mediated tumbling response in UU2612 had a lower threshold for O2, and the response curve was shifted to the left (compare the solid black curves in Figs. 2 and 3A). The reasons for this are currently unclear, because the steady-state accumulated levels of Aer-2 were similar in these and the other isogenic adaptation mutants (data not shown). Nonetheless, in both BT3388 and UU2612, Aer-2 induced a strong tumbling response at high O2 concentrations, and a smooth swimming response in the absence of O2. In contrast, the Aer-2-mediated response to O2 was greatly diminished in strains lacking CheR [cheR (UU2611: tar, tsr, trg, tap, aer, cheR) and cheR cheB (UU2610: tar, tsr, trg, tap, aer, cheR, cheB), Fig. 3A]. This result suggested that Aer-2 is methylated by E. coli CheR. This was confirmed by blocking protein synthesis with chloramphenicol, incubating cells with 3H-methionine, and visualizing methylated proteins after SDS-PAGE. As shown in Fig. 3B, Aer-2 was labeled (methylated) in the CheR+ CheB+ strain (UU2612), but not in the cheR cheB strain (UU2610).
FIG. 3.
Aer-2-mediated behavior in E. coli in the presence or absence of the E. coli adaptation enzymes CheR and CheB. A. Steady-state behavior of isogenic E. coli strains (CheR+ CheB+ strain UU2612, cheR cheB strain UU2610, cheR strain UU2611, and cheB strain UU2632) expressing WT Aer-2 (with 200 µM IPTG) at O2 concentrations between 20.9% and 1%. Error bars represent the standard deviation from multiple titration experiments. B. Methylation of Aer-2 in the CheR+ CheB+ strain UU2612 and the absence of methylation in the cheR cheB strain UU2610 (upper panel), at similar levels of Aer-2 accumulation (lower panel, HisProbe Western blot).
To examine the influence of CheB on Aer-2-mediated responses, we expressed Aer-2 in a chemoreceptor-less E. coli cheB strain (UU2632: tar, tsr, trg, tap, aer, cheB). In air, cheB cells expressing Aer-2 had a high tumbling bias and the O2 response profile mirrored that of CheR+ CheB+ cells (Fig. 3A). Thus, CheB, unlike CheR, appeared to have a minimal influence on Aer-2 signaling in E. coli. However, cheB cells expressing Aer-2 also showed a significantly delayed smooth-swimming response when the gas in the chamber was switched directly from 20.9% to 0% O2 (data not shown), suggesting that Aer-2 may be deamidated and/or demethylated by E. coli CheB, although not very efficiently. Overall, the Aer-2-mediated tumbling response to O2 was dependent on the extent of receptor methylation, but the direction of the response was the same whether or not Aer-2 was methylated (Fig. 3A). This result indicates that the Aer-2 response was not inverted due to diminished CheB activity, as was previously observed for Tsr-mediated aerotaxis in E. coli (Dang et al., 1986).
The Aer-2 PAS domain binds b-type heme
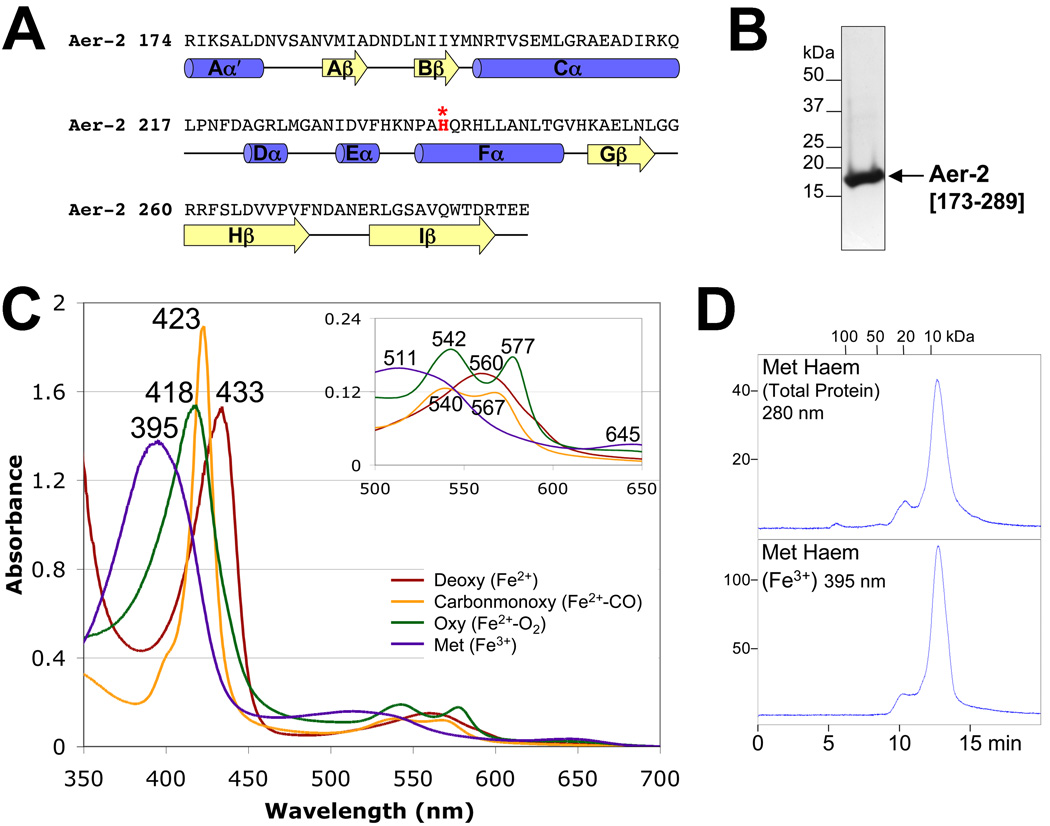
The E. coli aerotaxis receptor Aer senses O2 indirectly via an FAD-binding PAS domain (Bibikov et al., 1997; Rebbapragada et al., 1997). Because Aer-2 also mediates a response to O2, we examined the possibility that the Aer-2 PAS domain (Fig. 4A) binds a co-factor, such as flavin or heme. To do this, we over-expressed a His6-tagged PAS domain (res. 173–289), purified the peptide to apparent homogeneity on a Ni-NTA column (Qiagen) (Fig. 4B), and scanned the UV/Vis spectrum for cofactors. The absorption spectra of the Aer-2 PAS domain were characteristic of heme-binding proteins (e.g., oxy Aer-2 had γ-, β- and α-maxima at 418 nm, 542 nm and 577 nm, respectively, Fig. 4C). Spectra were similar for full-length Aer-2 (res. 1–679), but unlike the PAS fragment, purification to apparent homogeneity was not possible because full-length Aer-2 readily aggregated in solution. In contrast to full-length Aer-2 and the isolated PAS domain, a C-terminal HAMP-kinase control domain fragment (res. 330–432) showed no evidence of cofactor binding (data not shown). Of published PAS-heme spectra, the Aer-2 PAS spectra were most similar to the spectra of the direct O2 sensor FixL from Bradyrhizobium japonicum (Gilles-Gonzalez et al., 1994). Deoxy Aer-2, like BjFixL, had a red-shifted Soret peak and a single broad band replacing the α/β-bands of oxy Aer-2 (Fig. 4C), which is typical of penta-coordinated b-type heme. Although the spectra of the Aer-2 PAS domain were similar to those of other PAS-heme domains, the primary sequence of Aer-2 does not easily align with the sequences of other heme-containing PAS domains, e.g., Escherichia coli DOS (EcDOS) (Delgado-Nixon et al., 2000), BjFixL (Gong et al., 1998), or Burkholderia xenovorans RcoM-2 (Marvin et al., 2008). This difference is partly due to an unusually long Cα helix predicted for Aer-2 (Fig. 4A). Nonetheless, Aer-2 does contain a histidine residue in the PAS Fα3 position (H239, Fig. 4A, highlighted red), which coordinates heme binding in the structures of DOS and FixL (Gong et al., 1998; Kurokawa et al., 2004; Park et al., 2004). Whether H239 coordinates heme in Aer-2 is unclear, and is currently under investigation.
FIG. 4.
Aer-2 PAS secondary structure, spectra and oligomeric state. A. Sequence and secondary structure of the Aer-2 PAS domain as predicted by PSIPRED (http://bioinf4.cs.ucl.ac.uk:3000/psipred) and a PAS sequence alignment created by L. Ulrich and I. Zhulin (personal communication). α-Helices and β-strands are shown as blue cylinders and yellow arrows, respectively. Aer-2 PAS contains a histidine in the Fα3 position (highlighted red), which coordinates heme in the structures of DOS and FixL. B. Coomassie-stained SDS-PAGE gel of purified Aer-2 PAS protein (res. 173–289, 16.3 kDa). C. Absorption spectra of purified Aer-2 PAS protein in the reduced (deoxy, dark red line), oxidized (met, purple line), carbonmonoxide-bound (carbonmonoxy, orange line) and oxygen-bound (oxy, green line) states. The absorbance maximum at each peak is indicated. The insert shows an expanded view of peaks between 500 and 650 nm. D. Elution profile of isolated Aer-2 PAS protein in its met-heme state during size-exclusion chromatography. The elution profile is shown in arbitrary units at 280 nm to reveal total protein content (top panel) and at 395 nm to detect the elution of met-heme (bottom panel). Fractions were removed and analyzed by Western blotting; in all cases, the Aer-2 PAS protein co-eluted with the heme (not shown).
Because PAS domains, including DOS and FixL, commonly form dimers in solution (Rodgers et al., 2001; Sasakura et al., 2002; Zhong, 2003), we employed size-exclusion chromatography to estimate the apparent molecular weight, and hence the oligomeric state of the non-denatured Aer-2 PAS domain (res. 173–289, 16.3 kDa). The Aer-2 PAS domain eluted from a size-exclusion column as a compact monomer in both its met (Fe3+)- and deoxy (Fe2+)-heme states (<10 kDa apparent molecular weight, TSK G2000 SW column, Fig. 4D, met form shown). The isolated Aer-2 PAS domain therefore appears to be monomeric, at least in the absence of ligands. In contrast, full-length Aer-2, like other chemoreceptors, is expected to dimerize via its HAMP and kinase control domains (Fig. 1).
Aer-2 can signal in response to O2, CO and NO
In addition to binding O2, heme-binding PAS proteins often bind and respond to the diatomic oxy-gases carbon monoxide (CO) and nitric oxide (NO) (Gilles-Gonzalez and Gonzalez, 2005). To investigate whether Aer-2 can respond to CO, we introduced CO gas into the perfusion chamber under anaerobic conditions (N2 perfusion) and monitored the swimming response of E. coli BT3388 cells expressing Aer-2 (after induction with 200 µM IPTG). Cells expressing Aer-2 tumbled instantly in response to CO, unlike cells containing the vector alone, which did not respond to CO. After CO was removed, Aer-2/BT3388 cells continued tumbling for approximately 30 s, then resumed smooth swimming. In comparison, cells expressing Aer-2 at the same induction level tumbled for approximately 10 s after O2 was replaced by N2, presumably because O2 dissociated more rapidly than CO.
To test for an NO response, we employed the NO donor Proli NONOate (Cayman Chemical). Two microliters of anaerobic 500 µM Proli NONOate were mixed with 7 µl of anaerobic E. coli BT3388/Aer-2 cells in the gas perfusion chamber. The cells tumbled almost instantly in response to NO release, suggesting that Aer-2 was activated. Aer-2 did not respond to 500 µM of anaerobic proline (a dissociation product of Proli NONOate), and 10 mM NaOH (used to solubilize Proli NONate) caused no immediate response, although NaOH did invoke a delayed tumbling response after several minutes. BT3388 cells containing pProEX did not respond to Proli NONOate or NaOH. These results indicate that, in E. coli, Aer-2 is able to sense and signal in response to O2, CO and NO, mediating repellent responses to all three gases.
N-terminal HAMP domains 2 and 3 are required for Aer-2 signaling
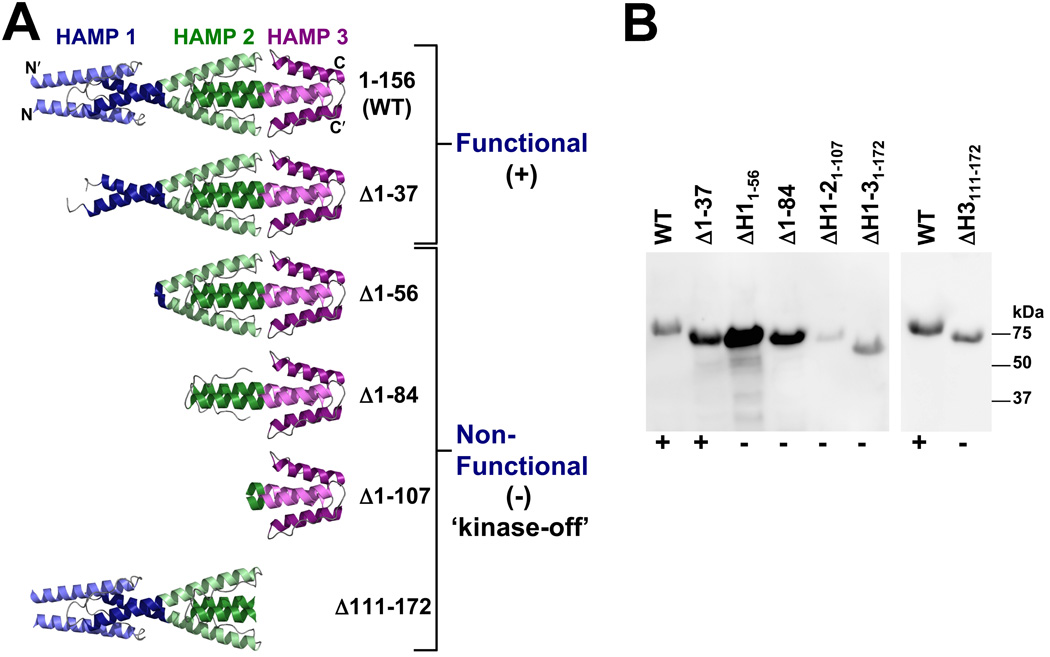
The region N-terminal to the Aer-2 PAS domain (res. 1–172) is a poly-HAMP chain, the structure of which was recently solved by crystallography [(Airola et al., 2010), Fig. 5A]. In this structure, HAMP 1 is separated from HAMP 2 by a helical linker, whereas HAMP 2 and 3 form a concatenated di-HAMP structure. To determine whether these HAMP domains influence the PAS signaling response, we created an N-terminal Δ1–172 truncation mutant lacking HAMP domains 1–3. In E. coli BT3388, this mutant expressed stable protein (Fig. 5B) but no longer responded to O2, CO or NO (i.e., smooth swimming persisted), indicating that the receptor was locked in the kinase-off state. To determine the influence of individual N-terminal HAMP domains, we constructed a series of HAMP truncation mutants (Fig. 5). A mutant that lacked HAMP 1 AS-1 and most of the HAMP 1 connector (Δ1–37, Fig. 5) behaved like WT Aer-2 in a temporal O2 assay, except that it generated a slightly lower tumbling bias at higher O2 concentrations (approx. 75% of cells tumbled in air versus 98% for WT). Thus, the integrity of HAMP 1 is not necessary for reversible signaling in Aer-2. In contrast, deletion mutants lacking the entire HAMP 1 domain (Δ1–56), HAMP 1 and part of HAMP 2 (Δ1–84), HAMP 1 and 2 (Δ1–107), or the HAMP 3 domain (Δ111–172) were all unresponsive to O2 and locked in the kinase-off state, even though these proteins were, for the most part, stably expressed (Fig. 5B). Residues 38–56 (encompassing HAMP 1 AS-2 and the HAMP 1–2 helical linker) therefore include a sequence that is necessary for reversible signaling by Aer-2. These residues may support the structural integrity of HAMP 2.
FIG. 5.
Influence of N-terminal HAMP domains 1, 2 and 3 on Aer-2 signaling. A. Structure of the N-terminal HAMP domains (Airola et al., 2010) and models of the truncation mutants that were tested for function in behavioral assays. The Δ1–37 mutant was the only truncated product that retained Aer-2 function in E. coli (+). The other truncation mutants all resulted in kinase-off phenotypes (−). See text for details. B. HisProbe Western blot showing size differences among the truncated Aer-2 products as well as variations in their steady-state accumulation levels in E. coli BT3388 after induction with 50 µM IPTG. Abbreviation: ΔH, ΔHAMP.
The N- and C-terminal HAMP domains have different roles
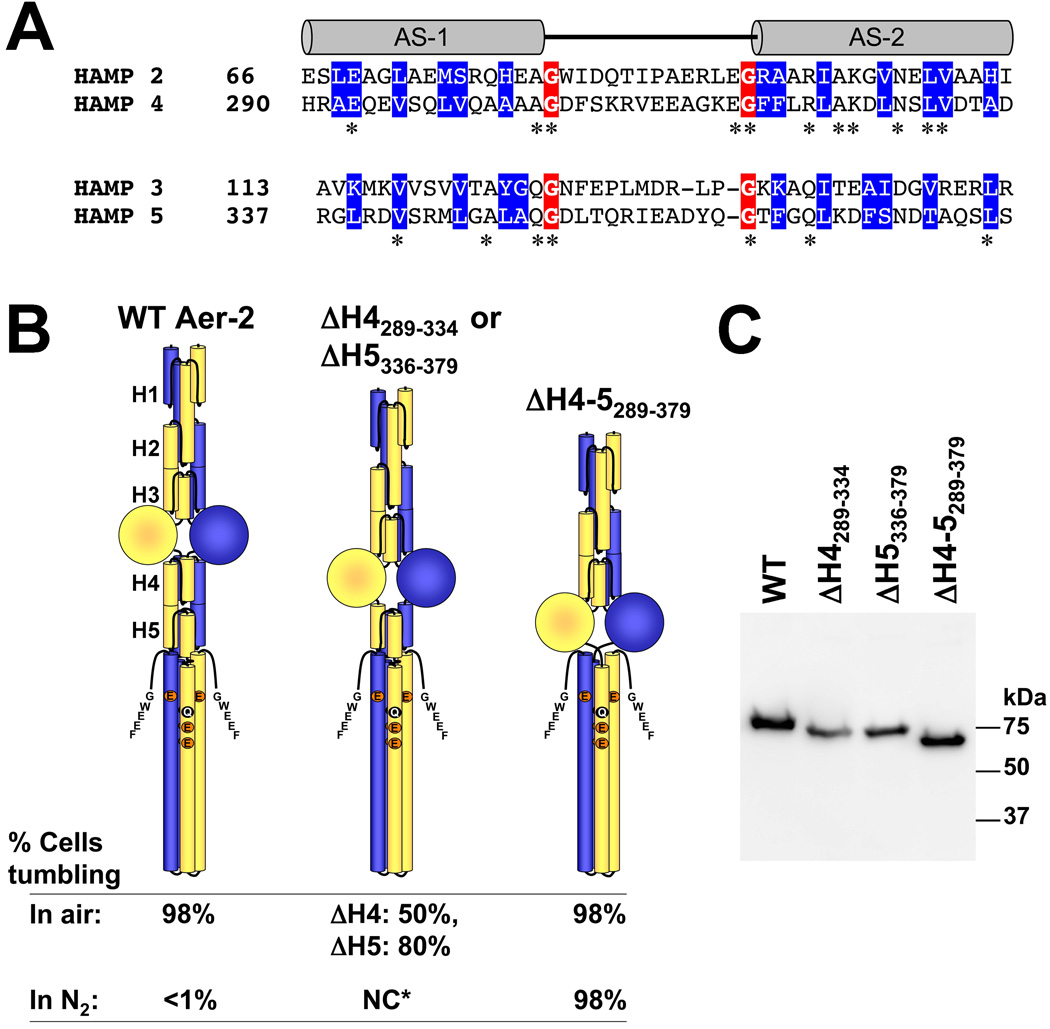
HAMP domains 4 and 5 lie between the PAS and kinase control domains of Aer-2 (Fig. 1) in a position analogous to the single HAMP domains in canonical chemoreceptors [see (Ma et al., 2005; Parkinson, 2010)]. The sequence of HAMP 4–5 aligns with that of HAMP 2–3 (Fig. 6A), suggesting that HAMP 4–5 may be an integrated di-HAMP unit similar to HAMP 2–3. Specifically, the two di-HAMP units share conserved, buried hydrophobic residues (based on the structure of HAMP 2–3), glycine residues at both ends of the AS-1-AS-2 connectors, and sequence stutters between the two HAMP domains of the di-HAMP unit (Airola et al., 2010). However, unlike the HAMP 2 and 3 deletions, which resulted in kinase-off receptors (Fig. 5), removing both HAMP 4 and 5 (Δ289–379) resulted in kinase-on receptors that were unresponsive to changes in O2 concentration (Fig. 6). Thus, HAMP 2 and 3 appear to promote a kinase-on phenotype, whereas HAMP 4 and 5 promote a kinase-off phenotype. Removing either HAMP 4 (Δ289–334) or HAMP 5 (Δ336–379) resulted in intermediate steady-state tumbling in air and N2 (approx. 50% and 80% respectively, Fig. 6B), indicating that the inhibitory effects of HAMP 4 and 5 are additive.
FIG. 6.
Influence of C-terminal HAMP domains 4 and 5 on Aer-2 signaling. A. Sequence alignment of di-HAMP 2–3 and di-HAMP 4–5. Connector-flanking glycine residues are shaded red, whereas buried core residues (based on the resolved structure of HAMP 2–3) are shaded blue. Stars indicate identical N- and C-terminal HAMP residues. B. HAMP 4 and 5 are required to maintain the kinase-off state. Aer-2 deleted for HAMP 4 and 5 (far right) exhibited maximal kinase-on activity in air (tumbling in E. coli BT3388), like WT (far left). However, unlike WT, the ΔHAMP 4–5 mutant remained kinase-on (no change, NC) in N2 (in the absence of a diatomic oxy-gas). Mutant proteins lacking either HAMP 4 or HAMP 5 exhibited intermediate kinase-on activities (center figure). The HAMP 5 deletion began at residue 336 because residue 335 forms part of HAMP 4 and HAMP 5. C. HisProbe Western blot showing size differences among the internally truncated Aer-2 products and variations in their steady-state accumulation levels in E. coli BT3388 after induction with 50 µM IPTG. Abbreviations: AS, amphipathic sequence; H, HAMP; NC*, no change for either mutant.
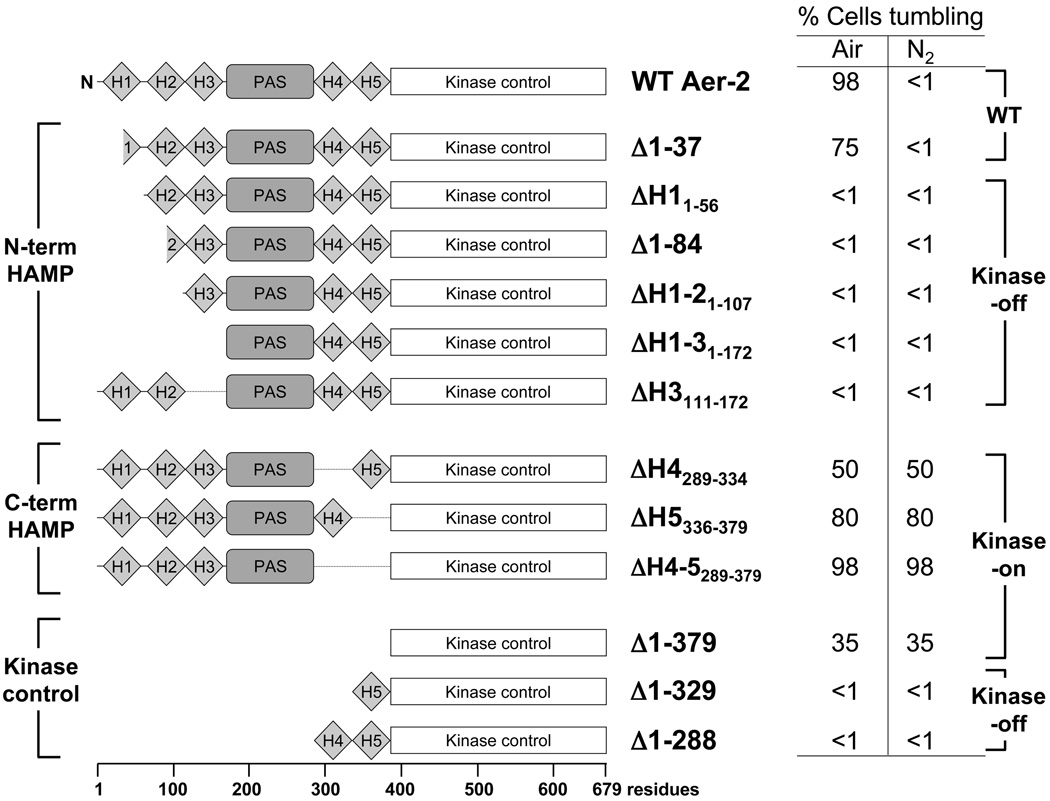
We next assayed the signal outputs of fragments of the C-terminal kinase control module, both with and without HAMP 4 or 5. By itself, the isolated kinase control module (Aer-2[380–679]) had a kinase-on bias and caused approximately 35% of cells to tumble in air and in N2 (Fig. 7). When the kinase control module was extended to include HAMP 5 (Aer-2[330–679]), or HAMP 4 and 5 (Aer-2[289–679]), tumbling was suppressed, and the fragments were locked in a kinase-off conformation (Fig. 7). This result again suggests that HAMP 4 and 5 work together to inhibit the output of the kinase control module.
FIG. 7.
Summary of Aer-2 deletions and the steady-state behavior mediated by these fragments in E. coli BT3388. Cells expressing WT Aer-2 (res. 1–679) or the Δ1–37 Aer-2 mutant tumbled in air and swam smoothly (<1% tumbling) in N2 (in the absence of a diatomic oxy-gas). Removing the N-terminal HAMP 2 and/or HAMP 3 domains resulted in a locked kinase-off phenotype, whereas removing C-terminal HAMP 4 and/or HAMP 5 resulted in a kinase-on phenotype. The default state of the isolated kinase control module was on, but it reverted to a kinase-off state in the presence of HAMP 5 or HAMP 4 and 5. All cells were induced with 200 µM IPTG for 45 min before monitoring their swimming behavior in a gas perfusion chamber. Mutants exhibiting less than 1% tumbling in air showed the same response after induction with 1 mM IPTG. Abbreviation: H, HAMP.
DISCUSSION
In this study, we determined that the Aer-2 receptor from P. aeruginosa can interact with the chemotaxis system of E. coli to direct repellent responses to O2, CO and NO. Thus, E. coli is a suitable model system for investigating sensory transduction by Aer-2. Aer-2 responses were presumably initiated by the PAS domain, which bound heme, but also required the N- and C-terminal poly-HAMP regions to modulate the signaling state of the kinase control module.
Inefficient adaptation of Aer-2 in E. coli
E. coli cells expressing Aer-2 as the sole receptor generated robust behavioral responses to O2, CO and NO, although the cells did not adapt to the stimuli and return to a pre-stimulus swimming bias. This inability to adapt was probably not due to inefficient CheR methyltransferase activity, as CheR was able to methylate Aer-2 and significantly alter behavioral responses (Fig. 3). In contrast, the presence or absence of the CheB methylesterase had little effect on Aer-2-mediated responses (Fig. 3), suggesting that CheB may not efficiently deamidate and/or demethylate Aer-2 in E. coli. Low CheB activity could be due in part to the unusual ‘GWEEF’ pentapeptide at the C-terminus of Aer-2 (Fig. 1). This sequence differs from that of the high-abundance E. coli chemoreceptors, which have an ‘NWETF’ pentapeptide sequence that is essential for maximal CheB binding and activity (Lai et al., 2006). Ineffective adaptation in E. coli may be compounded by the absence of CheD, which is normally expressed with Aer-2 in P. aeruginosa (Fig. 1). In other systems, CheD deamidates, and sometimes demethylates, chemoreceptors (Chao et al., 2006; Kristich and Ordal, 2002). Thus, it is possible that Aer-2 would adapt in E. coli if it was co-expressed with the P. aeruginosa Cluster II adaptation enzymes CheB2 and CheD.
The PAS-heme domain of Aer-2
When compared to other PAS-heme domains, the UV/Vis spectra of the Aer-2 PAS domain were most similar to spectra previously reported for the PAS domain of FixL (Gilles-Gonzalez et al., 1994), indicating that Aer-2, like FixL, binds penta-coordinated b-type heme. The PAS domains of FixL and DOS are both physiologic O2 sensors (Gilles-Gonzalez and Gonzalez, 2005), although several PAS-heme CO sensors have also been reported, including the human neuronal PAS domain protein 2 (NPAS2) (Dioum et al., 2002) and the PAS protein Regulator of CO Metabolism (RcoM) from B. xenovorans (Kerby et al., 2008). PAS-heme sensors often bind O2, CO and NO, but unlike Aer-2, which responded to all three gases, they typically respond to only one. Thus, FixL binds more tightly to CO and NO than to O2, but only responds to O2 (Dunham et al., 2003; Gilles-Gonzalez et al., 1994). To resolve the physiologically significant ligand of Aer-2, it will be helpful to determine the binding affinity between reduced heme and O2, CO, NO (and possibly other small ligands). These affinities can then be correlated with ligand availability in the P. aeruginosa environment during stationary phase, and compared with the level of activation at those ligand concentrations.
The roles of the two poly-HAMP regions in Aer-2
The unusual domain arrangement of Aer-2 (Fig. 1) differs from most well-studied chemoreceptors, which contain a single HAMP domain immediately following a membrane-spanning segment (Hazelbauer et al., 2008). Such canonical HAMP domains invariably contain an N-terminal ‘P[…]DExG’ capping motif that has been associated with transmembrane signaling (Dunin-Horkawicz and Lupas, 2010). All five HAMP domains of Aer-2 lack this capping motif and are considered to be divergent and characteristic of the HAMP domains found in poly-HAMP units (Dunin-Horkawicz and Lupas, 2010). We investigated the functional contribution of each of the poly-HAMP regions by deleting specific HAMP domains and gauging their effects on the behavior of Aer-2 in E. coli (Fig. 7).
In the absence of ligand, the default state of full-length Aer-2 in E. coli was kinase-off. Aer-2 was poised to respond to changes in the concentration of diatomic ligands, and was activated to the kinase-on state when cells were aerobic, or when they were exposed to CO or NO. HAMP deletions had diverse effects on the steady-state tumbling frequency, and provided some clues to the different roles of the proximal and distal HAMP domains. Although the integrity of HAMP 1 was unnecessary for Aer-2 function, Aer-2 peptides lacking either HAMP 2 and/or HAMP 3 did not respond to diatomic ligands and were locked in the kinase-off state (Fig. 7). This result suggests that HAMP 2 and 3 together promote a kinase-on state. In contrast, when the HAMP 4 and 5 domains were deleted from Aer-2, the cells had a tumbling phenotype with additive effects from the removal of HAMP 4 and HAMP 5. Thus, these Aer-2 peptides were locked in a kinase-on conformation, suggesting that HAMP 4 and HAMP 5 promote a kinase-off state, with HAMP 5 having a greater effect than HAMP 4 (Fig. 7). These results indicate that four of the five HAMP domains are required for reversible Aer-2 signaling, and that the proximal and distal di-HAMP units influence Aer-2 signaling in opposing ways.
Because deletions of HAMP 4 and 5 resulted in kinase-on phenotypes (Fig. 7), the HAMP 4–5 unit may play a similar role to the single HAMP domains of canonical chemoreceptors, whose function is to override the default kinase-on state of the kinase control module (Parkinson, 2010; Swain et al., 2009; Zhou et al., 2009). We found that when the C-terminal kinase control fragment of Aer-2 was expressed in E. coli, the cells had a tumbling bias, indicating that the default state of this region is kinase-on (Fig. 7). However, when the C-terminal fragment was extended to include HAMP 5, or HAMP 4 and 5, tumbling was suppressed and the fragments were locked in a kinase-off conformation. Notably, the PAS domain alone could not override the influence of HAMP 4–5 on the kinase control module, but required HAMP 2 and 3. Thus, the HAMP 4–5 unit imposes restraints on the signaling state, and ligand-bound PAS/HAMP 2–3 is required to override these restraints. Overall, these data suggest a model in which Aer-2 PAS, its ligand, and the N-terminal HAMP 2–3 domains act together to relieve HAMP 4–5 inhibition of the kinase control module, resulting in the kinase-on state of the receptor. Whether the PAS domain physically interacts with both of the di-HAMP units, or whether the two di-HAMP units signal in a concatenated unit, remains to be determined. The latter possibility is supported by the results of one recent study, which suggested that alternating poly-HAMP interactions might control the signaling state of a yeast histidine kinase (Meena et al., 2010).
The role of Aer-2 in P. aeruginosa
Conformational changes within Aer-2 are expected to be similar whether Aer-2 is expressed in E. coli or P. aeruginosa, but the downstream responses are apparently different. Aer-2 was originally reported as an aerotaxis receptor (Hong et al., 2004), but we (K. Watts and M. Gabra, unpublished) and others (Ferrandez et al., 2002; Garvis et al., 2009; Guvener et al., 2006) have observed no Aer-2- or Cluster II-mediated swimming responses in P. aeruginosa. The original report of Aer-2-mediated aerotaxis was based on a well-chamber assay, in which Aer-2 directed the migration of cells from an anaerobic to an aerobic chamber. However, this response did not require other Cluster II products (Hong et al., 2004), and was in fact opposite to the direction of the response we observed in E. coli, which was air-avoidance, rather than air-seeking. These differences are difficult to reconcile, unless the output of the Cluster II system is different from the E. coli chemotaxis system. In P. aeruginosa, Aer-2 and Cluster II are expressed in stationary phase cells by the alternative σ factor RpoS, which also regulates the expression of quorum-sensing and virulence genes (Hong et al., 2005; Schuster et al., 2004). This finding suggests that Aer-2 and Cluster II might have an important role during infection, a notion that was supported by a recent study showing that CheB2 from Cluster II is required for P. aeruginosa infection in a mouse-lung model (Garvis et al., 2009). Because Aer-2 is thought to be the principal, if not the only chemoreceptor associated with the Cluster II proteins (Guvener et al., 2006), it is possible that Aer-2 is a principal target of CheB2 and does not adapt in a cheB2 mutant. Clearly, to determine the real role of Aer-2, it may be important to consider which ligands are sensed during infection.
In summary, the role of Aer-2 and Cluster II in P. aeruginosa remains unclear, but by using an E. coli model system, we found that Aer-2 can signal in response to various diatomic oxy-gases. The PAS-heme domain likely initiates these responses, although signaling also required the two poly-HAMP regions that sandwich the PAS domain and control the activity of the kinase control module. In E. coli, Aer-2 is stable, soluble, and amenable to structural and functional analyses, and this system thus provides an excellent tool to study PAS/HAMP and poly-HAMP signaling mechanisms.
EXPERIMENTAL PROCEDURES
Bacterial Strains
E. coli BT3312 [tsr, aer (Repik et al., 2000)] and chemoreceptor-less BT3388 [tar, tsr, trg, tap, aer (Yu et al., 2002)] were used for most Aer-2 expression and behavioral assays. For methylation experiments, an isogenic set of chemoreceptor-less E. coli adaptation mutants were kindly provided by J.S. Parkinson and consisted of a CheR+ CheB+ strain, UU2612 (tar, tsr, trg, tap, aer), a ΔcheRB strain, UU2610 (tar, tsr, trg, tap, aer, cheR, cheB), a ΔcheB strain, UU2632 (tar, tsr, trg, tap, aer, cheB), and a ΔcheR strain, UU2611 (tar, tsr, trg, tap, aer, cheR).
Cloning and Mutagenesis
The aer-2 gene (PAO176) was amplified from PAO1 DNA and cloned into the NcoI and SalI sites of pProEXHTa (Invitrogen, Carlsbad, CA) to create pLH1, which expresses Aer-2 attached to an N-terminal His6 tag. To create N-terminal truncations, DNA fragments were PCR-amplified from pLH1 with PfuUltra (Agilent Technologies, Santa Clara, CA), and then ligated into the NcoI and SalI sites of pProEX. To create internal Aer-2 deletions, the entire pLH1 plasmid, except for the coding segment to be removed, was amplified. The resulting PCR product was treated with Dpn1 (New England Biolabs, Ipswich, MA) to remove template strands, and then the ends were phosphorylated and ligated to recircularize the plasmid. All new plasmids were introduced into E. coli BT3388 and Aer-2 expression was induced with 200 µM IPTG. Products of the correct size were confirmed by Western blotting with HisProbe™-HRP (Thermo Scientific, Rockford, IL). All genes and mutations were confirmed by sequencing the entire coding sequence of aer-2.
Monitoring Cellular Aer-2 Accumulation
Aer-2 was expressed in E. coli from pLH1 and pLH1-based plasmids with 0 to 1000 µM IPTG induction. Cellular accumulation was analyzed by Western blotting with HisProbe, except when estimating the induction necessary to match the copy number of native E. coli chemoreceptors (200 µM IPTG). In this case, Aer-2 levels were estimated in BT3312 with antisera against the highly conserved region of Tsr (common to all chemoreceptors), and compared to levels of chromosomally expressed Tar. The abundance of Tar relative to the total chemoreceptor pool was estimated using values reported by (Li and Hazelbauer, 2004). To compare steady-state levels of Aer-2 mutants with those of WT Aer-2, expression was induced with 50 µM IPTG for 3 h. Bands were visualized on Western blots and quantified on a BioSpectrum® digital imager (UVP, Upland, CA).
Behavioral Assays
For behavioral assays, cells were grown at 30°C in tryptone broth supplemented with 0.5 µg ml−1 thiamine and appropriate antibiotics. For temporal assays, E. coli cells were placed into a gas perfusion chamber and toggled between humidified air and N2 as described previously (Rebbapragada et al., 1997; Taylor et al., 2007). All temporal assays were repeated two or more times on at least two separate days. For O2 titrations, the total gas flow rate was held constant as the percentage of O2 in the gas proportioner was decreased stepwise from 20.9% to 1% (v/v) [see (Taylor et al., 2007) for set-up]. The concentration of O2 in the perfusion chamber was determined using a Hudson RCI® O2 sensor and monitor (Research Triangle Park, NC). To determine the proportion of cells tumbling or swimming, fields of view were examined that contained an average of 30 cells at any one time. Two individuals always viewed the results on a video monitor, and the experiments were repeated on at least two, but usually three or more, days. For determining responses to CO, CO gas (>99% purity, Sigma-Aldrich, St. Louis, MO) was administered through a tube inserted into the open end of the gas perfusion chamber. At the same time the chamber was perfused with N2 from the opposite end. For determining responses to NO, the NO donor Proli NONOate (Cayman Chemical, Ann Arbor, MI) was resuspended to 500 µM in fresh 10 mM NaOH and used immediately. Two microliters of anaerobic 500 µM Proli NONOate were mixed with 7 µl of N2-perfused cells in the gas perfusion chamber using an anaerobic stick. For controls, cells in the chamber were also tested with 500 µM of anaerobic proline in 10 mM NaOH and anaerobic 10 mM NaOH.
Methylation Assay
Cultures of UU2612 and UU2610 cells containing pLH1 were induced for 2 h with 200 µM IPTG, after which time the cells were centrifuged, washed twice and resuspended in chemotaxis buffer (Taylor et al., 2007). Methylation was carried out as described by (Kort et al., 1975) with modifications. Protein synthesis was inhibited by adding 200 µg ml−1 chloramphenicol, and methylation was initiated by adding 7.25 µCi ml−1 L-(methyl-3H)methionine (83 Ci mmol−1, GE Healthcare, Piscataway, NJ). Reactions were stopped with formaldehyde. After SDS-PAGE, gels were soaked for 30 min in Fluoro-hance™ (RPI Corp., Mt. Prospect, IL), then dried and exposed to autoradiography film at −80°C for at least five days.
Protein Purification
Aer-2 was over-expressed from pLH1-derived plasmids by inducing its expression in BT3388 cells with 600 µM IPTG. To improve heme incorporation, 25 µg ml−1 of 5-aminolevulinic acid (Sigma-Aldrich) was added to the cultures. After 3–5 hr of induction, the cells were centrifuged and resuspended to 1.5% of their original volume in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, and 10 mM imidazole) containing 0.3 mg ml−1 of lysozyme, 1 µg ml−1 of DNase I, and 100 µl of Protease Inhibitor Cocktail for His-tagged proteins (Sigma-Aldrich). The cells were disrupted by freeze-thawing five times, followed by sonication. Soluble protein was acquired by removing cell debris at low speed (10,000 × g for 20 min) and the membrane fraction at high speed (485,000 × g for 1 hr). The high-speed supernatant was directly applied to a Ni-NTA agarose column (Qiagen, Valencia, CA) and allowed to percolate by gravity flow. The column was washed with buffer containing 50 mM Tris, pH 7.5, 500 mM NaCl, and 20 mM imidazole and Aer-2 was eluted in the same buffer, except with 250 mM imidazole. The concentration of eluted protein was determined using a BCA™ Protein Assay (Thermo Scientific), and the quality of the sample was determined by staining SDS-PAGE gels with Coomassie brilliant blue.
Size-Exclusion Chromatography
His-tagged Aer-2 peptides were purified on Ni-NTA columns and filtered though 0.2 µM centrifugal filters (Millipore, Billerica, MA). Filtered samples (100 µl, 150–200 µg of protein) were separated on a Progel™-TSK G2000 SW size-exclusion column (Sigma-Aldrich) in 50 mM NaPO4, pH 7.0, 300 mM NaCl at a flow rate of 1 ml min−1. The spectrum of the eluant was monitored in real time (time constant 0.64 sec) from 200 and 500 nm with a diode-array detector (Shimadzu model SPD-M10A, Columbia, MD). Under these conditions, imidazole dissociated from Aer-2, leaving the met-heme form. To analyze deoxy-heme samples, 100 ml of the above buffer was bubbled with N2 in a semi-closed container for 15 min, after which time 35 mg of sodium dithionite was added, and the buffer was bubbled for an additional 5 min. The size-exclusion column was pre-equilibrated with this buffer before a reduced Aer-2 sample was loaded. To reduce Aer-2, a few grains of sodium dithionite were added to the sample immediately before loading. The spectrum of this eluant was monitored to ensure that the heme in the sample remained reduced. For all samples, 1 ml fractions were collected and analyzed for the presence of Aer-2 peptide by Western blotting. To determine apparent molecular weights, peaks corresponding to Aer-2 peptides were compared with the peaks of protein size standards from Schwarz/Mann Biotech (Cleveland, OH) and Sigma-Aldrich.
Absorption Spectra
His-tagged Aer-2 peptides were purified on Ni-NTA columns, loaded into Slide-A-Lyzer® dialysis cassettes (3,500 MW cut-off, Thermo Scientific), and then dialyzed against 50 mM Tris, pH 7.5, 500 mM NaCl buffer. To create a deoxy-heme sample, the dialyzed sample was placed into an anaerobic hood (Coy Laboratory Products, Grass Lake, MI) and reduced with several grains of sodium dithionite. As dithionite masks the UV region of the spectrum, it was removed by centrifuging the reduced sample through a Micro Bio-Spin® 6 column (Bio-Rad, Hercules, CA). The sample was then placed into a covered cuvette before removing it from the anaerobic hood for spectral measurements. To create carbonmonoxy- and oxy-heme samples, dialyzed protein was reduced with dithionite and bubbled with either CO or O2. In both cases, dithionite was removed from the sample on a Micro Bio-Spin column. To create met-heme samples, dialyzed protein was oxidized with an equimolar amount of potassium ferricyanide and purified on a Micro Bio-Spin column. For each sample, the spectrum was scanned between 350 and 700 nm on a Beckman DU® 650 spectrophotometer (Beckman Coulter, Brea, CA) and overlaid by zeroing at 700 nm.
ACKNOWLEDGEMENTS
We would like to thank Lorraine Hernandez, Darysbel PÈrez, Kris-Ann Robertson, Danielle Monahan, and Magi Ishak-Gabra who assisted with experiments while participating in the National Center on Minority Health and Health Disparities (NCMHD) program at Loma Linda University (NIH/NCMHD award 5P20MD001632 to M. De Leon). We would also like to thank Lauren Abraham for technical assistance, Daniel Salcedo for help with gas titration experiments, Sandy Parkinson for strains and Tsr antisera, Luke Ulrich and Igor Zhulin for providing an unpublished PAS sequence alignment, and Brian Crane, Michael Airola, Andrei Lupas, Caroline Harwood and Marie-Alda Gilles-Gonzalez for helpful discussions. This work was also funded by a grant from the National Institute of General Medical Sciences (GM029481) to B.L.Taylor.
REFERENCES
- Airola MV, Watts KJ, Bilwes AM, Crane BR. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure. 2010;18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- Baraquet C, Theraulaz L, Iobbi-Nivol C, Mejean V, Jourlin-Castelli C. Unexpected chemoreceptors mediate energy taxis towards electron acceptors in Shewanella oneidensis. Mol Microbiol. 2009;73:278–290. doi: 10.1111/j.1365-2958.2009.06770.x. [DOI] [PubMed] [Google Scholar]
- Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Simon MI. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- Bren A, Eisenbach M. The N-terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- Campbell AJ, Watts KJ, Johnson MS, Taylor BL. Gain-of-function mutations cluster in distinct regions associated with the signalling pathway in the PAS domain of the aerotaxis receptor, Aer. Mol Microbiol. 2010;77:575–586. doi: 10.1111/j.1365-2958.2010.07231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Niwano M, Ryu J, Taylor BL. Inversion of aerotactic response in Escherichia coli deficient in CheB protein methylesterase. J Bacteriol. 1986;166:275–280. doi: 10.1128/jb.166.1.275-280.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Dunham CM, Dioum EM, Tuckerman JR, Gonzalez G, Scott WG, Gilles-Gonzalez MA. A distal arginine in oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure. Biochemistry. 2003;42:7701–7708. doi: 10.1021/bi0343370. [DOI] [PubMed] [Google Scholar]
- Dunin-Horkawicz S, Lupas AN. Comprehensive analysis of HAMP domains: Implications for transmembrane signal transduction. J Mol Biol. 2010;397:1156–1174. doi: 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Elliott KT, Dirita VJ. Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol Microbiol. 2008;69:1091–1103. doi: 10.1111/j.1365-2958.2008.06357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B, Filloux A. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000540. e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G, Perutz MF, Kiger L, Marden MC, Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci U S A. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2004;231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- Hong CS, Kuroda A, Takiguchi N, Ohtake H, Kato J. Expression of Pseudomonas aeruginosa aer-2, one of two aerotaxis transducer genes, is controlled by RpoS. J Bacteriol. 2005;187:1533–1535. doi: 10.1128/JB.187.4.1533-1535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Robinson J, Shimkets LJ. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol. 2001;183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerby RL, Youn H, Roberts GP. RcoM: a new single-component transcriptional regulator of CO metabolism in bacteria. J Bacteriol. 2008;190:3336–3343. doi: 10.1128/JB.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort EN, Goy MF, Larsen SH, Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975;72:3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Ordal GW. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J Biol Chem. 2002;277:25356–25362. doi: 10.1074/jbc.M201334200. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Lee DS, Watanabe M, Sagami I, Mikami B, Raman CS, Shimizu T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–20193. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
- Lai WC, Barnakova LA, Barnakov AN, Hazelbauer GL. Similarities and differences in interactions of the activity-enhancing chemoreceptor pentapeptide with the two enzymes of adaptational modification. J Bacteriol. 2006;188:5646–5649. doi: 10.1128/JB.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Johnson MS, Taylor BL. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J Bacteriol. 2005;187:193–201. doi: 10.1128/JB.187.1.193-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin KA, Kerby RL, Youn H, Roberts GP, Burstyn JN. The transcription regulator RcoM-2 from Burkholderia xenovorans is a cysteine-ligated hemoprotein that undergoes a redox-mediated ligand switch. Biochemistry. 2008;47:9016–9028. doi: 10.1021/bi800486x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally DF, Matsumura P. Bacterial chemotaxis signaling complexes: formation of a CheA/CheW complex enhances autophosphorylation and affinity for CheY. Proc Natl Acad Sci U S A. 1991;88:6269–6273. doi: 10.1073/pnas.88.14.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena N, Kaur H, Mondal AK. Interactions among HAMP domain repeats act as an osmosensing molecular switch in group III hybrid histidine kinases from fungi. J Biol Chem. 2010;285:12121–12132. doi: 10.1074/jbc.M109.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophilasingle-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Park H, Suquet C, Satterlee JD, Kang C. Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH) Biochemistry. 2004;43:2738–2746. doi: 10.1021/bi035980p. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repik A, Rebbapragada A, Johnson MS, Haznedar JO, Zhulin IB, Taylor BL. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers KR, Tang L, Lukat-Rodgers GS, Wengenack NL. Insights into the signal transduction mechanism of RmFixL provided by carbon monoxide recombination kinetics. Biochemistry. 2001;40:12932–12942. doi: 10.1021/bi011237q. [DOI] [PubMed] [Google Scholar]
- Sasakura Y, Hirata S, Sugiyama S, Suzuki S, Taguchi S, Watanabe M, Matsui T, Sagami I, Shimizu T. Characterization of a direct oxygen sensor heme protein from Escherichia coli. Effects of the heme redox states and mutations at the heme-binding site on catalysis and structure. J Biol Chem. 2002;277:23821–23827. doi: 10.1074/jbc.M202738200. [DOI] [PubMed] [Google Scholar]
- Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- Sherris D, Parkinson JS. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981;78:6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RV, Schuster SC, Simon MI. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry. 1993;32:7623–7629. doi: 10.1021/bi00081a004. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Watts KJ, Johnson MS. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of Aer using in vivo disulfide cross-linking. Methods Enzymol. 2007;422:190–232. doi: 10.1016/S0076-6879(06)22010-X. [DOI] [PubMed] [Google Scholar]
- Watts KJ, Ma Q, Johnson MS, Taylor BL. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J Bacteriol. 2004;186:7440–7449. doi: 10.1128/JB.186.21.7440-7449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts KJ, Johnson MS, Taylor BL. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J Bacteriol. 2008;190:2118–2127. doi: 10.1128/JB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Ulrich LE, Zhulin IB, Alexandre G. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc Natl Acad Sci U S A. 2010;107:2235–2240. doi: 10.1073/pnas.0910055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HS, Saw JH, Hou S, Larsen RW, Watts KJ, Johnson MS, Zimmer MA, Ordal GW, Taylor BL, Alam M. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol Lett. 2002;217:237–242. doi: 10.1111/j.1574-6968.2002.tb11481.x. [DOI] [PubMed] [Google Scholar]
- Zhong X, Hao B, Chan MK. Structure of the PAS fold and signal transduction mechanisms. In: Crews ST, editor. PAS Proteins. Regulators and sensors of development and physiology. Kluwer Academic Publishers; 2003. pp. 1–16. [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]