Abstract
Prp8 is the largest and most highly conserved protein of the spliceosome, encoded by all sequenced eukaryotic genomes but missing from prokaryotes and viruses. Despite all evidence that Prp8 is an integral part of the spliceosomal catalytic center, much remains to be learned about its molecular functions and evolutionary origin. By analyzing sequence and structure similarities between Prp8 and other protein domains, we show that its N-terminal region contains a putative bromodomain. The central conserved domain of Prp8 is related to the catalytic domain of reverse transcriptases (RTs) and is most similar to homologous enzymes encoded by prokaryotic retroelements. However, putative catalytic residues in this RT domain are only partially conserved and may not be sufficient for the nucleotidyltransferase activity. The RT domain is followed by an uncharacterized sequence region with relatives found in fungal RT-like proteins. This part of Prp8 is predicted to adopt an α-helical structure and may be functionally equivalent to diverse maturase/X domains of retroelements and to the thumb domain of retroviral RTs. Together with a previously identified C-terminal domain that has an RNaseH-like fold, our results suggest evolutionary connections between Prp8 and ancient mobile elements. Prp8 may have evolved by acquiring nucleic acid–binding domains from inactivated retroelements, and their present-day role may be in maintaining proper conformation of the bound RNA cofactors and substrates of the splicing reaction. This is only the second example—the other one being telomerase—of the RT recruitment from a genomic parasite to serve an essential cellular function.
Keywords: reverse transcriptase, bromodomain, spliceosome, Prp8, prokaryotic retroelement, origin of mRNA splicing
INTRODUCTION
Removal of introns from the pre-mRNAs in the nucleus of eukaryotic cells consists of two consecutive transesterification reactions, catalyzed by a large macromolecular ribonucleoprotein (RNP) complex, the spliceosome. Mass-spectrometric analysis indicates that there are more than 100 different proteins associated with the spliceosome throughout most of its functional life, and at least 80 of these proteins are conserved in eukaryotes from yeast to humans (Wahl et al. 2009). Notably, neither of the two RNA transesterification reactions should strictly require any of these spliceosomal proteins, as the same chemistry can be achieved by the RNA of group II introns, the mobile elements encoded by genomes of prokaryotes and of eukaryotic organelles, which can self-splice in vitro in the presence of divalent cations but without any proteins. In fact, many structural and functional similarities between some of the small nuclear RNAs (snRNAs), which are integral components of the spliceosome, and group II intron ribozymes suggested that the two types of introns share common evolutionary origins (for reviews, see Cech 2009; Toor et al. 2009).
Among the spliceosomal proteins, Prp8 is the largest—about 2400 amino acids in most species—and most highly conserved in evolution, with >60% identity between fungi and mammals and between fungi and plants. As a stable component of the U5 small nuclear RNP (snRNP), Prp8 participates in multiple interactions with other protein and RNA components throughout the assembly, catalytic phase, and disassembly of spliceosome (Grainger and Beggs 2005). Importantly, Prp8 physically interacts with all three substrate sites directly involved in catalysis: the 5′ splice site (5′SS), branch site (BS), and 3′ splice site (3′SS). In addition to extensive contacts with the U5 snRNA with which it forms a complex, Prp8 also makes direct contacts with U2 and U6 snRNAs, thus interacting with all RNA components of the spliceosomal catalytic center (Grainger and Beggs 2005; Boon et al. 2006; Turner et al. 2006). It is not clear, however, whether the role of Prp8 is to serve as a landing pad and scaffold for other, catalytically active components of the spliceosome or perhaps to play a more direct role in catalysis of one or both of mRNA transesterifications that together constitute the splicing event (Abelson 2008).
For a long time, comparison of the Prp8 sequence to the databases of sequences and of the known protein domains did not reveal any conserved regions beyond the set of Prp8 full-length orthologs, except for an occasional intein-encoding insert in some fungal Prp8 genes (Butler et al. 2001). In the last decade, computational analyses revealed more distant similarities and suggested putative functions for several regions in Prp8. First, the domain of 250–300 amino acids at the extreme C terminus of Prp8 was found to belong to the JAB1/MPN family of metalloproteases (Anantharaman et al. 2002; Maytal-Kivity et al. 2002). This similarity can be verified by standard database search methods such as PSI-BLAST (Altschul et al. 1997); for example, when the complete sequence of yeast Prp8 is used as a query, fungal JAB1/MPN family members, annotated as STAM-binding proteins or endosome-associated ubiquitin isopeptidases AmsH, appear at the second iteration with low E-values of 10−11–10−12, indicating with confidence the common evolutionary origin of these sequences. Sequence comparisons with catalytically active and inactivated homologs have suggested that, similarly to many other JAB1/MPN domains, the C-terminal region of Prp8 is not an active hydrolase, because some of the zinc-chelating histidines, as well as the catalytic acidic residue, are not conserved in Prp8. High-resolution three-dimensional structures of this region from budding yeast and nematode Caenorhabditis elegans confirmed the JAB1/MPN fold (Pena et al. 2007; Zhang et al. 2007) and showed that the mutated isopeptidase catalytic center not only lacks any metal-coordinating moieties but is also precluded by a β-protrusion from binding the substrate. Instead of acting as an enzyme, this domain is thought to mediate protein–protein interactions within the spliceosome. Indeed, the JAB1/MPN domain in Prp8 interacts with ubiquitin (Bellare et al. 2006) and with the ubiquitinated form of Prp3 (Song et al. 2010), and Prp8 itself is ubiquitinated (Bellare et al. 2008). The JAB1/MPN domain also interacts with two factors involved in remodeling of the U5 complex—helicase Brr2 and GTPase Snu114.4 Interestingly, some forms of hereditary retinitis pigmentosa in humans are associated with an array of mutations in this region of human Prp8 ortholog (Towns et al. 2010).
Another putative conserved region, thought to be related to the ubiquitous RNA-recognition motif (RRM), has been identified in the middle part of Prp8 (amino acids 1059–1152), thus suggesting an RNA-binding function for this region (Grainger and Beggs 2005). Extensive cross-linking studies identified contacts between U5 snRNA and regions flanking RRM, yet there were no direct interactions between RRM itself and any of the RNA components of the spliceosome (Turner et al. 2006). The large size of Prp8 suggests that it contains several discrete functional modules, and this hypothesis was tested by random transposons insertion (Boon et al. 2006). The study revealed that the yeast Prp8p can be dissected at three positions so that pairs of separate polypeptides interact with each other and achieve in trans complementation. Importantly, a large central region between residues 770 and 2173 was resistant to dissection, an indication that this part of Prp8 most likely functions as a unit (Boon et al. 2006).
The size of Prp8 makes it a difficult target for crystallization, so limited proteolysis experiments were used recently in an attempt to isolate autonomous globular domains, some of which turned out to be amenable to structural analysis. In that way, the predicted structure of the JAB/MPN domain was confirmed, and important mechanistic aspects, not easily seen from the sequence analysis, were verified (see above). Surprisingly, the direct determination of the structure of the discrete globular region that is located just upstream of the JAB1/MPN domain in yeast and human Prp8 (Fig. 1A) showed that it adopts a modified RNase H fold, which was not obvious from sequence similarity searches (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). In this domain, too, some of the conserved charged residues conferring catalytic activity to the RNase H enzymes are replaced with noncharged residues, and one potentially catalytic aspartic acid residue is hydrogen-bonded to a distal arginine residue, making it unlikely that the two-metal catalytic mechanism could be effected by this domain. It was noted that structurally similar arrangements were observed in the catalytic center of the Piwi domain, which has Slicer activity and adopts an RNase H–like fold (Ritchie et al. 2008) but otherwise shares no discernible sequence similarity with Prp8.
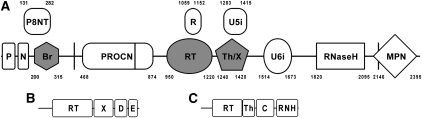
FIGURE 1.
(A) Diagram of conserved structural and functional domains in yeast Prp8p. P indicates proline-rich region (amino acids 5–78); N, nuclear localization signal (amino acids 81–120) (Boon et al. 2007); P8NT, PRO8 N-terminal domain (Staub et al. 2004); Br, bromodomain (this work); PROCN, PRO8 central domain (Staub et al. 2004); R, RNA recognition motif (Grainger and Beggs 2005); RT, reverse transcriptase-like palm-and-fingers domain (this work); Th/X, conserved domain in Prp8 and a subset of fungal RT-like proteins, located at the same position as “maturase-specific” X/thumb domain (this work); U5i, U5-interacting domain (Turner et al. 2006); U6i, U6-interacting domain (Turner et al. 2006); RNase H, RNase H–like domain (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008); and MPN, metalloprotease-like domain (Pena et al. 2007; Zhang et al. 2007). Approximate boundaries of each domain are indicated by numbers. Domains described in this work are shaded, and they overlap with previously known domains, which are raised above for clarity while preserving their relative positions and sizes. Three vertical lines indicate approximate positions where Prp8p can be split so that resulting pieces are able to complement in trans (Boon et al. 2006). (B) General domain organization of group II introns (data adapted from Lambowitz and Zimmerly 2004). RT indicates reverse transcriptase–like palm-and-fingers domain; X, maturase-specific X domain thought to be related to thumb domains (Blocker et al. 2005); D, DNA-binding domain; and E, DNA endonuclease domain. Not all members of this group have D and E domains (Lambowitz and Zimmerly 2004). (C) General domain organization of eukaryotic retroviruses (data adapted from Kohlstaedt et al. 1992). RT indicates reverse transcriptase-like palm-and-fingers domain; Th, thumb domain; C, connection domain; and RNH, RNase H domain.
In this work, we used computational analysis of protein sequence and structure to gain more insight into molecular organization of Prp8. We show that the N-terminal region of Prp8 contains a bromodomain-like structure and that the central region has sequence similarity to the main catalytic domain of reverse transcriptases (RTs) and also to another uncharacterized domain found in fungal RT-like proteins. In agreement with the general theme of rearranged catalytic centers of the known enzymes, the homolog of the RT palm domain in Prp8 does not seem to preserve the complete set of residues implicated in the two-ion catalytic mechanism of RTs and, therefore, is not likely to perform processive addition of nucleotides to DNA or RNA. Our analysis suggests that the evolutionary connections between spliceosome and group II introns are not limited to the similarities in RNA components but extend to the crucial spliceosomal protein Prp8, which may have evolved from the RT encoded by an ancestral retroelement.
RESULTS
Protein family databases such as Pfam (Finn et al. 2010) contain information about domains within Prp8 that have been defined before by analysis of sequence conservation (Staub et al. 2004; Grainger and Beggs 2005), genetic screens (Boon et al. 2006; Turner et al. 2006), and more recent structural studies (Pena et al. 2007, 2008; Zhang et al. 2007; Ritchie et al. 2008; Yang et al. 2008). Each of these domains is schematically drawn in Figure 1A, along with numbers indicating the amino acid positions of their boundaries in yeast Prp8p. One notable disagreement between different studies is in the PROCN domain, which was designated a compact unit by sequence analysis (Staub et al. 2004) yet tolerates the splitting and in trans complementation according to transposon-based screening (Boon et al. 2006). Some of the domain assignments made in our study agree with database annotations of Prp8 domains, and in other cases, domain borders were redefined and new domains were recognized. In Figure 1A three new domains are shaded, and in each case, they supersede other known domains that are shown above them. Newly defined protein domains within the Prp8 family are discussed in more detail below.
Bromodomain in the N-terminal region
In the comparison using the HHPred programs (Söding 2005), part of the PRO8NT region (Staub et al. 2004) gave several matches of moderate significance (lowest P-value was 1.7 × 10−5) to bromodomain-containing proteins. This region spans Prp8 residues 200–315 and aligns with several bromodomains without any insertions or deletions despite low sequence identity (14% or less). The bromodomains recognize acetyl-lysines in histones and many other proteins and consist of four α-helices and two loops (Fig. 2, ZA and BC; Wu and Chiang 2007). Multiple alignments of Prp8 proteins (Fig. 2, upper part) and several different classes of bromodomains (Fig. 2, lower part) show that several residues thought to be important for lysine recognition and for stabilization of the bromodomain's four-helix bundle are well-preserved. Red circles in Figure 2 indicate residues that are important for binding pocket formation, while the red triangle marks the asparagine residue (N294 in yeast Prp8p) that is universally found in the BC loop of bromodomain proteins and makes direct contact via a hydrogen bond with the acetylated lysine (Wu and Chiang 2007; Vollmuth et al. 2009). Since the recognition of acetyl-lysine alone cannot confer the required binding specificity, the residues in and around the binding pocket must contribute by recognizing the particular environment in which the modified lysine is found. Therefore, these residues may not be perfectly conserved between the Prp8 bromodomain and chromatin-binding bromodomains, because they most likely recognize acetyl-lysines in different sequence and structural contexts. It has been proposed that interspersed charged residues in the ZA loop form the electrostatic surface that ensures the specificity of histone recognition (Vollmuth et al. 2009). We notice that there are three stretches of positive and negative residues (identified by red lines above the alignment in Fig. 2) within or adjacent to the ZA loop of Prp8 proteins. These charged patches are highly conserved within Prp8 proteins but not in chromatin-binding bromodomains, and we hypothesize that they are important for recognition of the Prp8-specific substrate by its bromodomain.
FIGURE 2.
Putative bromodomain in Prp8. Prp8 proteins from 10 different species are grouped in the top part of the figure. Several different classes of chromatin-binding bromodomains are aligned in the bottom part of the figure. Residues in chromatin-binding bromodomains that are important for binding pocket formation and direct acetyl-lysine recognition are indicated by red circles and a red triangle, respectively. Function of the conserved glutamate marked by a question mark is unclear. Secondary structure of the bromodomain 1 of mouse Brd4 (Vollmuth et al. 2009) is shown with H marking residues in α-helices. ZA and BC loops are indicated on the secondary structure line. Three stretches of charged residues within or around the ZA loop of Prp8 proteins are indicated by horizontal red lines above the alignment. Lowercase letters preceding Prp8 or bromodomain names stand for the following species: h, Homo sapiens; x, Xenopus laevis; d, Drosophila melanogaster; w, Caenorhabditis elegans; n, Nematostella vectensis; t, Trichoplax adhaerens, a, Arabidopsis thaliana; o, Ostreococcus tauri; p, Paramecium tetraurelia; and y, Saccharomyces cerevisiae. Underline followed by a number at the end of protein names indicates the numerical order of bromodomains for proteins that have multiple copies. Aligned ranges of sequences are shown on each line. Capital letters on the consensus line mean that a single-residue is conserved in at least 90% of sequences. The meaning of lowercase letters on the consensus line is as follows: h, hydrophobic; b, big; and s, small.
We built a structural model of this region (provided in Supplemental Material) using the standard Modeller algorithm (Fiser and Sali 2003) and refined it with Rosetta (Das and Baker 2008). Energy evaluation by ProSa (Wiederstein and Sippl 2007) gave the Z-score of −4.55, which for protein models of about 100 residues is indicative of globally correct fold. It should be emphasized, however, that the modeling was done using a low-identity template (11% identity based on HMM-HMM comparison) and should not be relied upon to predict the atomic-level details of structure and protein interaction.
Reverse transcriptase
Excluding the JAB1/MPN domain, the highest similarity between the full-length Prp8 family model and any database domain was to RTs encoded by various prokaryotic mobile elements. For example, Prp8 matched the prokaryotic RT-like family (cd01709) with a P-value of 1.9 × 10−10, and Pfam RT model (PF00078) was matched with a P-value of 8.2 × 10−8. Family models that were obtained from the alignments of prokaryotic RTs, such as group II intron RTs and bacterial retron RTs, had higher similarity to Prp8 and lower P-values than the models derived from retroviruses and retroelements of eukaryotes, suggesting closer relationship to prokaryotic retroelements than to the known eukaryotic RT domains. The location of the match was in the central segment of Prp8 (amino acids 950–1220), and within the RTs, the aligned region nearly completely covered the catalytic nucleotidyltransferase palm domain and also included parts of the fingers domain.
Multiple sequence alignment of Prp8, of bacterial-type RTs and the more distantly related eukaryotic telomerases, as well as RTs of eukaryotic viruses confirms the presence of the core set of α-helices and β-strands that form the palm and finger domains (Fig. 3A). The alignment, nonetheless, shows that the catalytic motif C (typically YUDD in cellular and viral RTs, where U is a hydrophobic residue), implicated in coordination of the two metal ions and in multiple interactions with the double-stranded or partially unpaired polynucleotide substrates, is only partially conserved in Prp8 (Fig. 3A; Supplemental Fig. 1C). Specifically, this motif in Prp8 is YUDx, with “x” only occasionally preserved as canonical aspartate residue, for example, in the Prp8 homolog of microsporidian Encephalitozoon cuniculi (data not shown), but typically replaced by a positively charged residue. Interestingly, the extended variant of motif C, in the form of RUUDx, is shared by Prp8, group II introns, and telomerases. Motif B, which is involved in nucleotide binding and in RTs also mediates interactions with the 5′-end of the RNA-templating region (Mitchell et al. 2010), displays high conservation of residues with small or kinky side chains (Fig. 3A, G, P, S, or A). Another residue important for catalysis, namely, the metal-binding aspartate in motif A, is not conserved in Prp8 (Fig. 3A). The highly conserved positively charged residue in motif D, for which a role in the protonation of pyrophosphate has been recently proposed (Castro et al. 2009), is not preserved in Prp8 either (data not shown).
FIGURE 3.
(A) The reverse transcriptase-like domain in Prp8. The alignment with selected prokaryotic and eukaryotic retroelements, as well as with retroviruses and telomerases, was made using the programs MACAW (Schuler et al. 1991) and MUSCLE (Edgar 2004). Positions of motifs A, B, and C are shown on the top line. Predicted secondary structures (SecStr Prp8 and SecStr GIIint) are shown when they could be predicted with confidence of 7 or higher (0–9 scale). 3KYL corresponds to Tribolium castaneum telomerase for which the secondary structure is known. Both for known and predicted secondary structures α-helices are marked by H and β-strands by S. Names of Prp8 proteins are the same as in Figure 2. To conserve the space in the legend and for figure clarity, the remaining sequences are grouped by similarity and are identified by their GI numbers. Aligned ranges of sequences are indicated on each line. Numbers in parentheses are in the regions of long insertions and deletions and show how many residues were omitted to make the alignment more compact. Columns are colored if at least 90% of sequences match the consensus except for two aspartates identified by first and third red circles on the line that reads Catalytic residues. Even though these residues are not conserved in Prp8 proteins and therefore do not qualify for 90% consensus, they are colored yellow for emphasis. Four acidic residues and one arginine that are well conserved only in Prp8 proteins and group II intron reverse transcriptases are shaded in green. Capital letters on the consensus line indicate single-residue conservation. The meaning of lowercase letters on the consensus is as follows: h, hydrophobic; b, big; and s, small. (B) The putative Th/X domain in Prp8. Names of Prp8 proteins are the same as in A and Figure 2. The remaining sequences are identified by their GI numbers. SecStr line shows secondary structure prediction for Prp8 proteins. Aligned ranges of sequences are indicated on each line. Capital letters on the consensus line indicate single-residue conservation for at least 90% of sequences. The meaning of lowercase letters on the consensus line is as follows: h, hydrophobic; +, positively charged; b, big; and s, small.
Next we searched for matches between the RT region of Prp8 and known protein structures. The best match was found with the Tribolium telomerase (Gillis et al. 2008; Mitchell et al. 2010); although the best available structural template, this is not the closest sequence relative (telomerase matches Prp8 with a P-value of 7.4 × 10−6). The structural similarity between Prp8 and Tribolium telomerase is nonetheless evident from the alignment that includes predicted and known secondary structures, respectively (Supplemental Fig. 2). Based on this alignment, we constructed a tentative homology model of the core of palm domain and the adjacent portion of the fingers domain of Prp8 (Supplemental Fig. 1). After Rosetta refinement (Das and Baker 2008), this model had a ProSa Z-score of −5.27, indicative of models with a correct overall fold. In this case, too, the model is approximate, as the structural template is not particularly close to Prp8 (13% HMM-HMM identity), and the native structure of a much more closely related group II intron RT domain is not yet available. The close-up of the active site in our model shows that only one of the three catalytic aspartates is present (D1166 in Supplemental Fig. 1C), while the other two catalytic aspartates are replaced by residues that do not coordinate Mg2+ (T1053 and R1167). A negatively charged glutamate (E1051) in the vicinity of the missing aspartate in motif A is placed too far for productive Mg2+ coordination with D1166 (Supplemental Fig. 1C). An area with positive electrostatic potential is predicted near the active site (Supplemental Fig. 1B), raising the possibility that this part of the molecule interacts with the negatively charged RNA backbone. Finally, we have examined the positions of Prp8 suppressor mutations within this domain that alleviate mutations or deletions in RNA and protein components of the spliceosome (for a complete list, see Grainger and Beggs 2005). These mutations do not appear to cluster in space except when found in consecutive residues, nor do they localize near the active site (data not shown).
Another region of significant similarity between Prp8 and the database domains in the database was detected next to the palm-fingers region (designated Th/X in Fig. 1A). Befittingly, this match was to another protein domain encoded by retroelements—in this case, to the region that is found in RT-like proteins encoded by nuclear genomes of fungi and by the green nonsulfur bacterium Herpetosiphon aurantiacus. This region (portion of cd01709, which also includes the RT-like domain itself) matches amino acids 1240–1420 of the Prp8 with P-value 1.5 × 10−5. This novel conserved domain has not been experimentally characterized, and the biological role of fungal and bacterial RT-like elements within which it is found has not been studied. Secondary structure prediction suggests that this region consists of long α-helices (Fig. 3B). Based on domain colinearity and similar secondary structure, we speculate that this may be a functional equivalent of the X/maturase domain in group II RTs (Mohr et al. 1993), an evolutionarily diverse domain that facilitates splicing of the introns in which it resides and is thought to be structurally and functionally equivalent to the nucleic acid–binding thumb domain in viral RTs (Blocker et al. 2005). The cross-linking data indicate that this region of Prp8 makes contacts with U5 snRNA (Turner et al. 2006), supporting its putative nucleic acid–binding function.
DISCUSSION
Why bromodomain?
Structure–function relationships of the N-terminal region of Prp8 have been examined recently by yeast two-hybrid analysis combined with chemical cross-linking and proteomics (Grainger et al. 2009). It has been shown that Snu114p, a GTPase component of U5 snRNP thought to be involved in RNA remodeling during the spliceosome cycle, interacts with the region 420–542 of Prp8, which is located near the bromodomain that we identified. This same region is also involved in several other protein–protein and protein–RNA interactions. In particular, there is an intramolecular interaction between amino acids 1–427 and amino acids 420–542 of Prp8; between the former region and proteins Brr2p, Prp39, and Prp40; and, within the complex of the four proteins, with U4/U6 snRNA (Kuhn and Brow 2000; Grainger et al. 2009). It has been shown that several spliceosomal proteins are acetylated at lysine residues (Choudhary et al. 2009), and the importance of this modification is further confirmed by spliceosome assembly defects caused by small-molecule inhibitors of acetylation and deacetylation (Kuhn et al. 2009). Given the available data, we propose that the function of the bromodomain region in Prp8 may be to facilitate the assembly of spliceosomal proteins by directly recognizing their acetylated lysines. Alternatively, Prp8's bromodomain could recognize the acetylated lysine within its own U6i domain (K1463 in human and K1535 in yeast homologs) (see Choudhary et al. 2009), thus bringing together two parts of the protein separated by more than 1000 amino acids. The conserved asparagine residue and stretches of charged amino acids in the ZA loop (Fig. 2) are prime targets for site-directed mutagenesis aimed at testing these possibilities.
What is the function of RT?
The region of similarity between Prp8 and RTs contains a shorter region suggested by Grainger and Beggs (2005) to be related to a RRM. The canonical RRM domains in RNA-binding proteins have a ferredoxin fold, which is also found, usually with modifications, in RTs and in many other RNA polymerases and DNA polymerases (for the hierarchy of superfamilies within this fold, see http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.e.bcj.html; for discussion of the evolutionary connection between RRM and the enzymes of nucleic acid biosynthesis, see Aravind et al. 2002; Anantharaman et al. 2010). Thus, the earlier observation of Grainger and Beggs suggested the overall structural fold of the central portion of Prp8, while our data now specifically identify the sequence superfamily within this fold from which Prp8 apparently evolved.
The molecular mechanisms of the two spliceosomal reactions that rearrange the phosphoester bonds, resulting in intron removal and a splicing event, remain unknown. Several lines of evidence suggest that the RNA components of the spliceosome may be primarily involved in both of these transesterifications. First, the set of RNA intermediates in the spliceosome pathway and in the group II intron self-splicing pathway is all but the same, including the 2′-5′-linked RNA lariat not found elsewhere in nature. Second, the nucleophile for the first reaction is in both cases represented by an adenosine residue bulging out of an RNA element—the branch site helix on the snRNA U2 at the spliceosomal catalytic center, or domain VI at the core of the group II ribozyme. Third, U2 and domain VI elements share additional conserved sequence motifs. Fourth, the two splicing reactions have the same stereochemistry and are similarly affected by phosphorothioate substitutions at the conserved nucleotide positions (for review, see Pyle and Lambowitz 2006). Finally, metal ions are required for group II intron self-splicing, as well as for spliceosomal activity, and the three nucleotides within Domain V that coordinate the magnesium ion are positioned very similarly to the bases in U6 that also appear to be able to ligate magnesium (Sashital et al. 2004; Toor et al. 2008; Mefford and Staley 2009; Lee et al. 2010).
Compelling as these arguments are, many mechanistic questions remain. Most notably, self-splicing of group II introns is inefficient and requires the presence of the intron-encoded protein both in vivo and in vitro, and the transesterification reactions mediated by the spliceosome have been achieved in vitro only with partially purified extracts containing dozens of proteins. Moreover, the magnesium ion position in group II introns is known only from the post-catalytic form, and positioning of metal ions in the assembled spliceosome remains obscure. The existence of the RT-like module in Prp8, with the palm-finger region more closely related to the corresponding domains in group II intron-encoded proteins than to other known classes of RTs, adds intrigue to this picture. In a general theme of sequence conservation in Prp8, there are substitutions in the putative active center of the RT palm domain, such as the replacement of two catalytic aspartate residues in motifs A and C (Fig. 3A; Supplemental Fig. 1). These substitutions are likely to diminish the metal-binding capacity of this site and render it incapable of processive nucleic acid synthesis. Nevertheless, the absolute conservation of the third catalytic aspartate (D1166 in yeast Prp8p) still leaves open the possibility that this modified active site is capable of chelating a metal ion and performing simpler reactions such as transfer of a nucleotidyl or phosphoryl group, or a hydrolysis of a phosphoester bond, either on its own or when in complex with the RNA components of the spliceosome. This coordinated action of Prp8 and snRNAs would require precise stereochemical interactions with single-stranded and double-stranded RNA components of the spliceosome, including the mRNA transcript.
“RNase H domain” in Prp8: function or fold?
The penultimate structurally characterized domain in Prp8 adopts an RNase H–like fold (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). Despite the name, this common fold is found not only in the RNase H–like superfamily of RNases or in the enzymes involved in RNA metabolism but also in proteins with other activities, for example, small-molecule kinases, ATPases, and DNA transposases (for the hierarchy of families and superfamilies within this fold, some of which have confusingly similar names despite different levels of sequence and structure conservation, see SCOP database; Andreeva et al. 2008). Understanding the evolutionary affinity of this domain in Prp8 is of interest in view of the RT similarities described above, as the juxtaposition of the RT and RNase H domains is found in eukaryotic retroviruses (Fig. 1C) and could suggest the specific connection between retroviruses and Prp8. Even though the RNase H–like domain of Prp8 is presently classified with several other protein families of the RNase H–like superfamily (Andreeva et al. 2008), this assignment is based primarily on structural considerations as HHpred (Söding 2005) and other sensitive database search and fold recognition engines, many of which can be simultaneously accessed through MetaServer portal (Ginalski et al. 2003), do not show a statistically significant sequence similarity between the Prp8 RNase H–like domain and any actual RNase H enzyme. In our sequence-based searches, the highest similarity, albeit with only a partial match and borderline significance (P = 2.5 × 10−4), was observed between Prp8 and Pfam's PF04937/DUF659, a group of widespread transposase-like proteins (data not shown). The same results were obtained after we deleted the amino acids corresponding to the β-hairpin, clearly a Prp8-specific addition to the basic fold.
Interestingly, the connection domain of retrovirus RTs (marked C in Fig. 1C) appears also to adopt the RNase H fold (Artymiuk et al. 1993) even though no separate catalytic activity is known to reside in this domain. Thus, virus RTs contain two copies of the RNase H–like fold, yet only one of them is known to be catalytically active. In addition, the retroviruses encode a separate integrase domain that also has the RNase H fold, while Prp8 contains just one copy of a domain with this fold. This further underscores significant structural and sequence deviation of Prp8 from the retrovirus domain organization.
The RNase H–like domain in Prp8 lacks a complete set of catalytically important acidic residues and may either be inactive or have modified activity compared to RNase H enzymes (Pena et al. 2008; Ritchie et al. 2008). Structural comparisons using the DALI approach (Holm and Rosenstrom 2010) show that the RNase H–like domain of Prp8 has a slightly closer, albeit still distant, structural relationship with viral integrases and cellular endonucleases (Z-scores in the range of 6.5–8.1) than with either of the two copies of the RNase H fold found in RTs (the highest Z-score of 5.5 is in the twilight zone of structural similarity). This range of Z-scores does not allow completely unambiguous assignment one way or the other regarding the question whether the C-terminal RNase H–like domain of Prp8 is directly related to the RNase H domain of eukaryotic retroviruses. A stronger case for the relationship between Prp8 and eukaryotic retroviruses could be made by identifying the sequence or structural similarity between the connection domain of retroviruses (Fig. 1C, C) and the equivalently positioned U6i domain in Prp8 (Fig. 1A). At present, however, we have no proof of that relationship at the sequence level, and secondary structure prediction indicates a much higher content of β-strands in the U6i domain than in the connection domain of retroviruses. In summary, we were unable to obtain any specific evidence that the C-terminal RNase H–like domain of Prp8 is directly related to the RNase H domain of eukaryotic retroviruses. While it is difficult to pinpoint the exact evolutionary origin of the RNase H–like domain in Prp8 given that it has undergone structural modifications (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008), it is likely that this domain functions by binding and/or cleaving nucleic acids.
Domain organization of Prp8 and its function
Based on domain dissection results (Boon et al. 2006), it appears that the whole central portion of Prp8 (residues 770–2173 between the second and third vertical lines in Fig. 1A) functions as a discrete unit. Given that virtually all known protein–RNA contacts map within this region of Prp8 (Turner et al. 2006), our results lend credence to the idea that this part of Prp8 was acquired as a single unit for RNA-related functions of the spliceosome. Subsequently, both termini of that primordial Prp8 gained other domains, such as bromodomain and JAB1/MPN, which added the ability to interact with other proteins. This modular organization ensures precise molecular choreography required to correctly position all spliceosomal proteins, RNA components, and regions of Prp8 itself in the spliceosome active center, thereby poising them for catalysis. This would make the largest spliceosome protein Prp8 an active player, in effect the apoenzyme, in the splicing reaction.
The evolutionary origin of Prp8
Regardless of the exact mechanism of spliceosomal intron removal, the evolutionary origin of spliceosome itself has now become clearer. At least one RNA moiety of the spliceosome, snRNA U6, is thought to have evolved from group II intron RNA on the basis of shared sequence elements and a common chemical reaction mechanism (see above), and we have shown that Prp8, the crucial protein in the spliceosome, contains a RT most similar to group II introns encoded by prokaryotic/organellar genomes. Perhaps the most natural way to interpret this observation is that the two crucial elements of the eukaryotic splicing reaction, namely, snRNAs and the core region of Prp8 that included the RT module, have been simultaneously recruited into the emerging spliceosome at the dawn of the eukaryotic lineage, when a bacterial endosymbiont invaded its host, perhaps related to the ancestor of the present-day Archaea. The symbiont, which gave rise to mitochondria, may have brought its retroelements into the emerging eukaryote ancestor, and these retroelements invaded the host genome. Recent quantitative analysis of the selective pressures associated with the increase of genome complexity in eukaryotes suggests that reduced selection under the condition of low effective population size may have been sufficient to offset the cost of maintaining the machinery for intron removal (Lynch 2002, 2006). A retroelement-encoded RT may have been particularly suitable as the main part in such machinery, as it already could recognize RNA and enhance the rate of transesterification reactions between distant parts of it. As mRNA splicing does not require the synthesis of DNA from the RNA template, or the degradation of the DNA–RNA hybrid, active centers of the corresponding enzymes (i.e., processive RT and RNase H) were not retained by Prp8.
An alternative evolutionary scenario, namely, that Prp8 has evolved from a retrovirus RT, appears less likely than the group II intron RT connection, despite the partially conserved domain order in Prp8 and virus RTs (Fig. 1A,C). First, the sequence of the RT domain in Prp8 is closer to group II intron RTs than to virus RTs. In addition to lower P-values, there are four acidic residues and an arginine (shaded green in Fig. 3A) that are only conserved between Prp8 and group II intron RTs. Second, even after using state-of-the-art methods of sequence and structure comparison and structure prediction, we were not able to show compellingly a close connection of the Prp8 RNase H–like region to the virus RNase H enzyme or other retrovirus-encoded domains with a RNase H–like fold; we instead found weak sequence similarity between the Prp8 domain and a group of putative DNA transposases. Third, prokaryotic retroelements, such as group II introns and retrons, were on hand even before the symbiosis event that is thought to have given rise to eukaryotes. Thus, the machinery suitable for directly evolving spliceosomal introns was already in place right at the point of the origin of eukaryotes, and the insertion of an additional viral ancestor into the evolutionary history of spliceosome seems redundant.
Specific sequence and structure similarities between prokaryotic group II intron–encoded RTs and eukaryotic telomerases—another RT-derived complex in which RNA and protein are both required for activity—have been noted before, but telomerases were thought to be the only example of complete cooptation of a RT from a genomic parasite by an eukaryotic cell to serve an essential, highly conserved cellular function. Prp8 now is shown to be another such RT-mediated radical innovation.
MATERIALS AND METHODS
Sequence searches and profile–profile comparisons
We used the modified SAM T2K procedure (Karplus et al. 2003) to find the full-length homologs of yeast Prp8 in the databases and to construct multiple alignment of the family. This automated pipeline begins with PSI-BLAST searches (Altschul et al. 1997) against the protein database, followed by an accurate alignment procedure that starts with close homologs and is iterated five times by adding more distant proteins. The full-length alignments or their subsets were converted into profile hidden Markov models (HMMs) using the programs from the HHpred package (Söding 2005) and compared to the library of HMMs (Söding et al. 2005) derived from the protein families that were represented in the NCBI CDD database. We also created an in-house database using the same SAM T2K procedure on a set of proteins with a known structure from the PDB database. This database was searched locally with the aid of the HHSearch/HHPred suite of programs (Söding 2005; Söding et al. 2005). When statistically significant similarity to any domain was observed, the termini of this domain were used to isolate the remaining portions of Prp8, and these regions were analyzed in the same fashion again, in order to evaluate weaker sequence similarities that may have been obscured by the higher-scoring domains. This recursive dissection continued until no new similarities could be found.
Three-dimensional modeling and model evaluation
HHSearch-derived alignments were used for structural modeling by Modeller (Fiser and Sali 2003). At least 10 models were generated for each alignment, and their quality was evaluated using ProSa (Wiederstein and Sippl 2007). Energy evaluation by ProSa is not meant to validate that the model is correct in terms of fine details, yet low ProSa Z-scores strongly indicate relatedness between the protein sequence being modeled and its template. Initial alignments were manually adjusted in regions ProSa deemed to be energetically unfavorable, and the model-building was repeated with new alignments until ProSa's Z-scores could not be improved. At this point, models were relaxed and refined using standard protocols implemented in Rosetta (Das and Baker 2008). Models were visualized using PyMol (http://www.pymol.org/) (DeLano 2010).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Magda Konarska for many encouraging discussions and for help with the early draft of this manuscript, Steve Zimmerly for critically reading the manuscript and prompting us to reinterpret the X/maturase domain connection, and three anonymous reviewers for many helpful comments. M.D. is supported in part by an NIH grant P20-RR024237. A.M. is supported by the Stowers Institute of Medical Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2396011.
All gene names in this study are from yeast Saccharomyces cerevisiae, and amino acid positions within Prp8 refer to yeast protein YHR165c, gi 6321959 (2413 amino acids long).
REFERENCES
- Abelson J 2008. Is the spliceosome a ribonucleoprotein enzyme? Nat Struct Mol Biol 15: 1235–1237 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res 30: 1427–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L 2010. Presence of a classical RRM-fold palm domain in Thg1-type 3′- 5′nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol Direct 5: 43 doi: 10.1186/1745-6150-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG 2008. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res 36: D419–D425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Mazumder R, Vasudevan S, Koonin EV 2002. Trends in protein evolution inferred from sequence and structure analysis. Curr Opin Struct Biol 12: 392–399 [DOI] [PubMed] [Google Scholar]
- Artymiuk PJ, Grindley HM, Kumar K, Rice DW, Willett P 1993. Three-dimensional structural resemblance between the ribonuclease H and connection domains of HIV reverse transcriptase and the ATPase fold revealed using graph theoretical techniques. FEBS Lett 324: 15–21 [DOI] [PubMed] [Google Scholar]
- Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ 2006. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA 12: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ 2008. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol 15: 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker FJ, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM 2005. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 11: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon KL, Norman CM, Grainger RJ, Newman AJ, Beggs JD 2006. Prp8p dissection reveals domain structure and protein interaction sites. RNA 12: 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD 2007. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 14: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MI, Goodwin TJ, Poulter RT 2001. A nuclear-encoded intein in the fungal pathogen Cryptococcus neoformans. Yeast 18: 1365–1370 [DOI] [PubMed] [Google Scholar]
- Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE 2009. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol 16: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR 2009. Crawling out of the RNA world. Cell 136: 599–602 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Das R, Baker D 2008. Macromolecular modeling with Rosetta. Annu Rev Biochem 77: 363–382 [DOI] [PubMed] [Google Scholar]
- DeLano WL 2010. The PyMOL user's manual. DeLano Scientific, San Carlos, CA [Google Scholar]
- Edgar RC 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374: 461–491 [DOI] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455: 633–637 [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L 2003. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19: 1015–1018 [DOI] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Barrass JD, Jacquier A, Rain JC, Beggs JD 2009. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. RNA 15: 2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus K, Karchin R, Draper J, Casper J, Mandel-Gutfreund Y, Diekhans M, Hughey R 2003. Combining local-structure, fold-recognition, and new fold methods for protein structure prediction. Proteins 53: 491–496 [DOI] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256: 1783–1790 [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Brow DA 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R 2009. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA 15: 153–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2004. Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lee CH, Jaladat Y, Mohammadi A, Sharifi A, Geisler S, Valadkhan S 2010. Metal binding and substrate positioning by evolutionarily invariant U6 sequences in catalytically active protein-free snRNAs. RNA 16: 2226–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M 2002. Intron evolution as a population-genetic process. Proc Natl Acad Sci 99: 6118–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M 2006. The origins of eukaryotic gene structure. Mol Biol Evol 23: 450–468 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH 2002. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem 3: 28 doi: 10.1186/1471-2091-3-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford MA, Staley JP 2009. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 15: 1386–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E 2010. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol 17: 513–518 [DOI] [PubMed] [Google Scholar]
- Mohr G, Perlman PS, Lambowitz AM 1993. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res 21: 4991–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC 2007. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 25: 615–624 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Luhrmann R, Wahl MC 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM, Lambowitz AM 2006. Group II introns: ribozymes that splice RNA and invade DNA. In The RNA world (ed. Gesteland RF et al. ), pp. 469–505 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE 2004. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11: 1237–1242 [DOI] [PubMed] [Google Scholar]
- Schuler GD, Altschul SF, Lipman DJ 1991. A workbench for multiple alignment construction and analysis. Proteins 9: 180–190 [DOI] [PubMed] [Google Scholar]
- Söding J 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951–960 [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M 2010. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E, Fiziev P, Rosenthal A, Hinzmann B 2004. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays 26: 567–581 [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Pyle AM 2009. Structural insights into RNA splicing. Curr Opin Struct Biol 19: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towns KV, Kipioti A, Long V, McKibbin M, Maubaret C, Vaclavik V, Ehsani P, Springell K, Kamal M, Ramesar RS, et al. 2010. Prognosis for splicing factor PRPF8 retinitis pigmentosa, novel mutations and correlation between human and yeast phenotypes. Hum Mutat 31: E1361–E1376 [DOI] [PubMed] [Google Scholar]
- Turner IA, Norman CM, Churcher MJ, Newman AJ 2006. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA 12: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmuth F, Blankenfeldt W, Geyer M 2009. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J Biol Chem 284: 36547–36556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ 2007. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282: 13141–13145 [DOI] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R 2008. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shen J, Guarnieri MT, Heroux A, Yang K, Zhao R 2007. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci 16: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]