Abstract
Long noncoding RNAs (lncRNAs) are increasingly recognized to play major regulatory roles in development and disease. To identify novel regulators in breast biology, we identified differentially regulated lncRNAs during mouse mammary development. Among the highest and most differentially expressed was a transcript (Zfas1) antisense to the 5′ end of the protein-coding gene Znfx1. In vivo, Zfas1 RNA is localized within the ducts and alveoli of the mammary gland. Zfas1 intronically hosts three previously undescribed C/D box snoRNAs (SNORDs): Snord12, Snord12b, and Snord12c. In contrast to the general assumption that noncoding SNORD-host transcripts function only as vehicles to generate snoRNAs, knockdown of Zfas1 in a mammary epithelial cell line resulted in increased cellular proliferation and differentiation, while not substantially altering the levels of the SNORDs. In support of an independent function, we also found that Zfas1 is extremely stable, with a half-life >16 h. Expression analysis of the SNORDs revealed these were expressed at different levels, likely a result of distinct structures conferring differential stability. While there is relatively low primary sequence conservation between Zfas1 and its syntenic human ortholog ZFAS1, their predicted secondary structures have similar features. Like Zfas1, ZFAS1 is highly expressed in the mammary gland and is down-regulated in breast tumors compared to normal tissue. We propose a functional role for Zfas1/ ZFAS1 in the regulation of alveolar development and epithelial cell differentiation in the mammary gland, which, together with its dysregulation in human breast cancer, suggests ZFAS1 as a putative tumor suppressor gene.
Keywords: noncoding RNA, snoRNA, tumor suppressor
INTRODUCTION
Recent high-throughput studies of gene expression have revealed there is far more genomic transcription than previously anticipated, with the majority of the genome being transcribed into non-protein coding RNAs (ncRNAs) (Kapranov et al. 2002; Carninci et al. 2005; Birney et al. 2007). Although the functional significance of various classes of small ncRNAs is becoming increasingly well-established, the functionality of the tens of thousands of long ncRNAs remains controversial (Babak et al. 2005; Brosius 2005; Struhl 2007), and their functions remain largely unknown. Large-scale studies of long ncRNAs (lncRNAs) have shown that many are dynamically regulated during differentiation and exhibit cell- and tissue-specific expression patterns (Ravasi et al. 2006; Kapranov et al. 2007a, 2007b; Mercer et al. 2008). Together with the finding that lncRNA sequences, splice sites, and promoters are subject to selection (Ponjavic et al. 2007), these observations present a compelling case that lncRNAs are generally functional. However, given the diverse nature of these transcripts, which have various genomic contexts (intergenic, overlapping, intronic, or antisense to protein-coding mRNAs), widely different lengths (ranging from ∼100 to ∼100,000 bases), and may be spliced or unspliced, polyadenylated or nonpolyadenylated, and nuclear or cytoplasmically located (Dinger et al. 2009; Mercer et al. 2009), it is likely that their functions and mechanisms will also be highly diverse. Indeed, among the increasing number that have been functionally investigated (Amaral and Mattick 2008; Amaral et al. 2010), lncRNAs have been shown to function in a variety of cellular processes including transcriptional regulation (Feng et al. 2006), splicing (Yan et al. 2005), translation (Wang et al. 2005), and structure and organization of cellular components (Sunwoo et al. 2009). Furthermore, different subsets of differentially expressed ncRNAs have been identified in embryonic stem cell differentiation (Dinger et al. 2008a), T-cell differentiation (Pang et al. 2009), oligodendrogenesis (Mercer et al. 2010), and keratinocyte differentiation (Mazar et al. 2010), suggesting that any given developmental system will employ its own distinct repertoire of lncRNAs.
Although lncRNAs are difficult to classify (Mercer et al. 2009), one definable subgroup comprises host genes for small nucleolar RNAs (snoRNAs). SnoRNAs are a functionally diverse group of 60–150-nt trans-acting ncRNAs that function as guides for the 2′-O-ribose methylation or pseudouridinylation of ribosomal and spliceosomal RNAs. In vertebrates, most snoRNAs are intron-encoded, and their processing typically involves splicing followed by exonucleolytic trimming of the 5′ and 3′ ends. Although some snoRNA host genes encode proteins, which are primarily involved in the translational apparatus (Dieci et al. 2009), many are spliced, polyadenylated lncRNAs, which are considered to serve solely as a vehicle for snoRNA production (Pelczar and Filipowicz 1998). Structurally, snoRNAs are categorized as C/D-box snoRNAs (SNORDs) and H/ACA-box snoRNAs (SNORAs). Both SNORDs and SNORAs are dysregulated in various types of cancer and have been implicated in the development and progression of human malignancy (Dong et al. 2009; Mourtada-Maarabouni et al. 2009).
The mammary gland is one of the few organs that undergoes cycles of development and regression throughout adult life, with the full development of the gland proceeding in delineated phases: embryonic, pubertal, pregnancy, lactation, and involution (Hennighausen and Robinson 2001). Lactational development occurs with distinct morphological and molecular changes of the epithelial cells and allows for the production and secretion of milk. The secretory alveolar cells represent the final cellular state of the differentiation processes within the mammary gland (Hennighausen and Robinson 1998). These differentiation steps taking place during pregnancy and lactation are defined and characterized by the sequential activation of genes encoding the milk proteins WDNM1, β-casein, whey acidic protein (WAP) and α-lactalbumin in mouse (Robinson et al. 1995).
Further understanding the cellular pathways inherent to normal mammary growth and development is a vital precursor to elucidating the mechanisms that lead to malignant transformation in mammary epithelium. Therefore, to gain a better understanding of the molecular mechanisms that underlie development of the mammary gland and potentially breast cancer and to investigate the role of ncRNA in these processes, we performed microarray analyses and identified 97 lncRNAs that were differentially expressed in primary mammary epithelial cells of pregnant, lactating, and involuting mice.
One of the identified transcripts, Zfas1, exhibited an ∼10-fold decrease in RNA level between pregnancy and lactation. The Zfas1 locus is host to three C/D-box snoRNAs, and its transcription initiates from the antisense strand near the 5′ end of the protein-coding gene Znfx1. We determined the expression patterns of Zfas1 and Znfx1 during mouse mammary gland development and mammary epithelial cell differentiation. In contrast to previous assumptions that noncoding snoRNA host transcripts occurred only as vehicles to generate snoRNAs, we found that knockdown of Zfas1 by RNA interference in a mammary epithelial cell line resulted in an increase in markers of proliferation and differentiation, despite snoRNA levels remaining relatively constant. Furthermore, we show that the three snoRNAs do not occur in equimolar ratios but rather that their relative levels can vary drastically in different conditions, suggesting their stability is tightly regulated. Given the differential expression of Zfas1 during mammary development, we compared the expression of its human ortholog, ZFAS1, in a panel of matched normal and invasive ductal carcinoma tumor tissue and found ZFAS1 is down-regulated in the tumor tissue. In summary, we propose a functional role for Zfas1 in the regulation of the intracellular pathway affecting proliferation and milk production in the mammary gland, which, together with its dysregulation in human breast cancer, suggests ZFAS1 as a possible tumor suppressor gene.
RESULTS
Identification of long ncRNAs that are dynamically regulated in the mouse mammary gland
To identify lncRNAs (>200 nt) involved in mammary gland development, we interrogated custom-designed microarrays with RNA extracted from primary epithelial cells isolated from mouse mammary glands at three distinct stages: 15-d pregnant, 7-d lactating, and 2-d involuting. The microarray contained probes that uniquely profile 8946 high-confidence long ncRNAs and 29,968 mRNAs (includes alternative isoforms) in mouse. Analysis of these data showed significant differential expression (B-statistic > 3; fold change > 4) of 388 mRNAs and 97 lncRNAs in developing mammary glands (Supplemental Fig. S1; Supplemental Table S1).
To validate our experimental model of the developing mammary gland, we first examined the differentially expressed coding mRNAs. The list of differentially expressed genes (Supplemental Table S1) agreed with previously reported data on mouse mammary development (Master et al. 2002) and was generally consistent with expectations of the developmental transitions under investigation (Metcalfe et al. 1999), including dynamic regulation of mRNAs with roles in cell proliferation, milk production, and apoptosis, such as Bcl2- and casein-family genes. Gene ontology (GO) analysis of differentially expressed mRNAs by Babelomics (Al-Shahrour et al. 2006) showed enrichment in genes associated with regulation of cell growth and size and response to hormone stimulus (Supplemental Fig. S2). Based on these observations, we anticipated that the differentially expressed lncRNAs should be similarly relevant to the biological processes underlying mammary development. Indeed, among the differentially expressed lncRNAs, we identified known lncRNAs, such as Dio3os, which has previously been associated with decreased proliferation and increased differentiation of precursor cells to mature adipocytes, analogous to the transition from pregnancy to lactation during mammary gland development (Hernandez et al. 2007).
Zfas1 is a highly expressed, spliced lncRNA that is regulated during mammary gland development
To identify lncRNAs for subsequent experimental examination, we ranked the list of significantly differentially expressed lncRNAs by fold change and absolute expression level. Next, on the basis that many lncRNAs originate from complex transcriptional loci (Engstrom et al. 2006) in which they may have a functional relationship with the nearby protein-coding genes, we further refined our list of lncRNAs by examining their genomic context. Using our previously described classifications of lncRNA loci (Dinger et al. 2009), we identified 15 cis-antisense, 3 nearby antisense, 37 intronic, and 42 intergenic transcripts (Supplemental Table S1). Because we were interested in lncRNAs that may impact on human mammary development, we further refined our list of candidates by considering only those for which there was transcriptional evidence at the syntenic genomic position in human. In total, 19 of the 97 transcripts had human transcripts that arose from syntenic locations.
Taken together, our criteria highlighted a previously uncharacterized lncRNA, which had been annotated in RefSeq as 1500012F01Rik (GenBank ID AK005231). Transcription of this spliced lncRNA initiates from the nearby antisense strand of the Znfx1 (zinc finger NFX-1-type containing) promoter region (Fig. 1A; Supplemental Fig. S3), which led to our naming of it as Zfas1 (zinc finger antisense). As Zfas1 is not transcribed from an ultraconserved region (Bejerano et al. 2004) and is located close to a protein-coding gene, it does not belong to the existing lncRNA subclasses of T-UCRs (transcribed ultraconserved RNAs) (Calin et al. 2007) or lincRNAs (long intergenic noncoding RNAs) (Guttman et al. 2009). From our list of differentially expressed lncRNAs, Zfas1 was the second most highly expressed (A-value ∼10) and had the second largest fold change (34-fold down-regulated from pregnant to lactating) (Supplemental Table S1; Supplemental Fig. S4). In human, the ZNFX1 locus also features an equivalently positioned spliced noncoding transcript, which is annotated in RefSeq Genes as NCRNA00275, a feature not shared by the other highly differentially expressed transcripts. Another interesting feature of this transcript is that it hosts three snoRNA genes, Snord12, Snord12b, and Snord12c, within sequential introns (Fig. 1A; Supplemental Fig. S3; Supplemental Table S2). The combination of these characteristics led us to pursue Zfas1 for further characterization in mammary development.
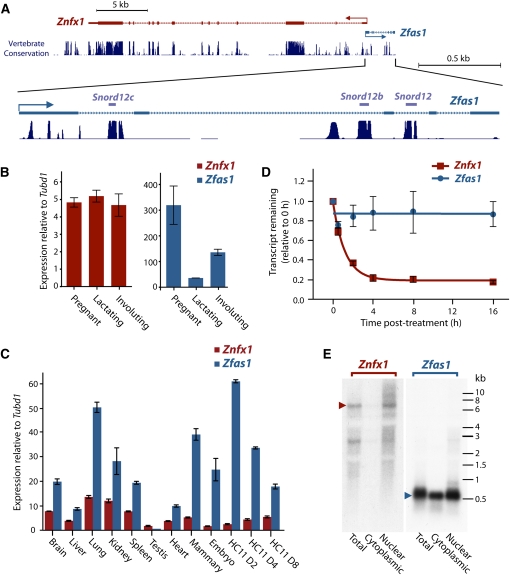
FIGURE 1.
Relationship and expression of Znfx1 and its associated ncRNA Zfas1. (A) Genomic context of Znfx1 and its associated ncRNA. The enlarged Zfas1 indicates the location of three snoRNAs derived from this gene. The high degree of conservation of these regions across mammalian species is indicated. (B) Relative expression of Znfx1 (left) and Zfas1 (right) in mammary epithelial cells during different developmental stages of mammary gland development to Tubulin delta 1 (Tubd1). Expression levels of three biological replicates for each stage were measured in triplicate by qPCR. (C) Relative expression profile of Znfx1 and Zfas1 to Tubd1 in different tissues by qPCR. Technical replicates were performed in triplicate for each sample. (D) Decay curve of Znfx1 and Zfas1 in N2A cells. Transcription was blocked by treatment with actinomycin D, and expression levels of three biological replicates were detected by qPCR. Error bars in B, C, and D are the standard error of the mean (SEM). (E) Northern blot analysis of Znfx1 and Zfas1 on RNA derived from total, cytoplasmic, and nuclear fractions of HC11 cells. Arrows indicate size of transcript for each gene.
Although our ncRNA annotation program had indicated that Zfas1 was noncoding, we noted that the sequence did contain a 79-amino acid open reading frame (ORF). Using BLASTP, we confirmed that this protein sequence was not conserved among mammals and did not contain any known protein motif. Moreover, the start and stop codons were not conserved in the human ortholog of Zfas1, and there was no evidence of any consensus ribosomal binding sequences. Analysis by the CRITICA algorithm (Badger and Olsen 1999) also indicated that the codon usage frequency of this ORF was inconsistent with other mouse genes. Querying PRIDE, a database of peptide sequences deduced from proteomic analyses (Vizcaino et al. 2009), showed no peptides have been identified that correspond to the Zfas1 ORF. Together, these observations led us to conclude that the transcript was unlikely to encode a protein.
Zfas1 expression is differentially regulated to Znfx1
As shown in Figure 1A, Zfas1 and Znfx1 are closely positioned in a head-to-head orientation and potentially share a bidirectional promoter. This raises the possibility that these genes may be coordinately regulated (Trinklein et al. 2004; Engstrom et al. 2006; Dinger et al. 2008a; Mercer et al. 2008). To investigate whether Zfas1 and Znfx1 are coregulated, we examined their expression profiles at different stages of mammary gland development by quantitative real time PCR (qPCR). Figure 1B illustrates the expression of these two genes relative to Tubulin delta 1 (Tubd1), a result in agreement with the microarray data (Supplemental Fig. S4). Although Zfas1 was significantly down-regulated (ninefold) between pregnancy and lactation and significantly up-regulated between lactation and involution (fourfold), Znfx1 did not change appreciably during these transitions. This, together with the finding that the ratio of Zfas1 to Znfx1 varies from 63:1 (in pregnancy) to 6:1 (in lactation) in different developmental stages of the mammary gland, suggests that the transcripts are independently regulated.
EST evidence from GenBank showed that Znfx1 and Zfas1 were expressed in many other tissues outside of the mammary gland, including kidney, brain, pancreatic bud, thymus, eye, heart, and embryo. To obtain a more comprehensive profile of the relative expression levels of Znfx1 and Zfas1 in mouse, we performed qPCR on a diverse range of tissues. Figure 1C shows the expression of Znfx1 is significantly lower than Zfas1 in each of the tissues examined (except testis). Although the expression profiles of Zfas1 and Znfx1 were positively correlated (Pearson co-efficient, R = 0.77), the ratios of Zfas1 to Znfx1 varied considerably, ranging from 1:2.6 in testis to 7.4:1 in mammary gland (Supplemental Fig. S5). The data shows that the highest level of expression of Zfas1 occurs in the lung followed by mammary gland and, in contrast, is almost undetectable in testis. Interestingly, the lung and mammary gland both have biologically similar alveolar structures, raising the possibility that Zfas1 may play a role in alveolar morphogenesis.
In addition to the various tissues, we also examined the relative expression of Zfas1 and Znfx1 in the murine mammary epithelial cell line HC11 during an 8-d in vitro assay. After an initial proliferative phase in the presence of epidermal growth factor (day 2), addition of lactogenic hormones to the HC11 cells at day 4 stimulates lactogenic differentiation in the formation of dome-like structures and the expression of milk proteins by day 8. As well as again showing discordant expression between Zfas1 and Znfx1, the results show that Zfas1 is both highly expressed in HC11 cells and is down-regulated upon differentiation (Fig. 1C; Supplemental Fig. S5). This is consistent with our observation of the differential expression of Zfas1 in the mammary gland and suggests that HC11 cells serve as a meaningful experimental model to explore Zfas1 function.
Given the overlapping promoter regions of Znfx1 and Zfas1, the highly discordant expression of these transcripts is surprising. One explanation for this observation is that Zfas1 is more stable than Znfx1, resulting in higher steady-state levels. To examine this hypothesis, we treated N2A cells with the general transcription inhibitor actinomycin D and quantified the expression levels after 0.5, 2, 4, 8, and 16 h (Fig. 1D). We calculated that Znfx1 had a mean half-life of 50 min (95% confidence interval; 41–65 min). In contrast, Zfas1 levels did not decline significantly after 16 h (relative to GAPDH), confirming that this transcript is indeed highly stable.
As a number of previously characterized lncRNAs have been shown to act in the nucleus (Wilusz et al. 2009), we performed Northern blot analysis on total, nuclear, and cytoplasmic RNA derived from HC11 cells using probes targeting Zfas1 and Znfx1 (Fig. 1E). The Northern hybridization for Zfas1 identified a single strong band of 0.5 kb, which is consistent with the length of the full-length cDNA clones of Zfas1. The Northern hybridizations revealed that Zfas1 was expressed in both cytoplasmic and nuclear fractions, while Znfx1 was highly enriched in the nucleus.
Zfas1 is expressed in the epithelial cells of the duct and alveoli of the mammary gland
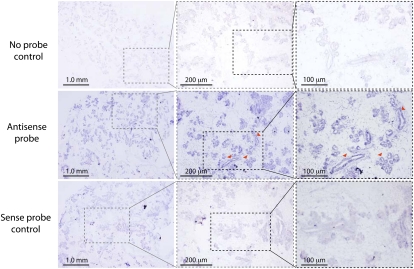
To further analyze Zfas1 expression, we performed section in situ hybridization (ISH) on mammary glands dissected from 15-d pregnant mice. DIG-labeled in vitro transcribed Zfas1 RNA was used to detect expression in section paraffin-embedded pregnant mammary gland. The staining revealed enrichment of Zfas1 expression in the epithelial cells of the ducts and alveoli of the pregnant mammary gland (Fig. 2) relative to the background. Together with the high expression of Zfas1 observed in mammary gland and lung generally, this result is consistent with the notion that Zfas1 is involved in alveolar development.
FIGURE 2.
ISH mammary gland sections from pregnant mice. Panels illustrate mammary gland sections hybridized with no probe (top; negative control), Zfas1 antisense probe (middle), and Zfas1 sense probe (bottom; negative control). Images in dotted boxed areas increase in magnification from left to right. The arrows show ductal and alveolar structure and the expression of Zfas1 within these structures. Scale bars in each panel are indicated. The genomic context of the ISH probe is shown in Supplemental Fig. S3.
Knockdown of Zfas1 results in an increase in markers of cell proliferation
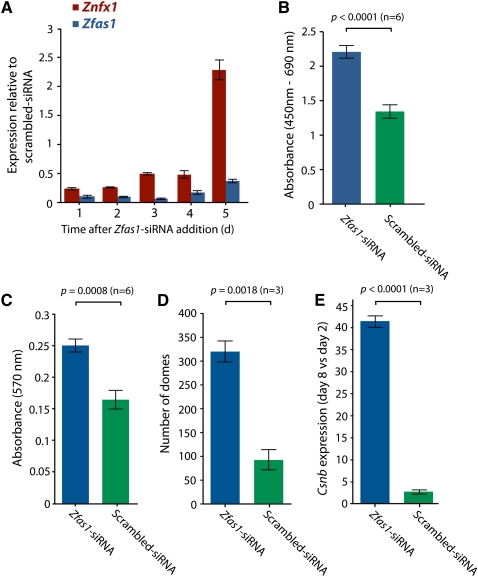
In light of the observation that Zfas1 expression was increased in the mammary gland during pregnancy when the cells are most proliferative, we hypothesized that Zfas1 may have a role in cell proliferation. To investigate the effect of Zfas1 on cell proliferation, we knocked down its expression in exponentially growing HC11 cells using several small interfering RNAs (siRNAs) specific to Zfas1. Using qPCR to measure the relative expression levels of Zfas1, we observed a reduction in expression of Zfas1 relative to a scrambled siRNA-transfected control of at least 80%, which was maintained for up to 4 d in the Zfas1-specific siRNA-treated cells (Fig. 3A; Supplemental Fig. S6). We also assayed Znfx1 expression to see if attenuation of Zfas1 expression had an effect on its protein-coding partner. As seen in Figure 3A, a similar but slightly offset change in expression was apparent for Znfx1 over the same period of analysis.
FIGURE 3.
Effect of Zfas1 knockdown by RNA interference. (A) The expression level, normalized to Tubd1, of Znfx1 and Zfas1 genes in HC11 cells transfected with Zfas1 siRNA relative to the respective expression of each gene in HC11 cells transfected with scrambled siRNA measured by qPCR 1–5 d after siRNA transfection. Technical replicates were performed in triplicate for each time point, with error bars indicating SEM. (B) Proliferation rates based on level of BrdU incorporation measured 48 h after cells were transfected with Zfas1 versus scrambled siRNA. Six technical replicates were performed with error bars indicating SEM. (C) MTT assay measuring the metabolic rate of HC11 cells transfected with Zfas1 versus scrambled siRNA. Six technical replicates were performed with error bars indicating SEM. (D) Effect of Zfas1 knockdown compared to the scrambled siRNA control on dome formation in differentiated HC11 cells measured on day 8. (E) Quantitative PCR, relative to Tubd1, of β-casein (Csn2) levels in differentiated (day 8) cells relative to undifferentiated (day 2) in HC11 cells transfected with Zfas1 or scrambled siRNA. The results in D and E represent data from three experiments, each with three technical replicates, with error bars indicating the SEM of the three experiments.
Cell proliferation rates were determined by quantifying incorporation of BrdU into DNA 48 h after cells were transfected by siRNA (Fig. 3B). We found that the Zfas1-knockdown cells displayed a higher rate of proliferation relative to the scrambled siRNA-transfected control. To further validate this effect, we also measured proliferation rates in Zfas1 knockdown and control cells using an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazol] assay (Mosmann 1983) to measure metabolic activity over a 4-h period. Consistent with the BrdU incorporation assay, cells transfected with siRNAs targeting Zfas1 showed increased proliferation (Fig. 3C). Together, these results suggest that Zfas1 may play a role in regulating cell proliferation.
Knockdown of Zfas1 induces β-casein expression and epithelial dome formation
To explore the hypothesis that Zfas1 has a regulatory role in alveolar development in the mammary gland, we examined the effect of knocking down Zfas1 during a dome formation assay. Formation of domes in cell culture, which can be induced in HC11 cells by addition of o-prolactin and dexamethasone, is considered to be a model for mammary epithelial cell differentiation (Zucchi et al. 2002). Relative to a scrambled siRNA control, HC11 cells that had been transfected with a Zfas1-targeted siRNA showed a marked increase in the number of domes formed following induction (Fig. 3D). O-prolactin- and dexamethasone-induced HC11 differentiation is also characterized by increased expression of β-casein. To further characterize the role of Zfas1 in this developmental model, we determined the response to Zfas1-knockdown in comparison to a scrambled siRNA-knockdown by measuring the expression level of Csn2 (β-casein) in differentiated (day 8) relative to undifferentiated (day 2) cells. We found that in Zfas1 knockdown cells, the expression of Csn2 increased over 40-fold in differentiated cells relative to undifferentiated cells, whereas it increased just twofold in cells transfected with the control siRNA (Fig. 3E). Together, these results suggest that Zfas1 may play a role in regulating HC11 differentiation.
SnoRNAs derived from Zfas1 are differentially expressed
Zfas1 is predicted to host three C/D box-containing homologous snoRNA genes, Snord12, Snord12b, and Snord12c, in consecutive introns (Fig. 1A). Intronic snoRNAs have been identified in all eukaryotic genomes and are frequently distributed in noncoding genes in this manner, with one snoRNA per consecutive intron (Huang et al. 2005). C/D box snoRNAs primarily guide the site-specific methylation of other RNAs, mainly ribosomal RNAs. Snord12 and Snord12b (previously referred to as MBII-99 and MBII-99B) are predicted to modify Gm3868 and Gm3878, respectively, in 28S rRNA (Huttenhofer et al. 2001; Yang et al. 2006). Snord12c (previously referred to as Snord106 or U106) contains antisense elements that match the G1536 and U1602 segments in 18S rRNA. However, as there is no evidence for methylation at these sites, Snord12c may function solely as an RNA chaperone or target chemical modifications in a nonribosomal transcript. The predicted size for Snord12, Snord12b, and Snord12c are ∼85, ∼87, and ∼93 nucleotides, respectively.
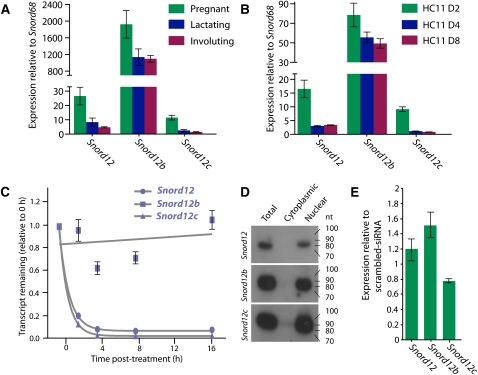
As the three Zfas1 snoRNAs had not previously been examined in mouse (Snord12b and Snord12c are unannotated in RefSeq and UCSC Known Genes), we designed primers to detect the expression of these SNORDs. Using cDNA prepared from mouse mammary epithelial cells, we were able to confirm the existence of all three snoRNAs. Next, we aimed to determine the relative expression levels of Snord12, Snord12b, and Snord12c in mammary epithelial tissue from pregnant, lactating, and involuting mice. Using qPCR (TaqMan), each of the snoRNAs was found to be most highly expressed during pregnancy (Fig. 4A), consistent with the higher expression of the host transcript Zfas1 in pregnancy. However, in contrast to Zfas1, which shows increased expression in involuting relative to lactating mammary epithelial cells, the SNORDs are expressed at similar levels at these developmental stages. Surprisingly, given that the three snoRNAs are derived from the same host transcript, Snord12b was expressed at up to 171-fold and 72-fold greater levels than Snord12c and Snord12, respectively. To examine whether the expression trends of Snord12, Snord12b, and Snord12c were similar in differentiating HC11 cells, we used qPCR to examine their expression levels in 2-, 4-, and 8-d differentiated HC11 cells (Fig. 4B). Although we found Snord12b was more highly expressed than Snord12 and Snord12c, the difference in expression was much less dramatic (approximately eightfold and approximately fivefold for Snord12c and Snord12, respectively). Similar to the expression trend of Zfas1, the snoRNAs were consistently most highly expressed in undifferentiated HC11 cells (day 2) and decreased significantly in differentiated (day 4 and 8) cells.
FIGURE 4.
Expression of snoRNAs that are intronic to Zfas1. (A) Relative expression (from left to right) of Snord12, Snord12b, and Snord12c to Snord68 during different mammary gland developmental stages. Expression levels of three biological replicates for each stage were measured in triplicate by qPCR. (B) Expression levels (from left to right) of Snord12, Snord12b, and Snord12c during HC11 cell differentiation relative to Snord68. Expression levels of two biological replicates for each stage were measured in triplicate by qPCR. Error bars in both A and B are SEM of the biological replicates. (C) Decay curve of Snord12, Snord12b, and Snord12c in N2A cells. Transcription was blocked by treatment with actinomycin D, and expression levels were detected in triplicate by qPCR. Errors are the standard error of the mean (SEM). (D) Northern blot analysis of Snord12, Snord12b, and Snord12c (see Fig. 1A and Supplemental Fig. S3 for genomic positions) expression in total, cytoplasmic, and nuclear RNA derived from undifferentiated HC11 cells. (E) Expression levels of Snord12, Snord12b, and Snord12c, normalized to Snord68, in undifferentiated (day 2) HC11 cells transfected with Zfas1 siRNA relative to expression levels of each normalized gene in HC11 cells transfected with scrambled siRNA. Technical replicates were performed in triplicate, with error bars indicating SEM.
One explanation for the highly differential molar ratios of Snord12, Snord12b, and Snord12c is that they have different stabilities. Using the same approach as described above to determine the stability of Znfx1 and Zfas1, we blocked transcription and determined the expression levels of the three SNORDs. We found that Snord12 and Snord12c levels decreased rapidly (with half-lives of 43 min and 37 min, respectively) following transcriptional inhibition, whereas Snord12b levels did not change appreciably even after 16 h (Fig. 4C). Although these snoRNAs are similar in sequenc and are accordingly considered to belong to the same family, we hypothesized that they may fold into different structures. To examine this hypothesis, we used MFOLD to predict the secondary structures of the three snoRNAs (see Supplementary Methods). Interestingly, Snord12b, which had exhibited much higher expression levels than Snord12 and Snord12c, folded into a distinct structure with an additional short hairpin in relation to Snord12 and Snord12c, which folded into the traditional secondary structure of C/D box snoRNAs (Supplemental Fig. S3).
As the Zfas1 transcript was detected in both nuclear and cytoplasmic fractions of HC11 cells, we examined the cellular localization of the snoRNAs. We prepared total RNA from HC11 cells and subsequently fractionated the cells into nuclear and cytoplasmic portions. Consistent with the qPCR data, Northern blot analysis indicated a high level of expression for all three Zfas1-derived snoRNAs, but unlike Zfas1, SNORD expression was specific to the nuclear fraction of these cells (Fig. 4D). The Northern analysis showed a single band of the predicted size for each snoRNA. The absence of smaller fragments in the Northern blot as well as examination of small RNA deep sequencing data (Taft et al. 2009) suggests that, unlike previous reports of other snoRNAs (Ender et al. 2008; Taft et al. 2009), Snord12, Snord12b, and Snord12c are not processed into smaller RNAs.
Although the siRNA-mediated knockdown of Zfas1 should not affect the expression of Snord12, Snord12b, and Snord12c, which we would expect to be spliced out of the Zfas1 prior to siRNA-directed breakdown of the host transcript, we nevertheless wished to confirm that the phenotypic changes observed during Zfas1-knockdown were not a direct consequence of reduced SNORD expression. Using qPCR, we found that there are only minor differences in the expression of Snord12, Snord12b, and Snord12c in the Zfas1-knockdown compared to the scrambled siRNA control (Fig. 4E). These small changes in expression, relative to the ∼80% knockdown of the host transcript, suggest that the phenotypic changes observed following Zfas1 knockdown are a consequence of the host transcript and that the mature form of Zfas1 functions intrinsically as an RNA.
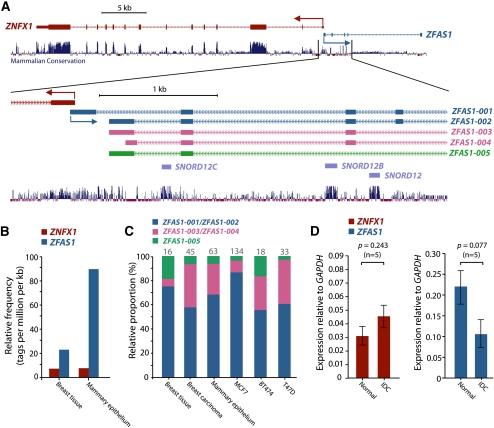
Human ZFAS1 transcript exists and undergoes regulated alternative splicing
The human ortholog of Zfas1, ZFAS1, is located on chromosome 20. In terms of the relative position of its transcription start site to ZNFX1 and the presence of intronic snoRNA genes (Fig. 5A), the ZFAS1 locus is similar to that in mouse. ZFAS1 is alternatively spliced with cDNA evidence indicating the presence of at least five different isoforms. Although the snoRNA genes (SNORD12, SNORD12B, and SNORD12C) are highly conserved between mouse and human (81.3%, 68.9%, and 71%, respectively), a comparison of the exonic regions of the most prevalent isoforms of ZFAS1 (isoforms 1-4 in Figure 5A; C20orf1999 uc002xuj.2, uc002xul.3, uc002xum.3, uc002xuo.2) with its mouse ortholog (NM_001081005.1) shows an average of only 43% identity. However, comparison of the secondary structure predictions of the mature Zfas1 and ZFAS1 transcripts revealed several highly structured regions of the transcripts were very similar despite lacking sequence identity (Supplemental Fig. S7).
FIGURE 5.
Expression analysis of human ZFAS1. (A) Genomic context of human ZNFX1 and ZFAS1. The 5′ ends of ZNFX1 and ZFAS1 are oriented head-to-head on opposite strands. The zoomed-in regions show five different ZFAS1 isoforms that are represented by ESTs. The positions of the intronically-derived snoRNAs—SNORD12, SNORD12B, and SNORD12C—are also shown with the degree of conservation across mammalian species indicated. (B) Comparative expression levels (tpm) of ZNFX1 and ZFAS1 based on RNA deep sequencing of human breast tissue and mammary epithelium. (C) Relative abundance of alternate isoforms of ZFAS1 in various human tissues and cell lines based on exon-exon junction spanning deep sequence tags. The number on top of each bar represents the number of informative tags. (D) Relative expression level of ZNFX1 and ZFAS1 to GAPDH in the five paired normal and invasive ductal carcinoma (IDC) samples detected by qPCR with technical replicates performed in triplicate. Error bars indicate SEM of the biological replicates.
To obtain an overview of ZNFX1 and ZFAS1 expression, we performed qPCR across a panel of 20 human tissues and the breast cancer cell lines MCF7, BT474, and T47D (Supplemental Figs. S8, S9). Similar to the mouse tissue data, the ratios of ZFAS1 to ZNFX1 varied across tissues (from 2:1 in testis to 12:1 in mammary tissue). However, overall there was a positive correlation (Pearson's correlation, R2 = 0.797, n = 20) between the expression profiles of ZNFX1 and ZFAS1 across the tissue samples (Supplemental Fig. S8). Analysis of publicly available transcriptomic deep sequencing data in normal human breast tissue and mammary epithelium (Wang et al. 2008) mirrored our expression analysis in mouse, showing that ZFAS1 is very highly expressed in mammary tissue (in the top 2%–5% of all genes) (Fig. 5B).
The deep sequencing data recapitulated the presence of at least three different isoforms of ZFAS1. Furthermore we were able to detect ZFAS1 isoforms in RNA isolated from MCF7, BT474, and T47D by using PCR primers designed to common exons (Supplemental Fig. S9). To determine whether different isoforms were alternatively regulated in different tissue types, we examined the relative proportions of the isoforms in the RNA deep sequencing libraries. Although the longer isoforms were predominant in each tissue type (ranging from ∼55% to ∼85% of the three distinguishable groups), the relative proportions of the isoform groups differed between tissue types, suggesting regulation of the alternative splicing (Fig. 5C; Supplemental Fig. S9).
Human ZFAS1 levels are reduced in ductal carcinoma relative to normal breast tissue
Given the role of Zfas1 in proliferation, we hypothesized that decreased ZFAS1 expression may be a marker for breast cancer and, moreover, that ZFAS1 may be a tumor suppressor gene. To investigate this hypothesis, we examined ZFAS1 expression in total RNA isolated from the epithelial cells isolated by micro-dissection from frozen sections of normal breast and invasive ductal carcinoma (IDC) tissue (five paired and seven unpaired samples). The result shows ZFAS1 expression is decreased (2.0-fold, p = 0.08, paired; 2.7-fold, p = 0.09, unpaired) in ductal carcinoma relative to normal epithelial cells (Fig. 5D; Supplemental Fig. S10). Taken together with the effects of Zfas1 knockdown on mammary epithelial cell proliferation and differentiation, our results suggest ZFAS1 as a novel human tumor suppressor gene in breast cancer and that its dysregulation may be useful as a marker for breast cancer.
DISCUSSION
Although the importance of lncRNAs in cell function is now becoming firmly established (Mercer et al. 2009), only a relatively small number have been shown to be involved in cancer (Huarte and Rinn 2010). In light of the potential value of lncRNAs as biomarkers and therapeutic targets (Huarte and Rinn 2010), as well as to further our understanding of the molecular mechanisms underlying cancer formation and development, we sought to identify lncRNAs involved in breast cancer. Under the hypothesis that lncRNAs involved in mammary development may be dysregulated in breast cancer, we examined the expression of 8946 lncRNAs at different stages of mouse mammary gland development and found a total of 97 that showed significant dynamic changes. We ranked the candidates on the basis of overall expression level, fold change, and conservation in humans. As a result, we selected the lncRNA Zfas1, which is positioned on the antisense strand at the 5′ end of the Znfx1 protein-coding gene and is host to three C/D box snoRNAs, for further functional examination.
Knockdown of Zfas1 in mouse mammary epithelial cells resulted in a significant increase in proliferation and metabolic activity. Examination of the expression of the intronically hosted SNORDs following knockdown showed that their expression was only minimally altered, suggesting that the processed Zfas1 transcript is itself functional. The high level of proliferation and low level of Zfas1 expression is analogous to our observations in human mammary tissues, where we observe a substantially decreased level of ZFAS1 in highly proliferative invasive ductal carcinoma cells compared to normal breast tissue. Collectively, these observations led us to propose ZFAS1 as a putative tumor suppressor gene.
The head to head arrangement between Zfas1 and the oppositely transcribed protein-coding gene Znfx1 occurs frequently in mammalian genomes (Trinklein et al. 2004; Engstrom et al. 2006). Znfx1 and Zfas1 share a CpG island, the methylation of which would be expected to similarly affect the expression of these transcripts. Although in some cases such bidirectional genes show concordant expression profiles, consistent with the notion of shared regulatory elements, others, as described below for Znfx1 and Zfas1, share more complex expression relationships (Dinger et al. 2008a; Mercer et al. 2008). In both mouse and human, the expression patterns of Znfx1 and Zfas1 were similar over a panel of tissues, with the exception of testis and mammary gland. In both species, relative to Znfx1, Zfas1 was considerably down-regulated in testis and up-regulated in mammary gland. The uncoupling of Znfx1 and Zfas1 expression was also evident both in the developing mammary gland, where Znfx1 remains relatively constant while Zfas1 undergoes significant dynamic changes, and during differentiation of the mouse mammary epithelial cell line HC11, where there was an inverse relationship in the expression of Zfas1 and Znfx1. Furthermore, knockdown of Zfas1 in HC11 cells results in a concomitant relative decrease in Znfx1 expression levels. However, as the levels of Zfas1 recovered following knockdown, there was an overcompensation of Znfx1, which increased to more than twofold that of normal cells. Together, these observations suggest that the expression of Znfx1 and Zfas1 is likely to be intertwined and therefore may participate in the same regulatory network. However, because the expression of these genes can be uncoupled in some conditions, it is likely that there are at least some independent regulatory controls underlying their expression and/or stability. Furthermore, the specific up-regulation of Zfas1 relative to Znfx1 in mammary gland suggests a specific role for Zfas1 in this organ, particularly during development.
Although the Znfx1 protein has not been previously studied, its predicted sequence contains an NFX-1 (nuclear transcription factor X-box binding) zinc finger domain. Despite the low homology between the NFX-1 binding domain and the corresponding region in Znfx1, (26% identity; 36% similarity over a 327-amino acids region), the critical cysteine residues that characterize the domain (Song et al. 1994) are highly conserved (36/40; 90%), suggesting that Znfx1 may also bind DNA. Interestingly, the human ortholog of Znfx1, ZNFX1 (previously referred to as KIAA1404 or MAD-Cap5), is specifically up-regulated in response to chemotherapeutic treatment in MCF7 and ZR-75-1 human mammary gland cell lines (Troester et al. 2004) and is also up-regulated in the serum of patients following treatment for prostate cancer (Dunphy and McNeel 2005). One conclusion drawn from these studies was that ZNFX1 might be involved in DNA repair. If Zfas1 indeed belongs to the same regulatory network as Znfx1, then we can speculate that Zfas1 may also have some role in a DNA repair pathway.
Alignment of mouse Zfas1 and human ZFAS1 reveals only moderate sequence conservation, with the notable exception of the snoRNA regions conserved in three consecutive introns. This evident conservation of Snord12, Snord12b, and Snord12c suggests the function of these transcripts is likely to be conserved across mammals. Interestingly, despite an apparent absence of alternative splice variants of Zfas1, the Zfas1-derived snoRNAs are not present in equal proportions. The highly differential ratios of the snoRNAs suggest the degradation and/or stability of the snoRNAs can vary considerably. Determination of the half-lives of the snoRNAs confirmed this hypothesis, showing that Snord12b was considerably more stable than Snord12 and Snord12c. Structural predictions reflected this differential stability, showing that Snord12b, the most highly expressed of the snoRNAs, has a structure consistent with increased stability. The differential stability/degradation of snoRNAs at the various stages of mammary development raises the hypothesis that the target region for these snoRNAs may be important during mammary gland development and, consequently, that the dysregulation of their expression levels may have important consequences in breast cancer etiology.
The moderate sequence homology of the evolutionarily conserved Zfas1 and ZFAS1 led us to look beyond nucleotide alignment of these orthologs. A number of ncRNAs have characteristic structures that are functional and hence are well conserved over evolutionary timescales. Most of the “classical” ncRNAs, including rRNAs, tRNAs, small nuclear RNAs (snRNAs), snoRNAs, as well as the RNA components of RNAse P and the signal recognition particle, show this evolutionary conservation of structure and function (Washietl et al. 2005). Comparison of the predicted secondary structures of the human and mouse forms of Zfas1 revealed several distinct regions that had almost identical structures, despite sharing minimal sequence identity over these areas. The remarkable stability of Zfas1 (half-life of >16 h), as well as the presence of conserved structures within Zfas1, implies that the RNA has functions beyond its role as a host for generating snoRNAs. This notion is supported by the observed lack of effect on snoRNA transcript levels upon siRNA-mediated Zfas1 knockdown. Thus, the phenotypic effects caused by the reduction of Znxf1-as indicate that, in this case, the snoRNA host transcript is itself functional. This may also be the case for many other noncoding host transcripts, as has been recently shown for the Gas5 SNORD host gene (Kino et al. 2010), as well as host transcripts for miRNA host genes, such as H19 (Gabory et al. 2010) and BIC (Eis et al. 2005).
In summary, we show that the mature spliced transcript of an RNA that harbors C/D-box snoRNAs can function independently of the snoRNAs. This RNA is highly regulated in the developing mouse mammary gland, acts as a repressor of proliferation and differentiation, and is dysregulated in human breast cancer. Our results highlight the importance of the largely unexplored population of non-protein-coding genes in understanding the molecular basis of disease and as sources of potential biomarkers and therapeutic targets.
MATERIALS AND METHODS
Animals and mammary epithelial cells isolation
All experiments were performed with Balb/c mice, which were maintained and handled according to Australian guidelines for animal safety. All experiments were approved by the Animal Research Ethics Committee of the University of Queensland. The mice were mated and then sacrificed at day 15 of pregnancy, day 7 of lactation, and day 2 of involution. Nine mice from each stage were sacrificed, and mammary glands were dissected. One thoracic gland from each mouse was fixed for in situ hybridization and remaining glands were pooled to create three pools for each developmental stage and processed for epithelial cell purification as described previously (Tan-Wong et al. 2008). For the adult mouse tissue expression analysis, brain, liver, lung, kidney, spleen, and testis were dissected from a single male mouse, the whole mammary gland tissue was derived from a virgin female mouse, and whole embryos were harvested from a single mouse 10.5 d post-coitum.
RNA extraction
Total cellular RNA from mammary gland epithelial cells or cultured cells or other tissues from mouse was purified using Trizol (Invitrogen) according to the manufacturer's instructions. For further detail on RNA extraction and qPCR analyses, see Supplemental Methods.
Microarray analyses
Custom-designed microarrays (ArrayExpress accession number A-MEXP-1958) were synthesized by NimbleGen, and experiments performed according to the manufacturer's instructions. The noncoding transcripts targeted by the custom microarray were identified using the CRITICA software, which uses a combination of statistical and comparative parameters, such as open reading frame (ORF) length, synonymous versus nonsynonymous base substitution rates, and similarity to known proteins (Badger and Olsen 1999; Frith et al. 2006). Although we cannot eliminate the possibility that small proteins or peptides are encoded by these transcripts (Dinger et al. 2008b), BLASTP searches of predicted ORFs indicated they did not contain any known protein motif and were not conserved in other species. Raw and normalized microarray data is available at the ArrayExpress Data Warehouse (EMBL-EBI; ArrayExpress Accession Number E-TABM-1106). For expression analyses, see Supplemental Methods.
In situ hybridization
In situ hybridization (ISH) was performed using digoxigenin (DIG)-labeled complementary RNA probes. In vitro transcription was performed according to the manufacturer's instructions using the Promega T7/SP6 reverse transcription kit to produce sense (control) and antisense ISH probes. Section ISH was performed on 5-mm sections of paraformaldehyde-paraffin-embedded 15-d pregnant mouse mammary glands (see Supplemental Methods for details).
Northern blot analysis
Northern blot analysis was performed as previously described (Amaral et al. 2009). The Zfas1 probe used for Northern blot analysis was the same antisense Zfas1 PCR product that was used for in situ hybridization. Both the Zfas1 and Znfx1 probe (see Supplemental Table S3 for primer sequences) were random-labeled (GE Healthcare) according to the manufacturer's instructions. The snoRNA probes were prepared by amplifying the respective genes using the primers listed in Supplemental Table S3 and were randomly labeled as above.
Cell lines
Mouse HC11 cells were cultured and induced to differentiation in an eight-day assay as previously described (Naylor et al. 2005). T47D, BT474, MCF7, and N2A were cultured as described previously (Soule et al. 1973; Keydar et al. 1979; Lasfargues et al. 1979; Georgopoulou et al. 2006).
RNA stability assay
N2A cells were grown to ∼50% confluence, before addition of 10 μg/mL actinomycin D to block RNA polymerase activity. RNA was extracted using RNeasy kits (Qiagen) from three biological replicates at 0, 0.5, 2, 4, 8, and 16 h after treatment. For Znfx1 and Zfas1, qPCR using random hexamers was used to quantify expression relative to GAPDH. Snord12, Snord12b, and Snord12c levels were determined from RNA isolated from 0, 2, 4, 8, and 16 h after treatment and quantified as described in Supplemental Methods. The control time point (t = 0) expression level was set to 100% and treated samples shown as a percentage of the control. A one-phase exponential decay curve and half-life value were calculated using nonlinear regression with a least squares fit by Prism 5 (plateau = unconstrained, k > 0). Where no decay was present, a linear line was calculated using nonlinear regression with a least squares fit.
RNA interference
Four pairs of siRNAs (see Supplemental Table S3) designed to knockdown Zfas1 expression and one pair of scrambled siRNAs were purchased from Sigma. Equal quantities of HC11 cells (5 × 105) were seeded per well in 12-well plates, and the siRNA knockdown was performed as described previously (Naylor et al. 2005). Three replicates per time point were performed for both the Znfx1/Zfas1 expression analysis (Fig. 3A) and β-casein expression assay (Fig. 3E), and quantitative PCR was performed as described above.
Proliferation assay
Quantification of cell proliferation based on the measurement of BrdU incorporation during DNA synthesis was performed on cells transfected with Zfas1 versus those transfected with scrambled siRNA using the cell proliferation ELISA, BrdU colorimetric immunoassay kit (Roche). Twenty-four hours after siRNA transfection, cells were trypsinized, and six replicates of 12 × 103 cells were seeded per well in a 96-well plate, with the no-cells well used as a blank. Twenty-four hours after the cells were seeded in 96-well plates, the cell proliferation assay was performed according to the manufacturer's instructions.
MTT assay
Twenty-four hours after HC11 transfection with Zfas1 versus scrambled siRNA, cells were trypsinized, and six replicates of 12 × 103 cells were seeded per well in 96-well plates, with no-cells wells used as blank. Proliferation was measured by an MTT (tetrazolium blue) conversion test and tritiated thymidine uptake (Sigma Aldrich). Briefly, 20 μL MTT (5 mg/mL) was added to each well and the cells grown at 37°C for 4 h. After addition of 100 μL of solubilization solution (10% SDS in 0.01 M HCl) cells were incubated at 37°C for a further 3 h. Specific optical density of all wells was then measured at 540 nm.
Dome formation assay
Twenty-four hours after HC11 transfection with scrambled versus Zfas1 siRNA, cells were seeded in six-well plates. Assays for dome formation were performed as documented previously (Naylor et al. 2005). Briefly, cell differentiation was induced by the addition of o-prolactin and dexamethasone. The number of domes in each well was counted. Results presented here are from three experiments with each individual assay performed in triplicate.
Statistical analyses
Two-tailed t-tests were performed for qPCR, proliferation, and dome formation assays. Standard error of the mean was calculated using Prism 5.0 (GraphPad Software, Inc.). Differential microarray expression analysis was performed by the LIMMA package using Bayesian statistics (B-statistics; posterior log odds) and Benjamini-Hochberg multiple testing adjustment (see Supplemental Methods).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTERESTS
A provisional patent for the use of ZFAS1 as a biomarker, therapeutic, and/or a therapeutic target has been filed.
ACKNOWLEDGMENTS
This work was supported by the National Health and Medical Research Council of Australia (Project Grant number 456080 and Career Development Award CDA631542; M.E.D.), the Australian Research Council (Federation Fellowship FF0561986; J.S.M.), a Queensland Government Department of Employment, Economic Development and Innovation Smart Futures Fellowship (M.E.D.), and a Clinical Fellowship from the Ludwig Institute for Cancer Research (A.C.V.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2528811.
REFERENCES
- Al-Shahrour F, Minguez P, Tárraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J 2006. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res 34: W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS 2008. Noncoding RNA in development. Mamm Genome 19: 454–492 [DOI] [PubMed] [Google Scholar]
- Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS 2009. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15: 2013–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS 2010. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39: D146–D151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak T, Blencowe BJ, Hughes TR 2005. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics 6: 104 doi: 10.1186/1471-2164-6-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger JH, Olsen GJ 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol Biol Evol 16: 512–524 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J 2005. Waste not, want not–transcript excess in multicellular eukaryotes. Trends Genet 21: 287–288 [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. 2007. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12: 215–229 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Dieci G, Preti M, Montanini B 2009. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 94: 83–88 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. 2008a. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS 2008b. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol 4: e1000176 doi: 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Mattick JS 2009. Pervasive transcription of the eukaryotic genome: Functional indices and conceptual implications. Brief Funct Genomics Proteomics 8: 407–423 [DOI] [PubMed] [Google Scholar]
- Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT 2009. Implication of snoRNA U50 in human breast cancer. J Genet Genomics 36: 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy EJ, McNeel DG 2005. Antigen-specific IgG elicited in subjects with prostate cancer treated with flt3 ligand. J Immunother 28: 268–275 [DOI] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci 102: 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, et al. 2006. Complex loci in human and mouse genomes. PLoS Genet 2: e47 doi: 10.1371/journal.pgen.0020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20: 1470–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Bailey TL, Kasukawa T, Mignone F, Kummerfeld SK, Madera M, Sunkara S, Furuno M, Bult CJ, Quackenbush J, et al. 2006. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol 3: 40–48 [DOI] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L 2010. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 32: 473–480 [DOI] [PubMed] [Google Scholar]
- Georgopoulou N, Hurel C, Politis PK, Gaitanou M, Matsas R, Thomaidou D 2006. BM88 is a dual function molecule inducing cell cycle exit and neuronal differentiation of neuroblastoma cells via cyclin D1 down-regulation and retinoblastoma protein hypophosphorylation. J Biol Chem 281: 33606–33620 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW 1998. Think globally, act locally: The making of a mouse mammary gland. Genes Dev 12: 449–455 [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW 2001. Signaling pathways in mammary gland development. Dev Cell 1: 467–475 [DOI] [PubMed] [Google Scholar]
- Hernandez A, Garcia B, Obregon MJ 2007. Gene expression from the imprinted Dio3 locus is associated with cell proliferation of cultured brown adipocytes. Endocrinology 148: 3968–3976 [DOI] [PubMed] [Google Scholar]
- Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH 2005. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA 11: 1303–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Rinn JL 2010. Large non-coding RNAs: Missing links in cancer? Hum Mol Genet 19: R152–R161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J 2001. RNomics: An experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 20: 2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007a. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR 2007b. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Keydar I, Chen L, Karby S, Weiss FR, Delarea J, Radu M, Chaitcik S, Brenner HJ 1979. Establishment and characterization of a cell line of human breast carcinoma origin. Eur J Cancer 15: 659–670 [DOI] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP 2010. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8 doi: 10.11126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasfargues EY, Coutinho WG, Dion AS 1979. A human breast tumor cell line (BT-474) that supports mouse mammary tumor virus replication. In Vitro 15: 723–729 [DOI] [PubMed] [Google Scholar]
- Master SR, Hartman JL, D'Cruz CM, Moody SE, Keiper EA, Ha SI, Cox JD, Belka GK, Chodosh LA 2002. Functional microarray analysis of mammary organogenesis reveals a developmental role in adaptive thermogenesis. Mol Endocrinol 16: 1185–1203 [DOI] [PubMed] [Google Scholar]
- Mazar J, Sinha S, Dinger ME, Mattick JS, Perera RJ 2010. Protein-coding and non-coding gene expression analysis in differentiating human keratinocytes using a three-dimensional epidermal equivalent. Mol Genet Genomics 284: 1–9 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long noncoding RNAs: Insights into function. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF 2010. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11: 14 doi: 10.1186/1471-2202-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe AD, Gilmore A, Klinowska T, Oliver J, Valentijn AJ, Brown R, Ross A, MacGregor G, Hickman JA, Streuli CH 1999. Developmental regulation of Bcl-2 family protein expression in the involuting mammary gland. J Cell Sci 112: 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T 1983. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods 65: 55–63 [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT 2009. GAS5, a non-protein-coding RNA, controls apoptosis and is down-regulated in breast cancer. Oncogene 28: 195–208 [DOI] [PubMed] [Google Scholar]
- Naylor MJ, Oakes SR, Gardiner-Garden M, Harris J, Blazek K, Ho TW, Li FC, Wynick D, Walker AM, Ormandy CJ 2005. Transcriptional changes underlying the secretory activation phase of mammary gland development. Mol Endocrinol 19: 1868–1883 [DOI] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS 2009. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol 182: 7738–7748 [DOI] [PubMed] [Google Scholar]
- Pelczar P, Filipowicz W 1998. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol Cell Biol 18: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GW, McKnight RA, Smith GH, Hennighausen L 1995. Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development 121: 2079–2090 [DOI] [PubMed] [Google Scholar]
- Song Z, Krishna S, Thanos D, Strominger JL, Ono SJ 1994. A novel cysteine-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and functions as a transcriptional repressor. J Exp Med 180: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule HD, Vazguez J, Long A, Albert S, Brennan M 1973. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst 51: 1409–1416 [DOI] [PubMed] [Google Scholar]
- Struhl K 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN varepsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA 2008. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci 105: 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM 2004. An abundance of bidirectional promoters in the human genome. Genome Res 14: 62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troester MA, Hoadley KA, Sorlie T, Herbert BS, Borresen-Dale AL, Lonning PE, Shay JW, Kaufmann WK, Perou CM 2004. Cell-type-specific responses to chemotherapeutics in breast cancer. Cancer Res 64: 4218–4226 [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L 2009. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9: 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, Hellen CU, Tiedge H 2005. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol 171: 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL 2009. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE 2005. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet 14: 1465–1474 [DOI] [PubMed] [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH 2006. snoSeeker: An advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res 34: 5112–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi I, Bini L, Albani D, Valaperta R, Liberatori S, Raggiaschi R, Montagna C, Susani L, Barbieri O, Pallini V, et al. 2002. Dome formation in cell cultures as expression of an early stage of lactogenic differentiation of the mammary gland. Proc Natl Acad Sci 99: 8660–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]