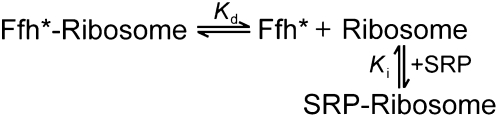Abstract
During cotranslational protein targeting by the Signal Recognition Particle (SRP), the correct cargo accelerates stable complex assembly between the SRP and SRP receptor (FtsY) by several orders of magnitude, thus enabling rapid and faithful cargo delivery to the target membrane. The molecular mechanism underlying this cargo-induced rate acceleration has been unclear. Here we show that the SRP RNA allows assembly of the SRP–FtsY complex to be specifically stimulated by a correct cargo, and, reciprocally, a correct cargo enables the SRP RNA to optimize its electrostatic interactions with FtsY. These results combined with recent structural work led us to suggest a “conformational selection” model that explains the synergistic action of the SRP RNA with the cargo in accelerating complex assembly. In addition to its previously proposed role in preventing the premature dissociation of SRP and FtsY, we found that the SRP RNA also plays an active role in ensuring the formation of productive assembly intermediates, thus guiding the SRP and FtsY through the most efficient pathway of assembly.
Keywords: Signal Recognition Particle, protein targeting, catalytic RNA, fluorescence, GTPases
INTRODUCTION
The Signal Recognition Particle (SRP) is a key cellular machinery responsible for the cotranslational targeting of proteins to their proper membrane destinations (Walter and Johnson 1994). SRP recognizes ribosome-nascent chain complexes (referred to as the RNC or cargo) carrying strong signal sequences (Pool et al. 2002; Halic et al. 2004, 2006; Schaffitzel et al. 2006) and delivers the cargo to protein translocation machineries on the target membrane (Gilmore et al. 1982a,b). The simplest SRP, found in prokaryotes, is a ribonucleoprotein complex comprised of the SRP54 protein (called Ffh in bacteria) and the 4.5S SRP RNA (Walter and Johnson 1994). Ffh and the SRP receptor (called FtsY in bacteria) each contains a conserved NG-domain, comprised of a GTPase G-domain and a helical N-domain (Freymann et al. 1997; Montoya et al. 1997). Direct interaction between the NG-domains of Ffh and FtsY (Egea et al. 2004; Focia et al. 2004) mediates the delivery of RNC to the target membrane. An additional M-domain in Ffh contains the binding sites for the SRP RNA and for the signal sequence emerging from the translating ribosome (Zopf et al. 1990; Keenan et al. 1998; Batey et al. 2000; Janda et al. 2010).
Previous kinetic and biophysical analyses showed that assembly of a stable SRP–FtsY complex is a dynamic process involving at least two distinct conformational stages (Zhang et al. 2008, 2009). Complex assembly initiates with the formation of a transient “early” intermediate, which forms quickly (kon = 5.6 × 106 M−1 sec−1) but is highly labile (koff = 62 sec−1); this intermediate has loose contacts between the G-domains and hence can form in the presence or absence of GTP (Zhang et al. 2008). Subsequently, extensive conformational changes are required to bring the G-domains of both proteins into close contact with one another and to allow their bound GTP molecules to directly hydrogen-bond across the dimer interface, thus giving a highly specific, GTP-dependent stable complex (Egea et al. 2004; Focia et al. 2004). Due to the extensive rearrangements required to form these specific and extensive interface contacts, assembly of a stable complex by the free SRP and FtsY is thermodynamically favorable but kinetically very slow (Peluso et al. 2001). In the presence of a correct cargo carrying a strong signal sequence, however, stable complex assembly between the SRP and FtsY is accelerated >102-fold (Zhang et al. 2009, 2010). This stimulation enables rapid delivery of the correct cargos to the target membrane and provides kinetic discrimination against the incorrect cargos to improve the fidelity of protein targeting (Zhang et al. 2009, 2010).
How does the cargo, whose signal sequence binds to the M-domain of the SRP, induce much more efficient assembly of the GTPase complex? The M-domain of SRP is connected to its NG-domain via a flexible linker, and no direct interaction has been detected between these two domains, making a direct communication via the M-domain less likely. The other essential component of the SRP, the SRP RNA, is a more likely candidate to mediate the allosteric communication between the cargo and the GTPases. The SRP RNA binds to a helix–turn–helix motif in the M-domain close to the signal sequence binding site (Batey et al. 2000), and cryo-EM and cross-linking analyses indicated that the SRP RNA forms close contacts with the ribosome (Gu et al. 2003; Ullers et al. 2003; Halic et al. 2006; Schaffitzel et al. 2006). On the other hand, the SRP RNA has also been shown to communicate with the GTPases, accelerating SRP–FtsY complex assembly ∼102-fold (Peluso et al. 2000, 2001). We recently showed that the conserved GGAA tetraloop of the SRP RNA makes a transient interaction with basic residues on the SRP receptor (primarily Lys 399 in Escherichia coli FtsY) (Shen and Shan 2010; Estrozi et al. 2011); this electrostatic interaction stabilizes the early intermediate and prolongs its lifetime, thus giving the GTPases a longer time window to rearrange to the stable complex. Importantly, the stimulatory effect of the SRP RNA was only observed in the presence of a signal peptide or a stimulatory detergent, Nikkol, that mimics the effect of signal peptide (Bradshaw et al. 2009), strongly suggesting that the SRP RNA can sense information about signal sequence binding in the M-domain and, in response, turns on its stimulatory activity on the GTPases. However, the mechanism by which the SRP RNA bridges between the cargo and the GTPases remains unclear.
Integrating the information from the recent cryo-EM structures of the RNC–SRP complex in the context of the biochemical results begins to shed light on this question. These structural analyses revealed extensive interactions between the RNC and the SRP including: (1) contacts of the tip of Ffh's N-domain with ribosomal proteins L23p and L29p; (2) interaction of the emerging signal peptide with the signal sequence binding groove in the Ffh M-domain; and (3) contacts of the SRP RNA with ribosomal proteins L17p and L18p (Halic et al. 2006; Schaffitzel et al. 2006). Importantly, these contacts position the GGAA tetraloop of the SRP RNA next to the NG-domain of the SRP, which would allow the electrostatic interaction between the GGAA tetraloop and the basic residues on FtsY to be more readily established (Estrozi et al. 2011).
In this study, we tested this hypothesis and probed the mechanism of action of the SRP RNA by systematically examining how the ribosome, the signal sequence, and the SRP RNA cooperate with one another to provide maximal acceleration of SRP–FtsY complex assembly. We showed that the SRP RNA allows assembly of the SRP–FtsY complex to be specifically stimulated by a correct cargo, and, conversely, a correct cargo enables the SRP RNA to optimize its electrostatic interaction with FtsY and thus maximize its stimulatory effect on complex assembly. Moreover, in addition to acting as a passive tether that holds the SRP and FtsY GTPases together, the SRP RNA plays an essential role in preventing the formation of non-productive intermediates, thus guiding complex assembly through the most efficient and productive pathway.
RESULTS
SRP RNA does not affect the cargo binding affinity of SRP
Previous cross-linking (Gu et al. 2003; Ullers et al. 2003) and cryo-EM (Halic et al. 2006; Schaffitzel et al. 2006) studies suggested that the SRP RNA makes extensive contacts with the ribosome, which raised the possibility that the SRP RNA could enhance the binding affinity between the SRP and the RNC. To test this hypothesis, we used fluorescence anisotropy to determine the binding affinity between Ffh and purified RNCs in the absence and presence of the SRP RNA (Fig. 1). Ffh was labeled with fluorescein at residue 421 in the M-domain, and the binding of RNC was detected as an increase in the fluorescence anisotropy of Ffh 421-fluorescein (Zhang et al. 2010). Based on this anisotropy change, equilibrium titrations were carried out to test the effect of the SRP RNA on two different ribosomal complexes: (1) RNCFtsQ, a correct cargo that carries the N-terminal 76 residues of FtsQ, a model SRP substrate (Fig. 1A); and (2) RNCLuciferase, an incorrect cargo that carries the N-terminal 50 amino acids of firefly luciferase, a cytosolic protein (Fig. 1B). Consistent with previous observations (Zhang et al. 2010), Ffh binds to RNCFtsQ with an equilibrium dissociation constant (Kd) of 0.87 nM, ∼102-fold lower than that to RNCLuciferase. Unexpectedly, neither of these binding affinities was substantially enhanced by the SRP RNA (Fig. 1A,B; summarized in Fig. 1F, Ffh vs. SRP). Thus, the SRP RNA does not strengthen the binding affinity between Ffh and its cargo.
FIGURE 1.
The SRP RNA does not strengthen the binding between SRP and the RNC. (A) Equilibrium titrations for the binding of Ffh (□ and dashed line) or SRP (○ and solid line) to RNCFtsQ, performed and analyzed as described in the Materials and Methods. (B) Equilibrium titrations for the binding of Ffh (□ and dashed line) or SRP (○ and solid line) to RNCLuciferase. (C,D) Binding of empty ribosomes to Ffh (C) or SRP (D), determined using equilibrium titrations and competition experiments, respectively, as described in the Materials and Methods. (E) Binding of Ffh to RNCFtsQ in the presence (○ and solid line) and absence (□ and dashed line) of Nikkol. (F) Summary of the Kd values obtained from A to E.
Because most of the contacts between the SRP RNA and RNC are mediated by the ribosome, we further tested the effect of the SRP RNA on the binding affinity of Ffh to empty ribosomes. Binding of ribosomes to Ffh induced a significant change in the fluorescence anisotropy of a coumarin dye labeled at residue 153 in the Ffh G-domain; based on this anisotropy change, the Kd value for Ffh-ribosome binding was determined to be 24 nM (Fig. 1C,F). In the presence of the SRP RNA, however, the binding of ribosomes did not induce a sufficiently large anisotropy change to allow for accurate equilibrium titrations; therefore, the binding affinity of SRP for ribosomes was determined by using unlabeled SRP as a competitive inhibitor for the binding between Ffh C153-coumarin and the ribosome (Fig. 1D; see Materials and Methods). This competition experiment gave a Kd value of 71 nM for SRP-ribosome binding, threefold weaker than in the absence of the SRP RNA (Fig. 1D,F). Together, these results indicate that the SRP RNA does not exert its stimulatory effect on the protein targeting reaction by helping to recruit Ffh to the ribosome. Instead, interactions of the SRP RNA with the ribosome may help position the SRP into a more active conformation that serves to stimulate subsequent steps in the pathway (see sections below).
It has been suggested that a stimulatory detergent included in previous biochemical assays, Nikkol, mimics the signal peptide and binds to the signal sequence binding groove of Ffh (Bradshaw et al. 2009). If this were the case, Nikkol might compete with the RNC for binding to Ffh. To test whether this possibility interferes with the SRP–RNC binding measurements above, we compared the binding affinity of Ffh for RNCFtsQ in the presence and absence of saturating (0.01%) Nikkol (Fig. 1E). No significant difference was observed in these binding affinities (Fig. 1E,F), suggesting that Nikkol does not directly or effectively compete with the RNC for binding Ffh.
A correct cargo enables the SRP RNA to most effectively stimulate SRP–FtsY complex formation
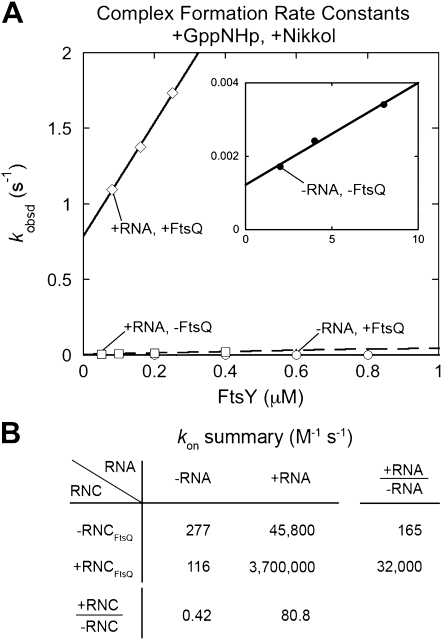
In the presence of the stimulatory detergent Nikkol, the SRP RNA has been shown to accelerate the assembly of a stable SRP–FtsY complex ∼100-fold (Peluso et al. 2000, 2001; Bradshaw et al. 2009). A correct cargo, such as RNCFtsQ, provides an additional 100–400-fold acceleration of complex assembly (Zhang et al. 2009). As the SRP has a limited time window to complete the protein targeting reaction (∼3–5 sec) (Siegel and Walter 1988; Zheng and Gierasch 1996; Flanagan et al. 2003), these rate accelerations are crucial for bringing the complex formation kinetics to values suitable for biological function. To test whether the SRP RNA is required for the cargo-mediated acceleration of complex assembly, we directly measured and compared the effect of these different activators using Forster Resonance Energy Transfer (FRET) between donor (DACM)- and acceptor (BODIPY-FL)-labeled Ffh and FtsY, respectively (Fig. 2). In the absence of RNCFtsQ, the SRP RNA induced a 165-fold stimulation of SRP–FtsY complex assembly (Fig. 2A, +RNA −FtsQ [□] vs. −RNA −FtsQ [○]; Fig. 2B), in agreement with previous studies (Zhang et al. 2009). RNCFtsQ had a marginal effect on the rate of complex assembly (approximately twofold) in the absence of the SRP RNA, but provided an additional 80-fold acceleration of complex assembly in the presence of the SRP RNA. Thus, the correct cargo enabled a much larger stimulatory effect of the SRP RNA, 32,000-fold (Fig. 2A, +RNA +FtsQ [⋄] vs. −RNA +FtsQ [○]; Fig. 2B).
FIGURE 2.
A correct cargo allows the SRP RNA to more strongly stimulate SRP–FtsY complex assembly. (A) SRP–FtsY complex formation rates in the presence and absence of the SRP RNA and RNCFtsQ, measured using the FRET assay as described in the Materials and Methods. All measurements in this figure were carried on in the presence of 0.01%–0.02% Nikkol. (B) Summary of complex formation rate constants from A.
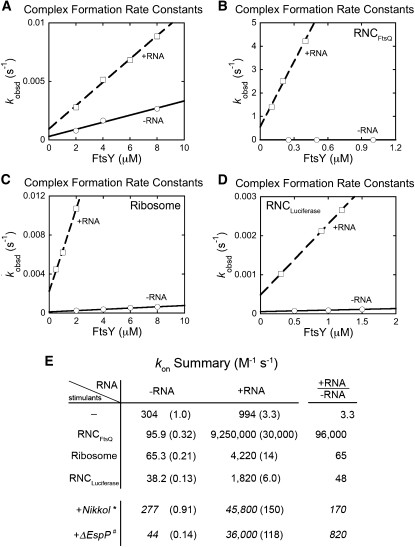
To dissect the contributions of the signal sequence and the ribosome to the stimulatory effect of cargo, we measured and compared the complex assembly rates when SRP is bound with three distinct complexes: the empty ribosome, RNCLuciferase, and RNCFtsQ. Furthermore, a recent study suggested that the detergent Nikkol partially mimics the effect of signal peptides to stimulate SRP–FtsY complex assembly (Bradshaw et al. 2009) and may thus mask the effect of the SRP RNA and cargo in stimulating complex assembly in previous studies (Zhang et al. 2010) and the above (Fig. 2) analyses. To uncover the full extent of stimulation provided by the cargo and the SRP RNA, we carried out these measurements in the absence of Nikkol (Fig. 3).
FIGURE 3.
Both the ribosome and a strong signal sequence are required to maximize the stimulatory effects of the SRP RNA on SRP–FtsY complex assembly. All experiments in this figure were performed in the absence of Nikkol. (A–D) SRP–FtsY complex assembly rates with (□ and dashed line) or without (○ and solid line) the SRP RNA, in the presence of (A) no stimulants, (B) RNCFtsQ, (C) empty ribosome, and (D) RNCLuciferase. (E) Summary of complex formation rates constants from A–D. The rate constants in the presence of Nikkol (*) are from Figure 2, and the data of ΔEspP (#) are from Bradshaw et al. (2009).
In the absence of the ribosome complexes or signal peptides, complex assembly between Ffh and FtsY is extremely slow, with rate constants of ∼3 × 102 M−1 sec−1, and the SRP RNA has only a small stimulatory effect on complex formation, ∼3.3-fold (Fig. 3A). In contrast, the correct cargo, RNCFtsQ, allows the SRP RNA to accelerate complex assembly 96,000-fold (Fig. 3B,E). Empty ribosomes and incorrect cargos such as RNCLuciferase allowed only partial stimulation by the SRP RNA, with the RNA providing rate enhancements of 65-fold and 48-fold, respectively (Fig. 3C,D). In comparison, a signal peptide ΔEspP used in a previous study (Bradshaw et al. 2009) and the signal peptide mimic Nikkol have been shown to provide partial rate accelerations by the SRP RNA of 160-fold and 820-fold, respectively (Fig. 3E; Bradshaw et al. 2009). Finally, both the cargo and the SRP RNA were required for the observed rate accelerations, as in the absence of the RNA, neither the ribosomal complexes nor signal peptides (or signal peptide mimics) stimulated complex assembly; but, rather, these components had a small inhibitory effect (Fig. 3E, −RNA column). Taken together, these results demonstrate that the SRP RNA is essential for the cargo to stimulate the SRP–FtsY interaction, and, reciprocally, a correct cargo enables the SRP RNA to maximize its stimulatory effect on assembly of the GTPase complex.
A correct cargo optimizes the electrostatic interaction of the SRP RNA with the SRP receptor
During complex formation between SRP and FtsY, a key electrostatic interaction between the GGAA tetraloop on the SRP RNA and Lys 399 on FtsY must be established (Shan et al. 2004; Siu et al. 2007; Zhang et al. 2008; Shen and Shan 2010). This interaction provides a transient tether that holds the GTPases together during the transition state of complex assembly and is essential for mediating the RNA-induced rate accelerations (Shen and Shan 2010). Because the cargo makes extensive interactions with the SRP RNA and the M- and NG-domains of Ffh (Halic et al. 2006; Schaffitzel et al. 2006), we hypothesized that a correct cargo could help position the SRP into a more active conformation that optimizes the electrostatic interaction between the RNA tetraloop and the incoming FtsY (Estrozi et al. 2011). If this were the case, then this electrostatic interaction would provide a much larger contribution to complex assembly with the correct than the incorrect cargos. To test this hypothesis, we used the FRET assay to measure and compare how the complex assembly rates were affected by either the mutant FtsY-K399A or an RNA tetraloop mutant, RNA-GAAU, which specifically disrupts this electrostatic interaction (Zhang et al. 2008; Shen and Shan 2010).
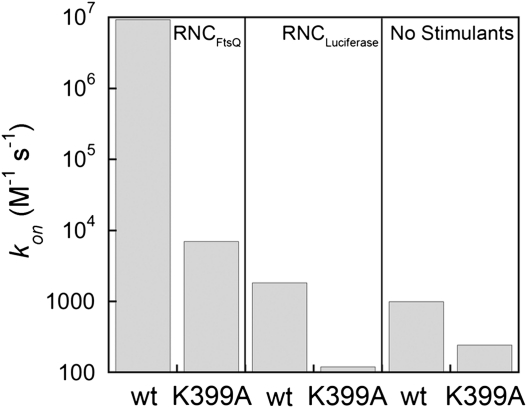
In the presence of RNCFtsQ, mutation of FtsY-Lys399 resulted in a 103-fold reduction in the rate constant of SRP–FtsY complex assembly (Fig. 4; Supplemental Fig. 1A,C). Mutation of the RNA tetraloop exhibited a similar phenotype (Supplemental Fig. 1A,C), and the combination of both mutations decreased the complex assembly rates another 10-fold, approaching the value observed in the absence of the RNA (Supplemental Fig. 1A,C). In contrast, in the presence of an incorrect cargo, RNCLuciferase, the FtsY-K399A mutation caused only an 18-fold reduction in complex assembly rates (Fig. 4; Supplemental Fig. 1B,C). In the absence of cargo or stimulatory reagents (Nikkol or signal peptides) that mimic the effect of cargo, this interaction contributed only fourfold to SRP–FtsY complex assembly (Fig. 4; Supplemental Fig. 1C). The reduced contribution of FtsY-Lys399 and RNA tetraloop to complex assembly in the absence of the correct cargo closely correlated with the reduced stimulatory effect of the SRP RNA under these conditions (cf. Fig. 4 with Fig. 3E). Together, these results suggest that the electrostatic interaction between the RNA tetraloop and FtsY-Lys399 is most effectively established in the presence of a correct cargo, whereas incorrect cargos are much less effective in positioning the SRP to make this interaction.
FIGURE 4.
A correct cargo optimizes the electrostatic interaction of the SRP RNA tetraloop with FtsY-K399 during SRP–FtsY complex formation. The contribution of this electrostatic interaction is compared in the presence of RNCFtsQ (left), RNCLuciferase (middle), and no stimulants (right). The rate constants are from the data in Supplemental Figure 1.
The SRP RNA ensures the formation of a productive early intermediate
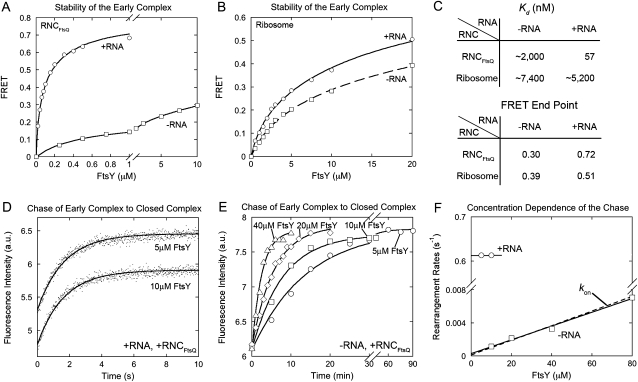
In the presence of the SRP RNA, assembly of a stable SRP–FtsY complex is preceded by a transient early intermediate (Zhang et al. 2008). In previous work, we found that the SRP RNA stabilizes the early intermediate, which provides a longer time window for the intermediate to undergo its subsequent rearrangement and thus accelerates the formation of the stable complex (Shen and Shan 2010). We therefore asked if correct cargos are also essential for the ability of the SRP RNA to stabilize the early intermediate. To this end, we isolated the early intermediate by leaving out GTP or non-hydrolyzable GTP analogs, which blocks the rearrangement of this intermediate to the subsequent stable complex. FRET between SRP and FtsY was used to measure the equilibrium stability of the early intermediate. When SRP was loaded with RNCFtsQ, SRP and FtsY formed a stable early complex in the presence of the SRP RNA, with a Kd value of 57 nM (Fig. 5A, ○; Fig. 5C), 150-fold more stable compared to that without RNC (8.8 μM) (Zhang et al. 2008; Shen and Shan 2010). In the absence of the SRP RNA, however, the early intermediate was considerably less stable, with a Kd value of ∼2 μM (Fig. 5A, □; summarized in Fig. 5C), indicating that the SRP RNA stabilizes the early intermediate ∼40-fold when the SRP is loaded with a correct cargo. In contrast, when the SRP is loaded with empty ribosomes, the SRP RNA has a negligible effect on the stability of the early intermediate (Fig. 5B,C). Thus, correct cargos bearing strong signal sequences also enable the SRP RNA to exert its stabilizing effect on the early intermediate.
FIGURE 5.
The SRP RNA ensures the formation of a stable and productive early intermediate. (A,B) Effects of the SRP RNA on the equilibrium stability of the early complex in the presence of RNCFtsQ (A) or the empty ribosome (B). (C) Summary of the Kd values and FRET end points of the early complex from A and B. (D,E) Rate constants for formation of the stable complex starting from a preformed cargo–Ffh–FtsY early complex in the presence (D) and absence (E) of the SRP RNA. (F) Dependence of the rearrangement rate constants on FtsY concentration, obtained from D and E, in the presence (○) and absence (□) of the SRP RNA. The dashed line shows the rate constant for complex assembly starting from free Ffh and FtsY.
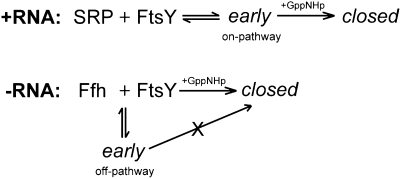
We also noted that in the absence of the SRP RNA, the FRET end point of the early intermediate at saturating FtsY concentrations was much lower (∼0.3) (Fig. 5A, □; Fig. 5C) than that in the presence of the SRP RNA (∼0.7) (Fig. 5A, ○; Fig. 5C). This indicated that the early intermediate formed in the absence of the SRP RNA has a different conformation from that formed in the presence of the RNA and raised the question of whether the former are productive, “on-pathway” intermediates, in other words, whether they can efficiently rearrange to form the stable complex. To address this question, we measured the rate constant of this rearrangement by preforming the early complex using saturating FtsY in the absence of nucleotides and triggered the formation of the stable complex from this intermediate by the addition of GppNHp. Stable complex formation was detected using an environmentally sensitive dye, acrylodan, labeled at residue 235 of Ffh, which specifically changes fluorescence upon formation of the stable complex (Zhang et al. 2008). If the early intermediate is productive and on-pathway, the kinetics of stable complex formation should be independent of FtsY concentration, reflecting the first-order rate constant for the early → stable rearrangement (Scheme 1, +RNA). This was indeed the case for the early complex formed with RNCFtsQ in the presence of SRP RNA: the stable complex was quickly generated with a rate constant of 0.61 sec−1, and this rate constant was independent of FtsY concentration (Fig. 5D,F, ○). In contrast, in the absence of the SRP RNA, the kinetics for attaining the stable complex was ∼104-fold slower (note the difference in time scales in Fig. 5D vs. E). Furthermore, the observed rate constants for formation of the stable complex became linearly dependent on FtsY concentration up to 80 μM and overlapped well with the second-order rate constants for assembly of this complex starting from free Ffh and FtsY (Fig. 5E,F, □). These results strongly suggest that in the absence of the SRP RNA, rearrangement of the early intermediate to the stable complex is extremely slow; instead, the GTPases must first dissociate into free Ffh and FtsY and then generate the stable complex via an alternative pathway (Scheme 1, −RNA). Thus, the SRP RNA not only stabilizes the early intermediate, but also ensures a productive conformation of this intermediate in the presence of a correct cargo.
SCHEME 1.
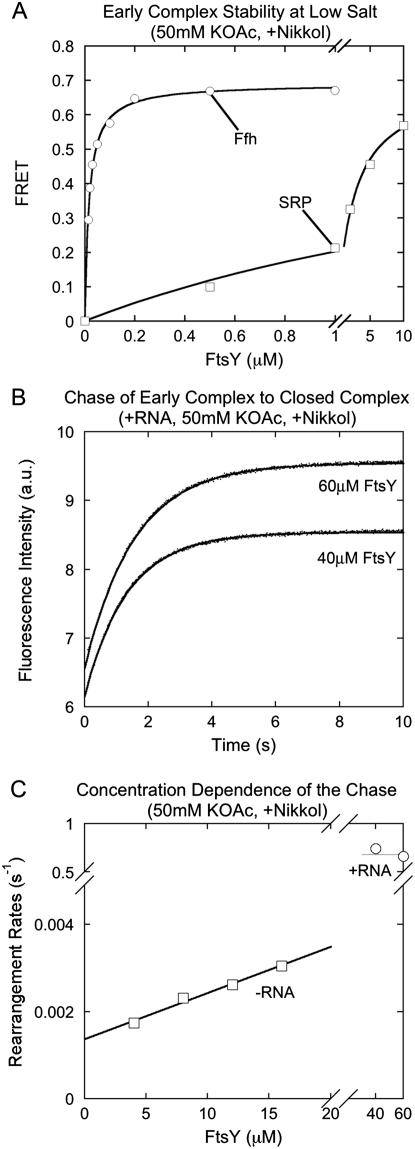
The notion that the SRP RNA can actively modulate the conformation of the early intermediate was further supported by observations under low salt (50 mM KOAc) conditions. Under these conditions, Ffh and FtsY assembled an extremely tight early complex (Fig. 6A, ○; Kd = 17 nM), presumably because the early encounter between these two GTPases is driven by electrostatic interactions (Zhang et al. 2011) that are stabilized by decreasing ionic strength. However, this did not lead to a corresponding acceleration of the assembly of the stable complex (Fig. 6C, −RNA; Shen and Shan 2010); furthermore, the early intermediate formed under these conditions reached the stable conformation slowly and with a strong FtsY concentration dependence (Fig. 6C, −RNA). These results indicate that the early intermediate formed under these conditions, although stable, is non-productive and off-the-pathway. Intriguingly, the SRP RNA reduced the affinity of the early complex formed under low salt conditions by 150-fold (Fig. 6A, □), in contrast to its stabilizing effect on the early complex at higher salt conditions (Fig. 5A; Zhang et al. 2008; Shen and Shan 2010). In addition, the presence of SRP RNA allowed the intermediate to quickly and directly rearrange to the stable complex (Fig. 6B,C, +RNA), suggesting that it also restored the productivity of the early intermediate under these conditions. Together, these results suggest that aside from preventing the premature disassembly of the early intermediate, the SRP RNA is essential for ensuring a productive conformation of this intermediate, such that it can readily rearrange to the subsequent conformations in response to its biological cues.
FIGURE 6.
SRP RNA prevents the formation of a non-productive early intermediate at low salt concentrations. (A) The stability of the early intermediate at 50 mM KOAc. The Kd values are 17 nM for Ffh (○) and 2.5 μM for SRP (□). (B) Rate constants for formation of the stable complex from a preassembled early intermediate in the presence of the SRP RNA. Exponential fits of data gave rearrangement rate constants of 0.74 sec−1 and 0.66 sec−1 at 40 and 60 μM FtsY, respectively. (C) FtsY concentration dependence of the rate constants for formation of the stable complex starting from the early intermediate in the presence (○) and absence (□) of the SRP RNA. Linear fit of the −RNA data gave a slope of 106 M−1 sec−1, consistent with the kon value of 159 M−1 sec−1 measured under the same conditions (Shen and Shan 2010).
DISCUSSION
Protein targeting by the SRP requires efficient and faithful delivery of translating ribosomes to the target membrane in response to correct signal sequences. Recent work strongly suggested that this is achieved, in part, by having an intrinsically slow rate of SRP–FtsY complex assembly that minimizes the delivery of incorrect cargos and allowing efficient complex assembly only when RNCs carrying strong signal sequences are loaded on the SRP (Zhang et al. 2010). In this study, we showed that the SRP RNA plays an essential role in enabling specific stimulation of SRP–FtsY complex formation by a correct cargo. The strong synergistic effect between the SRP RNA and the cargo, in combination with recent structural work, suggests a potential mechanism for how a correct cargo stimulates SRP–FtsY complex assembly. Finally, the SRP RNA actively modulates the conformation of the SRP–FtsY complex to guide complex assembly through the most efficient and productive pathway.
Previous work has reported that the cargo and the SRP RNA each accelerates SRP–FtsY complex assembly ∼102-fold (Peluso et al. 2000, 2001; Zhang et al. 2008, 2009). A recent study, however, suggested that the full extent of their stimulatory effects might be masked because a detergent Nikkol used in previous studies mimics the effect of signal peptides (Bradshaw et al. 2009). Indeed, omission of this detergent revealed much larger stimulatory effects from both the cargo and the SRP RNA, 104-fold (herein; Zhang et al. 2010) and 105-fold (herein), respectively. These results emphasize the essential role of the SRP RNA in cotranslational protein targeting and the specificity that it confers on the correct cargo during SRP–FtsY complex assembly.
Comparison of the complex assembly rates when the SRP is loaded with the correct or incorrect cargos illustrates the kinetic discrimination that the SRP RNA provides. The intrinsic complex assembly rate between Ffh and FtsY is extremely slow, on the order of 102 M−1 sec−1, and is only marginally (threefold) stimulated by the SRP RNA. In the presence of the SRP RNA, complex assembly is accelerated ∼104-fold by the correct cargo but only ∼10-fold by empty ribosomes or incorrect cargos. This allows the correct cargo to gain a 103-fold kinetic advantage over incorrect cargos during its delivery to the target membrane. In contrast, in the absence of the SRP RNA, none of the ribosomal complexes provide any rate acceleration and are, in fact, slightly inhibitory. The low efficiency of complex formation in the absence of the SRP RNA is accompanied by a loss of specificity. Thus, the SRP RNA enables Ffh to sense the cues from correct signal sequences and to kinetically discriminate against incorrect signal sequences during the cargo delivery step, introducing both efficiency and specificity.
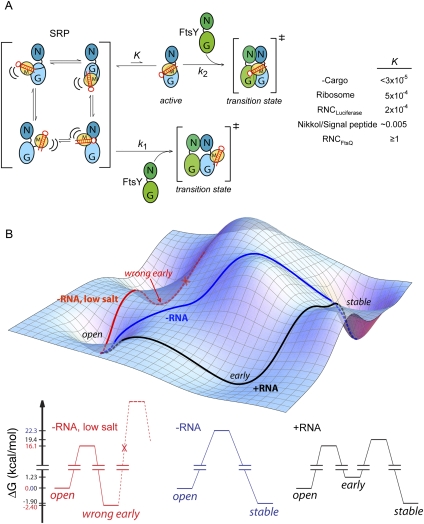
How is SRP–FtsY complex assembly specifically stimulated by a correct cargo? Recent biochemical and structural work has provided important clues. First, previous biochemical analyses and structural probing experiments have established an important role of the electrostatic interaction between the SRP RNA's GGAA tetraloop and FtsY-Lys399 in facilitating SRP–FtsY complex assembly (Siu et al. 2007; Zhang et al. 2008; Shen and Shan 2010). The analyses herein showed that a correct cargo maximizes the stimulatory effect of the SRP RNA on complex assembly, and this is paralleled by an optimization of the electrostatic interaction between the RNA tetraloop and FtsY-Lys399 in the presence of the correct cargo. Second, cryo-EM analyses of the RNC–SRP and RNC–SRP–SR early complexes have revealed extensive interactions of the Ffh M- and NG-domains with the nascent polypeptide and ribosomal proteins L23/ L29, respectively (Halic et al. 2006; Schaffitzel et al. 2006; Estrozi et al. 2011); these interactions help position the SRP RNA's tetraloop adjacent to the Ffh NG-domain, allowing it to be poised for contacting the incoming FtsY. In contrast, the free SRP has been found in at least four different conformations (Keenan et al. 1998; Rosendal et al. 2003; Buskiewicz et al. 2005; Spanggord et al. 2005); in each of these structures, the SRP RNA was oriented differently with respect to the NG-domain, and in most structures, the RNA tetraloop pointed away from the Ffh–FtsY interface (Fig. 7A, bracket). Given these observations and the flexibility of the linker connecting the Ffh M- and NG-domains, we speculate that free SRP has a relative flat conformational space and is able to adopt a variety of “latent” conformations in which the RNA tetraloop is not well positioned (Fig. 7A, bracket). The action of the RNC could be likened to a conformational selection process (Fig. 7A): A correct cargo selectively stabilizes the rare but active conformation of SRP in which the RNA tetraloop is optimally positioned to interact with FtsY, thereby activating the SRP RNA to achieve efficient SRP–FtsY complex assembly.
FIGURE 7.
Model for how the SRP RNA and cargo act synergistically to stimulate complex assembly. (A) The SRP exists in an equilibrium (K) between an active conformation, in which the SRP RNA tetraloop is positioned to contact the incoming FtsY, and an ensemble of latent conformations (bracket on left) in which the RNA is not properly oriented. The observed rate constant for complex assembly is a weighted average of complex assembly rates from these different conformations. Using this model, the effects of different stimulatory factors on the conformational equilibrium of SRP were estimated from their respective complex assembly rate constants, as described in the Materials and Methods. (B) The SRP RNA guides complex assembly through a more efficient pathway. In the presence of the SRP RNA, efficient complex assembly occurs through formation of a productive and stabilized early intermediate, which can readily rearrange to the stable complex (black). Without the SRP RNA, however, the early complex is unstable and nonproductive; hence stable complex assembly likely proceeds through an alternative pathway that bypasses this intermediate (blue). Under low salt conditions, Ffh and FtsY could be trapped in a tight but nonproductive early complex (red) without the SRP RNA. All three cases refer to reactions with cargo (black or blue) or Nikkol (red) present.
Using this model and the observed SRP–FtsY complex assembly rate constants, a rough estimate could be made for how the ribosome, signal peptides, and different cargos affect the conformational equilibrium of SRP (Fig. 7A, right panel). As the complex assembly rate constant levels off at ∼9 × 106 M−1 sec−1 for RNCs with increasingly strong signal sequences, this value likely represents the rate of complex assembly from the “active” SRP molecules (Fig. 7A, k2), and the SRPs bound to a correct cargo such as RNCFtsQ are primarily in this conformation (Fig. 7A, K ≥ 1). With free SRP, however, only a small fraction of the molecules exist in the “active” conformation (K ≤ 3 × 10−5). The ribosome and signal peptides each shifts the conformational equilibrium of SRP toward the active state by ∼10-fold and ∼102-fold, respectively, but the SRP would still be primarily in the “latent” conformations in the presence of these isolated components of the cargo. This analysis also shows that the additive effect of the ribosome and signal peptides (∼103-fold) is at least an order of magnitude lower than that provided by a correct cargo (≥3 × 104-fold), suggesting a modest but significant degree of cooperativity between the different components of the RNC.
In previous studies, the catalytic effect of the SRP RNA was attributed to its role as a transient “tether”: By holding the Ffh and FtsY GTPases together in the early intermediate, the SRP RNA prevents the premature disassembly of this intermediate and thereby increases the probability of obtaining the stable complex (Peluso et al. 2000; Shen and Shan 2010). In addition to this role, several observations in this study indicate that the SRP RNA also actively modulates the conformation of the early intermediate. First, the early intermediate formed in the absence of the SRP RNA has a much lower FRET value than that formed in the presence of the RNA. Second, the early intermediate formed in the presence of the SRP RNA directly and rapidly rearranges to the stable complex (0.5–1 sec−1), whereas in the absence of the SRP RNA, the intermediate is off-the-pathway, and complex assembly has to go through an alternative pathway that bypasses this intermediate (Fig. 7B, blue lines). Third, under low salt conditions, Ffh and FtsY are trapped in a stable but off-pathway intermediate (Fig. 7B, red lines), and the SRP RNA destabilizes this intermediate while restoring its kinetic competence to rearrange to the stable complex. Together, these results strongly suggest that the SRP RNA is essential for ensuring a productive conformation of the early intermediate and actively guides complex assembly through the most efficient and productive pathway (Fig. 7B, black lines).
MATERIALS AND METHODS
Materials
E. coli Ffh, FtsY, and SRP RNA were expressed and purified using established protocols (Peluso et al. 2001). Mutant proteins and SRP RNA were constructed using the QuickChange mutagenesis protocol (Stratagene) and were purified as described previously (Peluso et al. 2001). Fluorescent dyes fluorescein, BODIPY-FL, DACM, and acrylodan were purchased from Invitrogen. 70S ribosome was purified as described (Moazed and Noller 1989). RNCs were prepared and purified as described (Schaffitzel and Ban 2007).
Fluorescence experiments
Single cysteine mutants of Ffh and FtsY were labeled using maleimide chemistry and purified as described (Zhang et al. 2008). Labeling efficiency was usually ≥95%. Fluorescence measurements were carried out on a FluoroLog-3-22 spectrofluorometer (Jobin-Yvon) in assay buffer [50 mM KHEPES at pH 7.5, 150 mM KOAc, 10 mM Mg(OAc)2, 2 mM DTT, with or without 0.01% Nikkol].
The binding affinities of SRP for RNCs or ribosomes were determined using fluorescence anisotropy measurements, using either one of the two procedures below. (1) Varying concentrations of RNCFtsQ or RNCLuciferase were added to a small, fixed amount of C421-fluorescein labeled Ffh or SRP. Anisotropy values were calculated and plotted against RNC (or ribosome) concentration, and quadratic fits of the data gave the Kd value of the complex (Zhang et al. 2010). (2) Varying amounts of unlabeled SRP were added as competitive inhibitors to a ribosome–Ffh complex formed by C153-coumarin-labeled Ffh (Scheme 2, Ffh*). Under conditions in which the concentration of labeled Ffh (Ffh*) was low (10 nM) compared to that of ribosome (≥50 nM), the observed anisotropy value (Aobsd) as a function of competitor concentration can be approximated by Equations 1 and 2, derived from Scheme 2, to obtain the Ki value:
 |
where
 |
In Equations 1 and 2, A1 is the anisotropy value of the Ffh*–ribosome complex, A0 is the anisotropy value of free Ffh*; [SRP]0 and [R]0 are the total concentrations of SRP and ribosome, respectively; and the equilibrium constants Ki and Kd are defined in Scheme 2.
SCHEME 2.
Association rate constants for SRP–FtsY complex formation were determined using FRET. In all cases, saturating concentrations of ribosome or ribosomal complexes were used so that ≥90% of SRP was bound with the different cargos. Complex assembly was initiated by mixing SRP with varying amounts of FtsY in the presence of 100 μM GppNHp, and the time course of fluorescence change was monitored using either a FluoroLog-3-22 spectrofluorometer (Jobin-Yvon) or an SF-2004 stopped-flow (KinTek). Linear fits of the observed rate constants as a function of FtsY concentration gave the second-order association rate constant (kobsd = kon[FtsY] + koff). Equilibrium titrations of the early intermediate were carried out using FRET as described previously (Zhang et al. 2008). Rate constants of stable complex formation from the early intermediate were measured using Ffh-C235 labeled with acrylodan. The reaction was initiated by mixing 500 μM GppNHp with a preformed early intermediate, assembled in the presence of saturating amounts of SRP/Ffh and FtsY with respect to the Kd value of the early intermediate. The time course of fluorescence change was fit to single-exponential functions to give the observed rate constants. For experiments concerning SRP or Ffh loaded with different ribosome complexes, ribosome or RNC concentrations fivefold to 100-fold above their respective Kd for Ffh were used to ensure >90% occupancy of SRP or Ffh by the cargo.
Estimation of the partition of SRP into the active conformation
The fractions of SRP molecules in the active conformation under different conditions in Figure 7A were estimated using the complex assembly rate constants determined here and previously (Zhang et al. 2010). As the observed complex assembly rate constant levels off at a value of 9 × 106 M−1 sec−1 with increasingly strong cargos, this rate constant likely approximates the value of k2 (Fig. 7A), and SRPs bound with strong cargos such as RNCFtsQ are primarily in the active conformation (Fig. 7A, K ≥ 1). In the absence of the SRP RNA, the observed complex assembly rate constant (3 × 102 M−1 sec−1) approximates the complex formation rate from the ensemble of inactive SRP conformations (Fig. 7A, k1). The observed complex assembly rate constants under different conditions are therefore a weighted sum of the contributions from the active and inactive SRP populations according to Equation 3,
from which the equilibrium to form the active conformation K can be calculated; the results are listed in Figure 7A.
Free energy diagram
The free energy surface in Figure 7B was constructed using Mathematica (Wolfram Research). The free energy profile in the lower panel of Figure 7B was constructed based on the experimentally determined rate and equilibrium constants using a standard state of 1 μM. ΔG = −RT ln K, in which R = 1.987 cal mol−1 K−1, T = 298.15 K, and K is the experimentally measured equilibrium constant. ΔG‡ = −RT ln(kh/kBT), in which R = 1.987 cal mol−1 K−1, T = 298.15 K, kB = 3.3 × 10−24 cal K−1, h = 1.58 × 10−34 sec, and k is the experimentally measured rate constant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Shan group for helpful comments on the manuscript. This work was supported by NIH grant GM078024 to S.S. S.S. was supported by a career award from the Burroughs Wellcome Foundation, the Beckman Young Investigator award, the Packard and Lucile award in science and engineering, and the Henry Dreyfus teacher-scholar award.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2610411.
REFERENCES
- Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287: 1232–1239 [DOI] [PubMed] [Google Scholar]
- Bradshaw N, Neher SB, Booth DS, Walter P 2009. Signal sequences activate the catalytic switch of SRP RNA. Science 323: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W 2005. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J Mol Biol 351: 417–430 [DOI] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM 2004. Substrate twinning activates the signal recognition particle and its receptor. Nature 427: 215–221 [DOI] [PubMed] [Google Scholar]
- Estrozi LF, Boehringer D, Shan SO, Ban N, Schaffitzel C 2011. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol 18: 88–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE 2003. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem 278: 18628–18637 [DOI] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM 2004. Heterodimeric GTPase core of the SRP targeting complex. Science 303: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P 1997. Structure of the conserved GTPase domain of the signal recognition particle. Nature 385: 361–364 [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P 1982a. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol 95: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G 1982b. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol 95: 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W 2003. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9: 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814 [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R 2006. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444: 507–511 [DOI] [PubMed] [Google Scholar]
- Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K 2010. Recognition of a signal peptide by the signal recognition particle. Nature 465: 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94: 181–191 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1989. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57: 585–597 [DOI] [PubMed] [Google Scholar]
- Montoya G, Svensson C, Luirink J, Sinning I 1997. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature 385: 365–368 [DOI] [PubMed] [Google Scholar]
- Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P 2000. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science 288: 1640–1643 [DOI] [PubMed] [Google Scholar]
- Peluso P, Shan SO, Nock S, Herschlag D, Walter P 2001. Role of SRP RNA in the GTPase cycles of ffh and FtsY. Biochemistry 40: 15224–15233 [DOI] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B 2002. Distinct modes of signal recognition particle interaction with the ribosome. Science 297: 1345–1348 [DOI] [PubMed] [Google Scholar]
- Rosendal KR, Wild K, Montoya G, Sinning I 2003. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc Natl Acad Sci 100: 14701–14706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C, Ban N 2007. Generation of ribosome nascent chain complexes for structural and functional studies. J Struct Biol 158: 463–471 [DOI] [PubMed] [Google Scholar]
- Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N 2006. Structure of the E-coli signal recognition particle bound to a translating ribosome. Nature 444: 503–506 [DOI] [PubMed] [Google Scholar]
- Shan SO, Stroud RM, Walter P 2004. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol 2: 1572–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Shan SO 2010. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci 107: 7698–7703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P 1988. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J 7: 1769–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu FY, Spanggord RJ, Doudna JA 2007. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA 13: 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanggord RJ, Siu F, Ke AL, Doudna JA 2005. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol 12: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Ullers RS, Houben EN, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J 2003. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J Cell Biol 161: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Johnson AE 1994. Signal sequence recognition and protein targeting to the endoplasmic-reticulum membrane. Annu Rev Cell Biol 10: 87–119 [DOI] [PubMed] [Google Scholar]
- Zhang X, Kung S, Shan SO 2008. Demonstration of a multistep mechanism for assembly of the SRP·SRP receptor complex: Implications for the catalytic role of SRP RNA. J Mol Biol 381: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Schaffitzel C, Ban N, Shan SO 2009. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci 106: 1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rashid R, Wang K, Shan SO 2010. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science 328: 757–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lam VQ, Mou Y, Kimura T, Chung J, Chandrasekar S, Winkler JR, Mayo SL, Shan SO 2011. Direct visualization reveals dynamics of a transient intermediate during protein assembly. Proc Natl Acad Sci (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Gierasch LM 1996. Signal sequences: The same yet different. Cell 86: 849–852 [DOI] [PubMed] [Google Scholar]
- Zopf D, Bernstein HD, Johnson AE, Walter P 1990. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J 9: 4511–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]