Abstract
Rice Hoja Blanca Tenuivirus (RHBV), a negative strand RNA virus, has been identified to infect rice and is widely transmitted by the insect vector. NS3 protein encoded by RHBV RNA3 was reported to be a potent RNAi suppressor to counterdefense RNA silencing in plants, insect cells, and mammalian cells. Here, we report the crystal structure of the N-terminal domain of RHBV NS3 (residues 21–114) at 2.0 Å. RHBV NS3 N-terminal domain forms a dimer by two pairs of α-helices in an anti-parallel mode, with one surface harboring a shallow groove at the dimension of 20 Å × 30 Å for putative dsRNA binding. In vitro RNA binding assay and RNA silencing suppression assay have demonstrated that the structural conserved residues located along this shallow groove, such as Arg50, His51, Lys77, and His85, participate in dsRNA binding and RNA silencing suppression. Our results provide the initial structural implications in understanding the RNAi suppression mechanism by RHBV NS3.
Keywords: Rice Hoja Blanca Tenuivirus NS3, RNA silencing suppression, dsRNA binding, crystal structure
INTRODUCTION
RNA silencing is a well-conserved eukaryotic post-transcriptional gene regulation mechanism that targets and degrades aberrant endogenous or exogenous RNA molecules (Siomi and Siomi 2009). This phenomenon was first reported in plants and consequently discovered in fungus, Caenorhabditis elegans, Drosophila, and mammals (Napoli et al. 1990; Romano and Macino 1992; Guo and Kemphues 1995; Fire et al. 1998; Hammond et al. 2000; Wianny and Zernicka-Goetz 2000).
In plants, RNA silencing is regarded as an important antiviral defense strategy triggered by a viral double-stranded RNAs (dsRNA) invasion (Ding and Voinnet 2007). Highly structured viral RNAs, dsRNA replication intermediates, or dsRNAs produced by RNA-dependent RNA polymerase are first recognized and processed by Dicer-like enzymes into small interfering RNA (siRNA) duplexes (Ding and Voinnet 2007). The siRNA duplexes are then incorporated into an RNA-induced silencing complex to target viral transcripts for degradation (Hutvagner and Simard 2008). As a counterdefense strategy, many plant viruses encode suppressor proteins to evade host silencing mechanisms by targeting different steps involved in the antiviral defense pathway (Ding and Voinnet 2007). Both proteins and dsRNAs have been reported as the targets for RNA virus suppressor proteins (Lakatos et al. 2006; Li and Ding 2006; Zhang et al. 2006b; Baumberger et al. 2007; Bortolamiol et al. 2007; Csorba et al. 2010).
An antiviral defense mechanism is not only limited in plant viruses but also discovered in other eukaryotic systems. Insect-infecting Flock House Virus (FHV) B2, Drosophila C virus DCV-1A, and cricket paralysis virus CrPV-1A proteins showed RNA silencing suppression activity in insect cells (Li et al. 2002; Chao et al. 2005; van Rij et al. 2006; Nayak et al. 2010), whereas human influenza A virus NS1 protein, human immunodeficiency virus type 1 Tat protein, and Ebola VP35 displayed RNA silencing suppression activity in cultured human cells (Bennasser et al. 2005; Haasnoot et al. 2007; Leung et al. 2010). Cross-kingdom suppression of RNA silencing was observed for the FHV B2 protein and human influenza NS1 in plants (Li et al. 2002; Bucher et al. 2004; Cheng et al. 2009; de Vries et al. 2009).
Rice Hoja Blanca Tenuivirus (RHBV) from the genus Tenuivirus has been reported to cause a viral disease that occurs in cyclical outbreaks affecting rice production in Tropical America and the Caribbean (Muñoz et al. 2004). RHBV is transmitted by insect vector and observed to be having life cycles in both plants and insects. RHBV nonstructural protein NS3, encoded by RNA3, has been reported to be an RNA silencing suppressor that functions to evade the antiviral silencing mechanism in plants, insect cells and mammalian cells (Bucher et al. 2003; Hemmes et al. 2007; Schnettler et al. 2008, 2009).
RHBV NS3 forms a dimer in solution and binds to dsRNA as a length preference mode (Hemmes et al. 2007) similar to the recognition mode displayed by Tomato aspermy virus 2b (TAV2b) and Tombusviral P19 (P19) (Chen et al. 2008; Ye et al. 2003; Vargason et al. 2003; Lingel et al. 2005), but different from the mode displayed by Flock House virus B2 (FHVB2) and human influenza NS1 (Chao et al. 2005; Cheng et al. 2009). However, secondary structure prediction indicated that RHBV NS3 is a multiple domain protein comprising four continuous α-helices in the middle, flanked by β-strands at both N-terminus and C-terminus. Therefore, the structural motif for dsRNA binding should be significantly different from a hook-like motif adopted by TAV2b (Chen et al. 2008) or “reading heads” motif displayed by p19 (Vargason et al. 2003; Ye et al. 2003).
To define the molecular insights into dsRNA binding by RHBV NS3 and compare dsRNA recognition modes displayed by different RNA silencing suppressors, we have determined the high resolution crystal structure of RHBV NS3 N-terminal domain (residues 21–114, RHBV NS3NTD). RHBV NS3NTD forms a dimer both in solution and in crystal, and the overall structure shares no structural similarity to any known structures. The dimerization of RHBV NS3 NTD is formed by two pairs of α-helices in an anti-parallel mode, with one of the dimerization interfaces harboring a shallow groove at the dimension of 20 Å × 30 Å for putative dsRNA binding. In vitro RNA binding assay and RNA silencing suppression assay demonstrated that the structural conserved residues located along this shallow groove, such as Arg50, His51, Lys77, and His85, play an important role for siRNA duplex binding and RNA silencing suppression.
RESULTS
RHBV NS3 N-terminal domain crystallizes readily
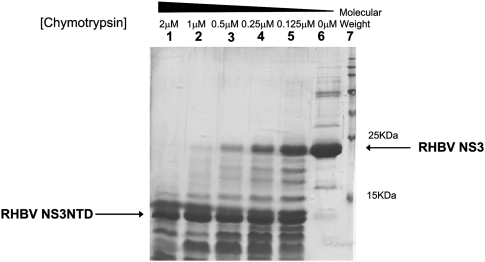
Full-length RHBV NS3 is not amenable to structural studies partially due to the low solubility and instability of the protein. We speculated that the presence of flexible regions in the protein probably causes instability that leads to precipitation. To overcome this, we performed limited proteolysis to define the minimal functional construct described in the literature (Wernimont and Edwards 2009). Initial truncations were identified by limited proteolysis by chymotrypsin treatment, and a fragment that was present in high abundance was consistently observed under different chymotrypsin concentrations used over different time scales (Fig. 1). Mass spectrometry analysis coupled with N-terminal sequencing analysis pointed to the same fragment (residues 21–114, hereafter referred to as RHBV NS3NTD). This fragment shows high resistance to enzyme digestion, suggesting that it could be an autonomously folded protein domain. The purified recombinant RHBV NS3NTD showed high stability and was readily concentrated to >12 mg/mL without aggregation or precipitation during the purification and concentration process. The Se–Met labeled protein crystal derived from this new construct diffracted up to 2.0 Å at the synchrotron radiation resource. The structure was determined by single wavelength anomalous dispersion (SAD) method with crystallographic statistics summarized in Table 1.
FIGURE 1.
Limited proteolysis analysis on RHBV NS3 treated by chymotrypsin. (Lanes 1–5) Full-length RHBV NS3 protein incubated in different concentrations of chymotrypsin varying from 2 to 0.125 μM for 5 min. (Lane 6) Full-length NS3 protein without subjected to enzyme digestion as a control. (Lane 7) Perfect protein marker.
TABLE 1.
Data collection, phasing, and refinement statistics
Overall structure of RHBV NS3 N-terminal domain (NS3NTD)
In the crystal, there are two pairs of RHBV NS3NTD dimers per asymmetric unit with solvent content ∼40%. Each monomer displays an almost identical structure (RMSD 0.2–0.7 Å, 84 Cα atoms for monomers) except for the N-terminal unstructured random coil.
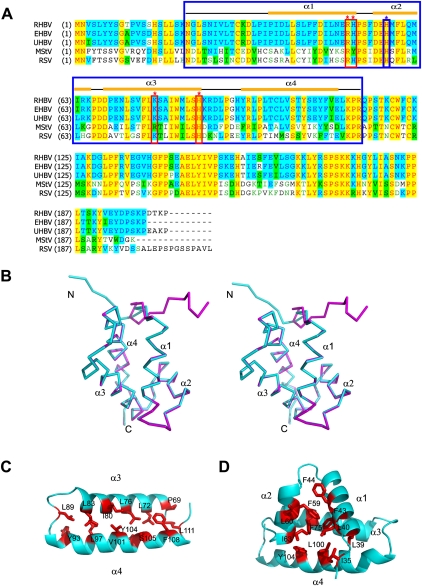
Within each monomer, RHBV NS3NTD starts with an unstructured random coil (residues 21–28) followed by a short putative β strand (residues 29–24) and four α-helices (α1: residues 36–51; α2: residues 55–65; α3: residues 69–85; α4: residues 93–110) (Fig. 2A,B). The four α-helices of NS3NTD are divided into two groups: α1, α2, and the well-ordered linker form one group resembling the architecture of Helix-turn-helix (HTH) motif (Fig. 2B), whereas anti-paralleled α3 and α4 form another group stabilized by hydrophobic residues from α3 (L72, L76, I80, and L83) and α4 (L97, V101, Y104/S105, and F108), respectively (Fig. 2B,C). These two groups are connected by a very short invariable link (residues 66PDD68), thereby α1 packs perpendicular to α3 with N-terminal putative β-strand and part of α4 in close proximity (Fig. 2B). The overall structure of RHBV NS3NTD is further stabilized by a hydrophobic core formed by I35 from the putative β-strand; L39, L40, and F43 from α1; F59 and I63 from α2; F75 from α3; and L100, Y104 from α4 (Fig. 2D). A structural similarity search of RHBV NS3NTD by DALI server (www2.ebi.ac.uk/dali) did not identify any similar architecture, suggesting that RHBV NS3NTD might adopt a novel fold.
FIGURE 2.
Crystal structure of Rice Hoja Blanca Tenuivirus NS3. (A) Sequence alignments of the NS3 protein from various organisms. Abbreviations are as follows: RHBV, Rice Hoja Blanca Tenuivirus; EHBV, echinochloa hoja Blanca virus; UHBV, urochloa hoja Blanca virus; MstV, maize stripe virus; and RSV, rice stripe virus. Secondary structural elements of RHBV NS3 are labeled at the top of the alignment. The key residues involved in both dsRNA binding and RNA silencing suppression are boxed in red and marked with red stars above the alignment, whereas the key residue involved in RNAi suppression only is boxed in blue and marked with blue star above the alignment. The NS3 fragment (NS3NTD) used for crystallization is boxed in blue. (B) Stereoview of overall structure of RHBV NS3NTD. The two monomers within one dimer colored in cyan and magenta, respectively, are superimposed. Secondary structural elements are indicated. (C) Ribbon view of hydrophobic interaction within α3–α4 subdomain with key residues indicated. (D) Ribbon view of hydrophobic core within each monomer with key residues indicated.
Putative dsRNA binding platform
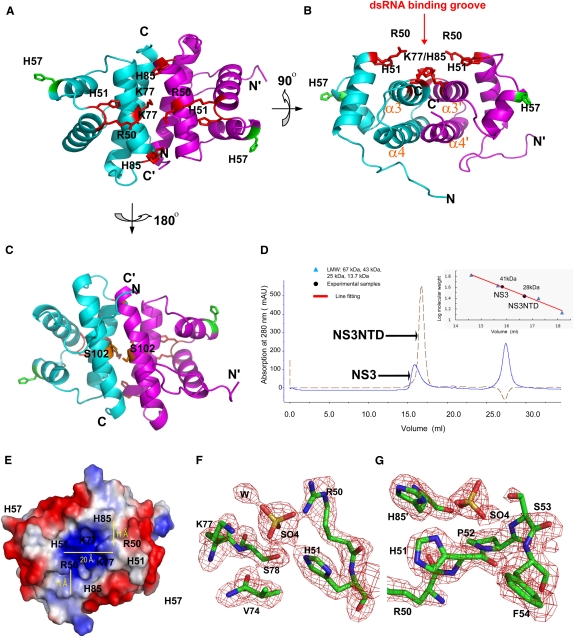
RHBV NS3NTD forms a dimer via closely packed four α-helices (α3–α4–α3′–α4′) in an anti-parallel arrangement with the α3–α3′ axis aligned ∼30° rotation from α4–α4′ axis (Fig. 3A,B). The dimer interface is further stabilized by hydrogen bonds formed by residue S102 from both monomers (Fig. 3C). The dimerization buries a large surface area (1600 Å2 or 30% of the total surface area) per monomer, suggesting that such a dimerization arrangement could be a physiological one. Consistent with this notion, size exclusion chromatography demonstrated that both full-length RHBV NS3 and RHBV NS3NTD form dimers in solution (Fig. 3D).
FIGURE 3.
Putative dsRNA binding platform. (A) Ribbon view of RHBV NS3NTD dimer with one monomer colored in cyan and the other monomer colored in magenta. Key residues (R50, H51, K77, and H85) involved in both dsRNA binding and RNA silencing suppression are indicated as red sticks, whereas key residues (H57) involved in RNA silencing suppression only are indicated as green sticks. (B) Ribbon view of RHBV NS3NTD rotating 90° along the x axis of A. Key residues involved in both dsRNA binding and RNA silencing suppression are clustered within a shallow groove above the dimerization interface for putative dsRNA binding. (C) Ribbon view of RHBV NS3NTD rotating 180° along the x axis of A. Hydrogen bonding involved in stabilization of dimer is highlighted. (D) Gel filtration analysis of full-length RHBV NS3 and RHBV NS3NTD. Full-length RHBV NS3 (total 203 a.a.) migrates as a dimer in solution to the apparent molecular weight of ∼41 kDa, whereas RHBV NS3NTD (total 94 a.a.) migrates as a dimer in solution to the apparent molecular weight of ∼28 kDa. (E) Electrostatic surface potential representation of RHBV NS3NTD dimer with the same view of A. Blue and red colors corresponding to positively and negatively charged patches, respectively. The dimension of the dsRNA binding groove is indicated by yellow lines. Pairs of H85/K77 and R50/H51 form the molecular ladder for putative dsRNA binding. (F,G) Simulated annealing omit map (Fo–Fc) contoured at the 3.5σ level around putative dsRNA binding groove. Two well-refined sulfate ions are coordinated by invariable residues R50, H51, and K77, or H85 and H51, respectively. Carbon atoms are colored in green, oxygen in red, nitrogen in blue, and sulfur in yellow.
The RHBV NS3NTD dimer displays a continuous elongated shallow groove (30 Å × 20 Å, length × width) at one surface formed by dimerization. Several conserved positively charged residues, such as Arg50/His51 from α1 and Lys77/His85 from α3, were found in this groove (Fig. 3E). Notably, two pairs of Lys77/His85 residues from both monomers form a molecular ladder separated by a spacing of ∼11 Å, which corresponds to the distance of phosphates across the major groove in an A-form dsRNA conformation (Fig. 3E). The conserved Arg50 and His51 from each monomer are located at the top of this shallow groove and probably recognize the other side of the binding dsRNA (Fig. 3B,E). In addition, several well-refined ammonium sulfate molecules form direct hydrogen bonds with the side chains of Arg50, His 51, Lys77, and His85 in our crystal structures (Fig. 3F,G), which suggests that Arg50, His51, Lys77, and His85 are probably directly involved in recognition of a phosphate–sugar backbone of the dsRNA stem.
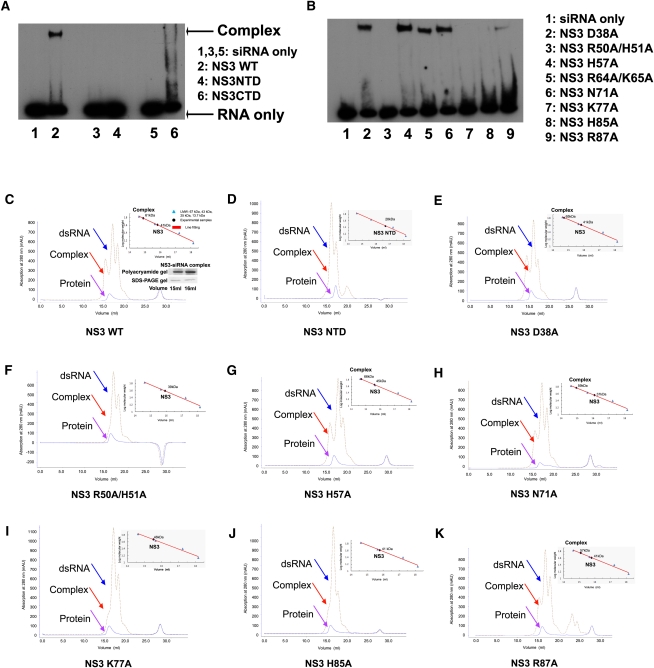
However, the limited size of this putative dsRNA binding platform (∼30 Å × 20 Å, length × width) is not able to accommodate the whole length of 21-nucleotide (nt) A-form siRNA duplex (∼60 Å in length) (Fig. 3E). Thus, the RHBV NS3NTD dimer alone is probably insufficient for dsRNA binding. Consistent with the structural observation, RHBV NS3NTD did not show any apparent binding affinity on siRNA duplex by either analytical gel filtration assay or electrophoretic mobility shift assay (EMSA) (Fig. 4A,D). By contrast, full-length RHBV NS3 displayed a robust high binding affinity to the siRNA duplex (Fig. 4A,C). Therefore, other residues/motifs at the C-terminal domain (residues 115–203), such as 173KKK175 motif, probably provide additional structural motifs for dsRNA binding and/or anchoring (Hemmes et al. 2009).
FIGURE 4.
Key residues involved in siRNA duplex binding. (A) Electrophoretic mobility shift assay on NS3 and its fragments binding to siRNA duplex. (Lanes 1,3,5) siRNA duplex only; (lane 2) NS3 WT; (lane 4) NS3 NTD; (lane 6) NS3 CTD. (B) Electrophoretic mobility shift assay on NS3 mutants binding to a siRNA duplex. (Lane 1) siRNA duplex only; (lane 2) NS3 D38A single mutant; (lane 3) NS3 R50A/H51A double mutant; (lane 4) NS3 H57A single mutant; (lane 5) NS3 R64A/K65A double mutant; (lane 6) NS3 N71A single mutant; (lane 7) NS3 K77A single mutant; (lane 8) NS3 H85A single mutant; (lane 9) NS3 R87A single mutant. (C–K) Superimposition of analytic gel filtration analysis on RHBV NS3 or its mutant with its siRNA duplex complex. (C) NS3 WT; (D) NS3 NTD fragment; (E) NS3 D38A single mutant; (F) NS3 R50A/H51A double mutant; (G) NS3 H57A single mutant; (H) NS3 N71A single mutant; (I) NS3 K77A single mutant; (J) NS3 H85A single mutant; (K) NS3 R87A single mutant. The individual peak corresponding to the dsRNA, complex and protein are indicated by arrows colored in blue, red, and pink, respectively. The molecular standard chart is shown at the top-right side of the picture. The Polyacrylamide and SDS-PAGE gels corresponding to RNA and protein peaks are shown at the bottom-right side of the picture at C.
To test if NS3 C-terminal domain (NS3CTD) alone is sufficient for dsRNA binding, NS3CTD was expressed for a dsRNA binding assay. An EMSA assay demonstrated that NS3CTD alone displays weak binding affinity to the siRNA duplex, as compared to full-length NS3, and this suggests that the C-terminal domain is not sufficient for dsRNA binding either (Fig. 4A).
To investigate if the conserved Arg50/His51/Lys77/His85 residues indeed play a role in dsRNA binding, a single alanine mutation on residue Lys77 or His85 or a double alanine mutation on residues Arg50/His51 was introduced to the full-length NS3. Both the analytic gel filtration assay and the EMSA assay showed that none of the full-length NS3 mutants displayed apparent binding affinity to the siRNA duplex (Fig. 4B,F,I,J). By contrast, single mutations on the residues clustered at the HTH-like motif, such as Asp38, His57, Arg64, Lys65, Asn71, or Arg87 did not show any impact on the binding affinity of the full-length NS3 to the siRNA duplex (Fig. 4B,E,G,H,K). These data strongly support the notion that the shallow groove provides the platform for dsRNA binding.
RNA silencing suppression by RHBV NS3
To test whether the key residues involved in siRNA duplex binding are also involved in host RNA silencing suppression, we performed in vitro RNA silencing suppression experiments using Agrobacterium co-infiltration assay in transgenic plants that express the green fluorescent protein (GFP). In these transgenic plants, GFP silencing can be triggered by transient GFP expression via Agrobacterium infiltration (Johansen and Carrington 2001). The presence of an RNA silencing suppressor, which initiates post-transcription gene silencing is able to rescue the transient expression of GFP in the infiltrated regions.
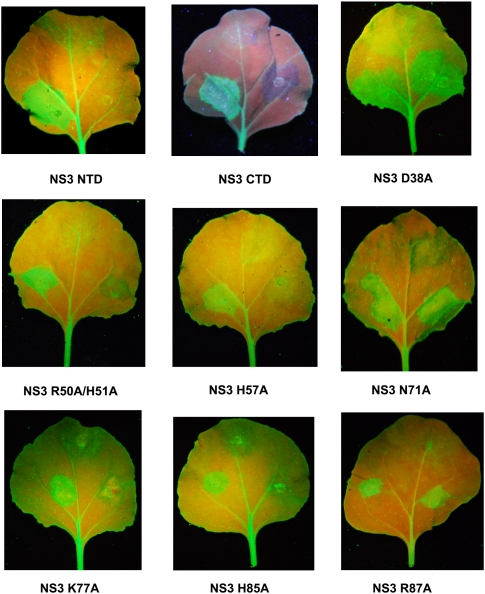
Consistent with the siRNA duplex binding assays, introduction of a single alanine mutation on Lys77 or His85, or double alanine mutations on residues R50/H51 to full-length NS3 showed a significant decrease in the GFP expression (Fig. 5). These results suggest that the conserved positive residues along the shallow groove not only play determination roles for dsRNA binding, but also play crucial roles for RNA silencing suppression. Alternatively, the introduction of the mutations at these positions may disrupt the stability of the NS3 proteins expressed in planta. By contrast, alanine mutations on residues Asp38, Lys64, His65, Asn71, and Arg87, located at the HTH-like motif showed complete activation of the GFP expression (Fig. 5; Supplemental Fig. S1). Interestingly, a single mutation on His57, which is also located at a HTH-like motif showed a dramatic decrease in GFP expression (Fig. 5, middle panel). Thus, although His57 has no impact on siRNA duplex binding in vitro, it might be involved in RNA silencing suppression through other mechanisms, probably by protein–protein interaction.
FIGURE 5.
RNA silencing suppression by RHBV NS3. The GFP-expressing Nicotiana benthamiana leaves (16C) were co-infiltrated with a mixture of two Agrobacterium tumefaciens strains. One directs the expression of RHBV NS3 WT together with GFP (lower-left spot on the leaf) and the other directs the expression of RHBV NS3 mutant (NS3 NTD, NS3 CTD, NS3 D38A, NS3 R50A/H51A, NS3 H57A, NS3 N71A, NS3 K77A, NS3 H85A, NS3 R87A, respectively) together with GFP (lower-right spot on the leaf). Agrobacterium tumefaciens strain with transformed GFP-expressing vector was infiltrated at the upper spot on the leaf as a negative control. The leaves were detached and photographed under UV illumination 6 d after infiltration.
In accordance with the structural observation and siRNA duplex binding assays, neither RHBV NS3NTD alone nor RHBV NS3CTD alone was sufficient to suppress RNA silencing (Fig. 5, panels 1,2). This observation is consistent with the previous report that C-terminal truncated RHBV NS3 (Δ167–176) lacks of RNA silencing suppression activity (Hemmes et al. 2009). By contrast, full-length C-terminal HA-tagged RHBV NS3 has efficiently rescued transient expression of GFP in the infiltrated regions (Fig. 5 right spot on each leaf). Our observation further confirms that RHBV C-terminal domain is crucial, but not sufficient, for both siRNA duplex binding and RNA silencing suppression (Hemmes et al. 2009).
DISCUSSION
Structural comparison with RNA silencing suppressor p19
RNA silencing suppressors encoded by different viruses have been identified to be diverse in their structures and suppression mechanisms (Yang and Yuan 2009). In addition to targeting protein components of small RNA processing machineries (Zhang et al. 2006b; Baumberger et al. 2007; Bortolamiol et al. 2007), viral suppressors also hijack RNA components along the small RNA processing pathway (Ding and Voinnet 2007).
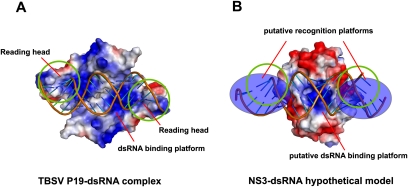
Among the known structures of viral RNA silencing suppressors, both p19 and RHBV NS3 were reported to recognize siRNA duplex at a size-dependent mode. (Silhavy et al. 2002; Vargason et al. 2003; Ye et al. 2003; Lakatos et al. 2006; Hemmes et al. 2007). The structure of a p19 homodimer is divided into two domains: a continuous eight-stranded β-sheet providing platform for siRNA duplex binding, and two α-helical “read heads” providing trypotophans for siRNA duplex bracketing (Fig. 6A). The β-sheet and α-helical “read head” are loosely linked by a disordered loop segment. Notably, neither β-sheet platform nor α-helical read head alone is sufficient for siRNA duplex binding (Vargason et al. 2003; Ye et al. 2003). Similar to p19, the full-length RHBV NS3 is also divided into two domains confirmed by limited proteolysis analysis: N-terminal dimerization interface providing a platform for siRNA duplex binding and C-terminal domain providing critical residues for siRNA duplex measurement. Strikingly, similar to p19, neither the NS3 N-terminal dimerization domain nor the NS3 C-terminal domain alone is sufficient for siRNA duplex binding. We speculate that highly conserved aromatic or positively charged residues located at the C-terminal domain might play a role in siRNA duplex measurement, thereby securing a tight binding (Fig. 6B). Future structural and functional analysis on full-length NS3 should provide the detailed information to address the recognition principles for small RNA binding by NS3.
FIGURE 6.
Comparison of different dsRNA binding modes between TBSV P19 and RHBV NS3. (A) Electrostatic surface potential representation of TBSV P19-siRNA duplex complex (PDBID: 1R9F). (B) Electrostatic surface potential representation of RHBV NS3-dsRNA hypothetical model (based on PDBID: 3AJF).
MATERIALS AND METHODS
Protein expression and purification
The RHBV NS3 gene was synthesized with optimized codon for Escherichia coli expression. Full-length RHBV NS3 (residues 1–203) and RHBV NS3NTD (residues 21–114) were generated by cloning into pET28b (Novagen) and pETSUMO (Invitrogen) vectors, respectively, with an N-terminal His-tag. Recombinant full-length RHBV NS3 was expressed in E. coli (BL21/DE3 strain) overnight at 20°C induced by 0.4 mM isopropyl β-d-thiogalactoside. Proteins were purified through a Ni2+ affinity column followed by a HiLoad Superdex S-75 26/60 column. Recombinant SUMO-fused RHBV NS3NTD was purified by the Ni2+ affinity column, followed by SUMO cleavage using protease Ulp1 and subsequently purified by the HiLoad Superdex S-75 26/60 column. Selenomethionine (Se-Met)-substituted RHBV NS3NTD was prepared by following the protocol described in the literature (Doublié 1997). Both full-length RHBV NS3 mutants and RHBV NS3NTD mutants were prepared using the QuikChange Site-Directed Mutagenesis Kit (www.stratagene.com), and the constructs were verified by sequencing. RNA oligoribonucleotides were purchased from Dharmacon without further purification.
Limited proteolysis analysis
Partially purified NS3 full-length protein with a concentration of 1 mg/mL was incubated with different concentrations of chymotrypsin varying from 2 to 0.125 μM at 4°C for 5, 15, 30 min, and overnight. Twenty μL of each sample was aspirated out, mixed with SDS and incubated at 100°C for 10 min. Subsequently, the treated samples were loaded onto a 15% SDS page gel and the gel was stained using coomasive blue. The desired digested fragments were subjected to MALDI TOF mass spectrometry analysis.
Crystallization data collection and structure determination
Crystals of RHBV NS3NTD were grown at 20°C by a mixture of 1.0 μL of a protein sample (12 mg/mL) with 1.0 μL of reservoir solution containing 1.44 M ammonium sulfate, 5% v/v MPD, and 0.1 M sodium cacodylate (pH 7.0), and equilibrated over 1 mL of reservoir solution for a course of 4 d to a dimension of 0.15 μm × 0.02 μm × 0.02 μm.
Crystals were flash frozen (100 K) in the above-mentioned reservoir solution supplemented with 30% glycerol prior to x-ray data collection. A total of 180 frames of 1° oscillation at wavelength 0.9792 Å were collected on a Se-Met crystal and processed by HKL2000 (www.hkl-xray.com). The structure was determined by the SAD method using SHARP (www.globalphasing.com). The model was built by using the program O (http://xray.bmc.uu.se/alwyn) and refined using REFMAC/CCP4 (www.ccp4.ac.uk) with the crystallographic statistics listed in Table 1. The model is comprised of residues 22–113 of the NS3 protein.
Analytical gel filtration
Full-length RHBV NS3 protein, RHBV NS3NTD and protein–RNA complexes were analyzed on Superdex 200 10/300 GL column with a flow rate of 0.5 mL/min and an injection volume of 0.1 mL. The experiments on the RHBV NS3NTD were performed in a buffer of 25 mM Tris base and 50 mM NaCl (pH 7.4), whereas the experiments on the full-length RHBV NS3 were performed in a buffer of 25 mM Tris base and 200 mM NaCl (pH 7.4). For the complex, NS3 and mutants were incubated with 21dsRNA at the molar ratio of (1:2) on ice for 10 min. The column was precalibrated by the low molecular weight gel filtration kit (GE), and the corresponding curves were established by OriginPro 7.5.
Electrophoretic mobility shift (EMSA) assay
The 21nt siRNA duplex was labeled with EZ-Link Psoralen-PEO3-Biotin (PIERCE). The labeled and purified nucleic acids were incubated at room temperature with RHBV NS3 protein and its mutants in the buffer containing 10 mM Tris pH7.5, 50 mM KCl, 1 mM DTT, 10 mM MgCl2, 0.1% NP-40, and 5% glycerol in 20 μL reaction. Following incubation of 20 min, the samples were immediately loaded onto 4% native poly-acrylamide gel with nondenaturating dyes. The resolved nucleic acids were electroblotted onto Hybond-N+ (GE Healthcare), and cross-linked. Blocking, detection, and washing of the membrane were performed using Chemiluminescent Nucleic Acid Detection Module Kit (PIERCE).
Agrobacterium co-infiltration assay
The coding sequences of RHBV NS3 and their mutants were cloned into the pBA vector with either a myc tag or a HA tag at either N-terminus or C-terminus to generate constructs for infiltration, as described in the literature (Voinnet et al. 2000; Guo and Ding 2002; Zhang et al. 2006a). These plasmids were then transformed into Agrobacterium tumefaciens strain EHA105 by electroporation and infiltrated into transgenic Nicotiana benthamiana expressing GFP transgene (line 16C). The GFP fluorescence expression at N. benthamiana leaves were visualized by long-wave UV lamp (SB-100P/F High intensity Ultraviolet lamp, Spectronics, USA) and photographed by a Nikon D-80 digital camera with a yellow filter.
ACCESSION NUMBERS
The coordinates have been deposited in the PDB under the accession code 3AJF.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank S. Huang and J. He at Shanghai Synchrotron Radiation Facility (SSRF) for assistance. This work was supported by funds from Singapore Ministry of Education (T208A3124), intramural funds from Temasek Life Sciences Laboratory, and funds from National University of Singapore undergraduate research programs.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2552811.
REFERENCES
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC 2007. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22: 607–619 [DOI] [PubMed] [Google Scholar]
- Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V 2007. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol 17: 1615–1621 [DOI] [PubMed] [Google Scholar]
- Bucher E, Sijen T, De Haan P, Goldbach R, Prins M 2003. Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J Virol 77: 1329–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher E, Hemmes H, De Haan P, Goldbach R, Prins M 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 85: 983–991 [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12: 952–957 [DOI] [PubMed] [Google Scholar]
- Chen HY, Yang J, Lin C, Yuan YA 2008. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep 9: 754–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Wong SM, Yuan YA 2009. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res 19: 187–195 [DOI] [PubMed] [Google Scholar]
- Csorba T, Lózsa R, Hutvágner G, Burgyán J 2010. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J 62: 463–472 [DOI] [PubMed] [Google Scholar]
- de Vries W, Haasnoot J, Fouchier R, de Haan P, Berkhout B 2009. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J Gen Virol 90: 1916–1922 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié S 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530 [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ 1995. par-1, A gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620 [DOI] [PubMed] [Google Scholar]
- Guo HS, Ding SW 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J 21: 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, De Vries W, Geutjes EJ, Prins M, De Haan P, Berkhout B 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3: e86 doi: 10.1371/journal.ppat.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hemmes H, Lakatos L, Goldbach R, Burgyán J, Prins M 2007. The NS3 protein of Rice hoja blanca tenuivirus suppresses RNA silencing in plant and insect hosts by efficiently binding both siRNAs and miRNAs. RNA 13: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmes H, Kaaij L, Lohuis D, Prins M, Goldbach R, Schnettler E 2009. Binding of small interfering RNA molecules is crucial for RNA interference suppressor activity of rice hoja blanca virus NS3 in plants. J Gen Virol 90: 1762–1766 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ 2008. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Johansen LK, Carrington JC 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol 126: 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington C, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 25: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, et al. 2010. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol 17: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ding SW 2006. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60: 503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW 2002. Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M 2005. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep 6: 1149–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M, Bolaños I, Arrieta-Espinoza G, Espinoza AM 2004. Expression of the rice hoja blanca virus (RHBV) non-structural protein 3 (NS3) in Escherichia coli and its in situ localization in RHBV-infected rice tissues. Rev Biol Trop 52: 765–775 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R 2010. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol 17: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N, Macino G 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6: 3343–3353 [DOI] [PubMed] [Google Scholar]
- Schnettler E, Hemmes H, Goldbach R, Prins M 2008. The NS3 protein of rice hoja blanca virus suppresses RNA silencing in mammalian cells. J Gen Virol 89: 336–340 [DOI] [PubMed] [Google Scholar]
- Schnettler E, de Vries W, Hemmes H, Haasnoot J, Kormelink R, Goldbach R, Berkhout B 2009. The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep 10: 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2009. On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyán J, Tanaka Hall TM 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Wernimont A, Edwards A 2009. In situ proteolysis to generate crystals for structure determination: an update. PLoS ONE 4: e5094 doi: 10.1371/journal.pone.0005094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2: 70–75 [DOI] [PubMed] [Google Scholar]
- Yang J, Yuan YA 2009. A structural perspective of the protein-RNA interactions involved in virus-induced RNA silencing and its suppression. Biochim Biophys Acta 1789: 642–652 [DOI] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 18: 874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH 2006a. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH 2006b. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]