Abstract
Numerous mechanisms have evolved to control the accuracy of translation, including a recently discovered retrospective quality control mechanism in bacteria. This quality control mechanism is sensitive to perturbations in the codon:anticodon interaction in the P site of the ribosome that trigger a dramatic loss of fidelity in subsequent tRNA and release factor selection events in the A site. These events ultimately lead to premature termination of translation in response to an initial miscoding error. In this work, we extend our investigations of this mechanism to an in vitro reconstituted Saccharomyces cerevisiae translation system. We report that yeast ribosomes do not respond to mismatches in the P site by loss of fidelity in subsequent substrate recognition events. We conclude that retrospective editing, as initially characterized in Escherichia coli, does not occur in S. cerevisiae. These results highlight potential mechanistic differences in the functional core of highly conserved ribosomes.
Keywords: eukaryotic ribosome, translation, fidelity, miscoding, termination
INTRODUCTION
The information contained in an organism's genome must be faithfully translated to produce functional proteins, which in turn allows the organism to grow and reproduce. Perfect fidelity, however, has a price: It would be too slow, and too costly in energetic terms, for a living organism (Thompson and Karim 1982; Ruusala et al. 1984). Hence, all branches of life have arrived at some compromise between speed and accuracy in transmission that allows for optimal survival.
This evolved level of fidelity during translation is not the same in all organisms; indeed, more complex organisms appear to have greater overall fidelity than simpler ones. For example, the in vivo miscoding frequencies in yeast appear to be between 1 × 10−4 and 5 × 10−4 misreading events per codon (Stansfield et al. 1998; Rakwalska and Rospert 2004; Salas-Marco and Bedwell 2005; Plant et al. 2007; Kramer et al. 2010), about 10-fold less frequent than the miscoding frequencies measured in vivo in Escherichia coli (Parker 1989; Kramer and Farabaugh 2007). In higher eukaryotes errors may be even less frequent (Martin et al. 1989).
These numbers provide a sense for the eventual outcome from whatever mechanisms exist to ensure this fidelity, but do not define the molecular process. For some time, much biochemical effort has been focused on understanding how the ribosome carefully selects the appropriate aminoacyl-tRNA (or release factor) during each round of elongation (or termination) (for review, see Zaher and Green 2009a). And, while this selection process appears to account for a majority of the discrimination at the level of elongation and termination in bacteria, there also appear to be molecular mechanisms in place, at least in certain bacteria, to retrospectively (after peptide bond formation) monitor (and increase) fidelity (Zaher and Green 2009b). This quality control mechanism monitors codon:anticodon interactions in the P and E sites of the ribosome, where the presence of mismatches appears to increase the promiscuity of interactions in the A site of the ribosome. The loss of A site fidelity manifests itself in one of two ways: Either another incorrect tRNA is selected, thereby iterating the miscoding error, or a release factor is selected at a sense codon, leading to premature termination. The ultimate consequence of this process is that protein synthesis is prematurely terminated in response to one or more miscoding errors (Zaher and Green 2009b). This retrospective quality control system is somewhat reminiscent of the proofreading activity of DNA polymerase in that both increase the fidelity of the process, though in the latter case the mistakes are actually repaired.
Here we explored the fidelity of codon selection by both tRNAs and release factors in our newly developed in vitro reconstituted yeast translation system. We focused our analysis on decoding events that occur on ribosome complexes with fully matched as well as mismatched P-site codon:anticodon helices. In broad terms, tRNA selection appears to share many similarities with the comparable events in E. coli, including, for example, increased miscoding and decreased release activity in the presence of aminoglycosides. Surprisingly, however, we saw only modest responses of the yeast ribosome to mismatches in the P site, and these responses would not obviously increase the overall fidelity of translation. Consequently, we conclude that retrospective quality control as initially defined in E. coli does not take place in yeast. It remains possible, however, that post-PT quality control does occur in eukaryotes, through a distinct mechanism, potentially involving extra-ribosomal factors.
RESULTS
Experimental system
For this study, we developed an in vitro reconstituted translation system with Saccharomyces cerevisiae components that could specifically follow the events of translation elongation and termination. Initiation complexes were prepared from purified yeast subunits (40S and 60S) and short unstructured mRNAs with a core set of recombinant initiation factors (eIF1, eIF1A, eIF5, eIF5B, eIF2, and charged in vitro transcribed initiator methionine tRNA) as previously described (Acker et al. 2007).
For elongation reactions, the translation factors eEF1A, eEF2, and eEF3 were expressed and purified as previously described (Jorgensen et al. 2002; Andersen et al. 2004). Commercially available tRNAPhe and in vitro transcribed tRNALys (see Materials and Methods) were chosen as elongation substrates, as both were efficiently aminoacylated by an S100 extract and incorporated by the ribosome when provided in a ternary complex with eIF1A•GTP.
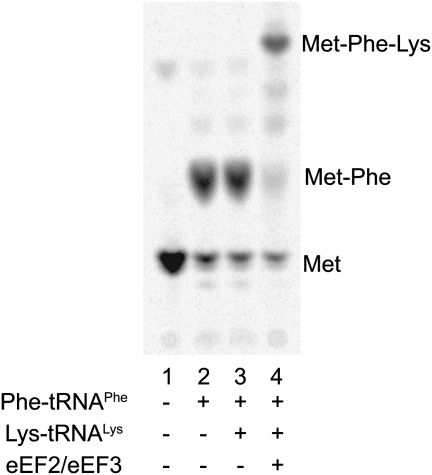
With our first set of experiments, we tested the behavior of the system during elongation. Initiation complexes were prepared on short mRNAs containing the coding sequence AUG UUC AAA, as described above, and the elongation cycle was performed. In the absence of ternary complex, only Met is observed (Fig. 1, lane 1). When Phe-tRNAPhe•eEF1A•GTP ternary complex is added, Met-Phe dipeptide is formed (Fig. 1, lane 2). When Lys-tRNALys•eEF1A•GTP ternary complex is included, synthesis of the tripeptide Met-Phe-Lys is only observed when both elongation factors eEF2 and eEF3 are also added (Fig. 1, lanes 3,4). The results from this staged reaction sequence indicate that our translation elongation system is appropriately responsive to the various components.
FIGURE 1.
Product analysis of reconstituted yeast translation elongation reactions. Initiation complexes were reacted with ternary complexes prepared with the indicated tRNAs and translocation factors.
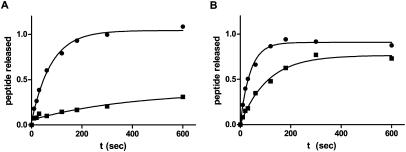
We next prepared reagents that would allow us to study translation termination in the yeast reconstituted system. N-terminally His-tagged versions of the class 1, eRF1, and class 2, eRF3, release/termination factors from S. cerevisiae were cloned, expressed in, and purified from E. coli. An N-terminally truncated version of eRF3 (ΔN165) (lacking a poorly behaved glutamine-rich domain) was utilized to increase solubility, as has been described in a reconstituted mammalian system (Alkalaeva et al. 2006). The activity of the termination factors was evaluated on ribosome complexes programmed with an mRNA carrying the coding sequence AUG UUC AAA UAA, Met-Phe-Lys-tRNALys poised in the P site, and the stop codon (UAA) in the A site. The purified release factors eRF1 and eRF3 together stimulated peptide release in the presence of GTP, while the non-hydrolyzable analog GMPPNP inhibited the release reaction (Fig. 2A). Additionally, eRF1 was able to stimulate peptide release alone, and the rate was substantially increased by the addition of eRF3 (Fig. 2B). These data are overall markedly similar to earlier observations in the reconstituted mammalian system (Alkalaeva et al. 2006). With these reagents in place, the reconstituted yeast system allows for detailed in vitro analysis of translation elongation and termination.
FIGURE 2.
Peptide release on authentic termination complexes. Representative curves show the fraction of Met-Phe-Lys tripeptide released over time; (A) by eRF1•eRF3ΔN165 and GTP (•) or GMPPNP (▪), (B) by eRF1•eRF3ΔN165•GTP (•) or eRF1 and GTP (▪).
Effects of paromomycin on miscoding
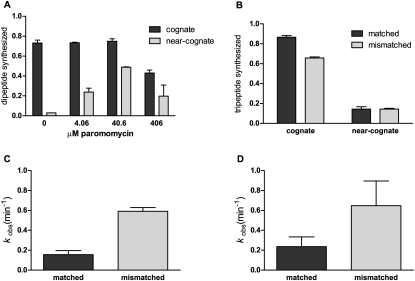
The aminoglycoside antibiotic paromomycin is known to promote translational miscoding in bacteria as a result of binding to a site in the highly conserved decoding center 16S rRNA (Moazed and Noller 1987; Carter et al. 2000). Similarly, paromomycin appears to stimulate miscoding in S. cerevisiae as determined with a series of reporter constructs (Fan-Minogue and Bedwell 2008; Kramer et al. 2010). Here, we evaluated the effects of paromomycin on tRNA and eRF selection in our in vitro reconstituted system. Initiation complexes were assembled on mRNAs with the coding sequence AUGAAA (MK, cognate for Met-Lys) or AUGAAU (MN, near-cognate for Met-Lys) and were reacted with Lys-tRNALys•eEF1A•GTP ternary complex at varying concentrations of paromomycin; the fraction of Met converted to Met-Lys dipeptide is shown in Figure 3A. As anticipated based on the in vivo studies, paromomycin stimulated the synthesis of Met-Lys dipeptide on MN mRNA without significantly affecting Met-Lys synthesis on MK mRNA (at least at lower concentrations of paromomycin). These results are consistent with published data showing that Lys-tRNALys can miscode on the AAU codon. In addition, paromomycin inhibited the RF-mediated peptide release reaction (data not shown), as previously reported in the bacterial system (Brown et al. 1993; Youngman et al. 2006). Our data suggest that yeast ribosomes qualitatively respond to paromomycin in the same manner as bacterial ribosomes, arguing for the presence of core structural features that dictate the events of translation elongation.
FIGURE 3.
Miscoding by yeast ribosomes during elongation is stimulated by paromomycin but not by P-site mismatches. (A) Met-Lys dipeptide synthesized on Met-Lys (dark bars) or Met-Asn (light bars) mRNA in the presence of increasing concentrations of paromomycin (indicated on the x-axis). (B) The extent of misreading (fraction dipeptide converted into tripeptide) is not substantially affected by P-site mismatches at moderate Lys-tRNALys ternary complex concentrations. Complexes were prepared with Met-Phe dipeptidyl-tRNA in the P site with either a matched Phe (UUC) (dark bars) or mismatched Leu (UUG) (light bars) codon in the P site with either a cognate Lys (AAA) or a near-cognate Asn (AAU) codon in the A site (x-axis). Rates of tripeptide synthesis by (C) cognate complexes at subsaturating Lys-tRNALys ternary complex concentration, and by (D) near-cognate complexes at saturating Lys-tRNALys ternary complex concentration. All data are presented as mean ± standard error of at least two measurements.
Effects of a P-site mismatch on tRNA selection (and peptidyl transfer)
We next evaluated the impact of a mismatch in the P-site decoding center on tRNA selection and subsequent peptidyl transfer (Zaher and Green 2009b). We prepared elongated, translocated dipeptidyl-tRNA-containing ribosome complexes, both with and without a mismatch in the P site (“matched” or “mismatched,” respectively) and with a cognate or near-cognate codon in the A site (“cognate” or “near cognate,” respectively). Given this terminology, there are four possible complexes that were analyzed: matched/cognate, matched/near-cognate, mismatched/cognate, and mismatched/near-cognate.
The ribosome dipeptidyl-tRNA (Met-Phe-tRNAPhe) complexes were assembled on the appropriate mRNAs where the first codon is AUG (M), the second codon is either UUC (F) or UUG (L), and the third codon is AAA (K) or AAU (N). Since we only use three tRNAs, tRNAiMet, tRNAPhe, and tRNALys, the matched complexes are formed on UUC encoding mRNAs, while the mismatched complexes are formed on UUG encoding mRNAs. In each case, either AAA or AAU is poised in the A site for cognate or near-cognate decoding by tRNALys, respectively. We confirmed the positioning of the mRNAs relative to the ribosome by toeprinting (Hartz et al. 1988) to rule out the possibility that mismatched complexes are prone to frameshifting (data not shown).
First, in a simple experiment where modest amounts of eEF1A•GTP•Lys-tRNALys ternary complexes were added, the cognate reactions went to completion, while little miscoding was observed with the near cognates, on both matched and mismatched complexes (Fig. 3B). We next asked whether there might be differences in the rates of these different reactions that might reflect changes in ribosome structure resulting from the perturbation of the P site. The cognate reactions were evaluated with reactions containing subsaturating amounts of Lys-tRNALys ternary complex (so called kcat/Km conditions) and were seen to be relatively similar to one another, with kobs values of 0.16 min−1 and 0.59 min−1 on matched and mismatched complexes, respectively (Fig. 3C). The slower near-cognate reactions were evaluated using saturating amount of Lys-tRNALys ternary complex (so called kcat conditions) and were again found to be relatively similar to one another, 0.24 min−1 and 0.65 min−1, on matched and mismatched complexes (Fig. 3D). Most significantly, the modest increases in rate seen on the mismatched complexes were about the same for the cognate and near-cognate reactions (3.7- and 2.7-fold, respectively).
Since the effects of the P-site mismatches on cognate reactions were performed at subsaturating concentrations of ternary complex, the observed effects may represent a combination of binding and catalytic defects. By contrast, since the effects of the P-site mismatch on near-cognate reactions were performed under saturating substrate concentrations, no binding component is included in the observed data. We note that these experiments were performed differently because the fast rates of tripeptide synthesis for cognate complexes require use of a quench-flow apparatus, and we are not yet able to generate the requisite quantities of material. We think it unlikely that the results would be qualitatively different if all measurements were made under kcat conditions. Our results with the release factors (below) suggest that binding to the A site is generally unperturbed in the mismatched complex. Together, these data suggest that while P-site mismatches do (modestly) accelerate tRNA selection in the A site for both cognate and near-cognate tRNAs, these effects are not specific for near-cognate tRNAs, and thus do not change the overall fidelity of the selection step.
To further test this hypothesis, we conducted tRNA competition experiments in which matched and mismatched dipeptidyl-tRNA complexes (with Met-Phe-tRNAPhe on AUGUUCAAA and AUGUUGAAA mRNAs, respectively) were reacted with total aminoacylated tRNA from a translating extract (Wu et al. 2007). The diverse reaction products were resolved on a two-dimensional TLC system where the first phase was a passive TLC separation using 70:20:10 EtOH:H2O:HOAc as the mobile phase, and the second phase was electrophoretic using the pyridine:acetate mobile phase described earlier (Zaher and Green 2009b). Overall, the patterns of product formation are quite similar for the matched and mismatched complexes (Supplemental Fig. 1); spots for Met, Met-Phe, and Met-Phe-Lys can be identified in each, in addition to several unidentified bands produced in approximately equal quantities by the distinct complexes. Consistent with our kinetic results, this competition experiment argues that mismatches in the P site do not promote iterated errors in tRNA selection in S. cerevisiae, as previously documented in E. coli (Zaher and Green 2009b).
Effects of a P-site mismatch on RF selection (and peptide release)
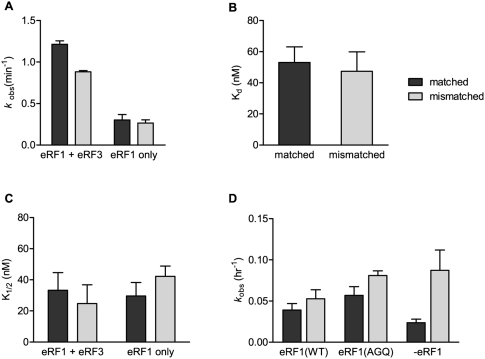
We next measured several parameters relevant to RF selection and subsequent peptide release on P-site matched and mismatched ribosome complexes. We began by measuring the rates of peptide release on matched and mismatched dipeptidyl-tRNA ribosome complexes (Met-Phe-tRNAPhe) containing a UAA stop codon in the A site (AUGUUCUAA and AUGUUGUAA coding sequence, respectively) and observed only small differences in the rates of catalysis in the presence of eRF1 alone or eRF1•eRF3 (indeed, the modest differences were in the opposite direction than anticipated) (Fig. 4A). We also directly measured the binding affinity between eRF1•eRF3 and the matched and mismatched ribosome complexes using a fluorescence assay where the P-site tRNAPhe is modified with a proflavin residue (Wintermeyer and Zachau 1979) and intrinsic fluorescence changes are followed (Zaher and Green 2010); here again we failed to observe any significant differences (Fig. 4B). Finally, K1/2 values for the canonical release reaction were determined as previously (Zaher and Green 2009b) for the matched and mismatched complexes; again we observed no differences in these values for the distinct ribosome complexes with either eRF1•eRF3 or eRF1 alone (Fig. 4C). These data are strikingly different than those reported in E. coli where even on stop codon containing complexes, K1/2 differences between matched and mismatched complexes were easily discerned (Zaher and Green 2009b).
FIGURE 4.
Kinetic and thermodynamic parameters of peptide release are not altered by a P-site mismatch. (A) Plot of observed rate constants [kobs(min−1)] for dipeptide release at a stop codon by eRF1•eRF3ΔN165•GTP or eRF1 alone. (B) Plot of measured Kd values of eRF1•eRF3ΔN165•GTP for dipeptidyl-tRNA ribosome termination complexes. (C) Plot of K1/2 values for dipeptide release on termination complexes by eRF1•eRF3ΔN165•GTP or eRF1 alone. (D) Plot of observed rate constants [kobs(hr−1)] for dipeptidyl-tRNA ribosome complexes with a UAC codon in the A site. Complexes were reacted with eRF1(WT)•eRF3ΔN165•GTP, eRF1(AGQ)•eRF3ΔN165•GTP, or no release factor as indicated on the x-axis. In all cases, dark bars correspond to matched complexes and light bars correspond to mismatched complexes. Mean values are reported for all measurements, with error bars indicating the standard error of at least three measurements.
We next measured the rate constants for peptide release on matched and mismatched complexes carrying a codon in the A site that is near-cognate to a stop codon, specifically the UAC “sense” tyrosine codon. As anticipated, the rates of release observed in both reactions were exceedingly slow, with half-lives on the order of 14 h (Fig. 4D). Indeed, the observed rates of peptide release in this system are not faster than the uncatalyzed rate of the reaction (with an inactive eRF1 variant [AGQ] or in the absence of release factors) where the loss of stability in the mismatched complexes contributes to slightly elevated rates of peptidyl-tRNA dissociation and solution hydrolysis. Again, these results provide no evidence to support a model where premature termination is triggered by mismatches in the P site, as previously documented in E. coli (Zaher and Green 2009b).
DISCUSSION
Our quantitative analysis of the events of decoding and termination in yeast both on normal (matched) and P-site mismatched elongation and termination complexes has revealed key similarities and differences between the molecular events of translation in bacteria and eukaryotes. To probe the biochemical features of eukaryotic translation elongation and termination, we developed an in vitro reconstituted translation system where various stalled ribosome complexes could be prepared and evaluated using presteady state kinetic approaches. As anticipated based on the known high level of translational fidelity in vivo (∼10−4) (Salas-Marco and Bedwell 2005; Kramer et al. 2010), the acceptance of aminoacyl-tRNAs and eRF1•eRF3 on near-cognate codons was minimal in the in vitro system. Moreover, as in bacteria, the aminoglycoside class of antibiotics stimulated misreading during tRNA selection and inhibited release factor function. These observations are consistent with the fact that the functional core of the ribosome is well conserved from bacteria to eukaryotes (Alksne et al. 1993; Gutell et al. 1994; Liebman et al. 1995).
Despite these similarities, we observed substantial differences in the response of the eukaryotic ribosome to mismatches in the P-site codon:anticodon helix relative to the dramatic response of E. coli ribosomes (Zaher and Green 2009b). While there were modest increases in the overall rates of peptidyl transfer on ribosome complexes carrying a mismatch in the P site (suggestive of communication between the P and A site), these effects were uniform for both cognate and near-cognate decoding events, thus not impacting the overall fidelity of the subsequent round of elongation. Secondly, the rates of peptide release, and the K1/2 and the Kd of release factors for stop and near-stop codon-programmed ribosome complexes, were unaltered in the presence of the P-site mismatch. Indeed, premature termination on a sense codon appears to be a particularly unlikely outcome in yeast, given the exceedingly slow rates that we observed for this reaction (6.5 × 10−4 min−1; Fig. 4D). Given these two different sets of observations, we argue that retrospective quality control (as mediated through iterated miscoding and premature termination following an initial misreading event) does not happen in yeast as it does in E. coli.
We are cautious in emphasizing that these results do not exclude the possibility that some form of retrospective monitoring of ribosome complex integrity does happen in yeast. For example, it is possible that our experiments lack a key factor that is present in vivo and is responsible for recognition of the P-site mismatched ribosome complexes, though we note that we did not observe any premature termination of matched or mismatched complexes when cellular extracts were added (data not shown).
It is noteworthy that eukaryotes exhibit greater overall translational fidelity, apparently without retrospective quality control that bacteria exhibit with it. Hence, our observations lead us to speculate about what might be the reason for such fundamental differences in quality control during translation in bacteria and eukaryotes. On the one hand, the phenomenon that we have characterized in bacteria appears to simply report on structural perturbations in the A site of the ribosome that result from triggering structural perturbations in the neighboring P (and E) site(s). These A-site perturbations are manifested as promiscuous tRNA and RF selection activity that ultimately leads to premature termination of protein synthesis. While the premature termination event in bacteria is unusually dependent on a class 2 release factor (RF3) for optimal catalysis, we argue that the phenomenon is fundamentally the result of conformational changes within the ribosome itself. This view is supported by clear differences in the chemical modification patterns of the core ribosome structure in P-site matched and mismatched complexes (H Zaher, unpubl.).
If the phenomenon is ribosome-triggered, we can ask what differences between yeast and bacterial ribosomes might result in such different behavior. Certainly the most crucial nucleotides in the decoding site and peptidyl transferase center are highly conserved, and their mutation is lethal in organisms in both kingdoms (Cochella et al. 2007; Fan-Minogue and Bedwell 2008). Still, yeast and bacterial ribosomes are not identical even in these functionally critical regions, and some of the molecular differences have significant consequences for ribosome function. For example, paromomycin resistance in yeast is mediated by a pair of nucleotides in helix 44 in the decoding site; yeast can be rendered sensitive to paromomycin by a pair of mutations A1754G and G1645A that restore the bacterial sequence to helix 44 (Fan-Minogue and Bedwell 2008). It seems possible that features of the ribosome that are important for retrospective editing may be missing from yeast ribosomes, either because these features arose late in evolution in bacteria or were lost from yeast after these branches of the tree diverged.
It is also possible that retrospective quality control in bacteria is promoted by special features of both the ternary complex and the release factors in recognizing mismatch-carrying ribosome complexes. Bacterial and eukaryotic release factors are completely different, independently evolved proteins and, as such, it is possible that they might respond differently to mismatches in the P site. In contrast, EFTu and eEF1A are homologous proteins that function similarly in tRNA selection in the two systems (for example, paromomycin promotes miscoding in both systems), making this factor unlikely to be the basis for the observed differences. Moreover, since commercially available yeast tRNAPhe was used for many of the experiments in both the bacterial (Zaher and Green 2009b, 2010) and the yeast experiments, tRNAs per se are unlikely to explain the different responses seen in bacteria and eukaryotes.
Another possible explanation for the lack of retrospective editing in yeast is that fidelity is more effectively imposed during the kinetically slower tRNA selection process (Mathews et al. 2007). Alternatively, since retrospective editing may be principally involved in the control of frameshifting in bacteria, it may be that spontaneous frameshifting is a sufficiently rare event in yeast as to require no additional control mechanisms. Given that the ORFs in yeast are generally longer than in E. coli, the yeast may have generally evolved higher overall levels of tRNA selection fidelity to ensure the production of accurate, full-length proteins.
In conclusion, we have shown that mismatched yeast ribosome complexes behave quite distinctly from their bacterial counterparts in vitro. The consequence of the different behaviors is that yeast do not exhibit the retrospective quality control mechanism as it exists in bacteria. That said, we certainly wonder whether mismatched ribosome complexes are targeted by non-core translation factors, as is the case in no-go decay (Shoemaker et al. 2010) and other mRNA decay pathways. Further work in extracts and in vivo may reveal the answers to these questions.
MATERIALS AND METHODS
Ribosome preparation
A small culture of the yeast strain YAS2488 (MATa leu2-3 112 his4-539 trp1 ura3-52 cup1∷LEU2/PGK1pG/MFA2pG) (Acker et al. 2007) was grown in YPD medium for 24 h at 30°C on a roller wheel. The starter culture was then diluted into 18 L of YPD, and the large cultures were grown at 30°C with shaking until they reached an OD600 of 1.0. The cells were then collected by centrifugation, washed, and resuspended in a 1/10th volume of lysis buffer (1× ribo buffer A, 1 mg/mL heparin, 2 mM DTT, 7.5 mM Mg(OAc)2, 400 mM KCl, with Roche EDTA-free Complete Protease Inhibitors). Ribo buffer A was prepared as a 10× stock of 1 M KOAc, 200 mM HEPES-KOH pH 7.4, and 25 mM magnesium acetate. The cell paste was frozen as droplets in liquid nitrogen and lysed by grinding under cryogenic conditions, using the model 6870 freezer/mill (SPEX SamplePrep). The frozen powder was stored at −80°C until use.
The lysate was thawed and clarified by centrifugation at 25,000 × g for 30 min at 4°C. The supernatant was layered on top of a sucrose cushion (1× ribo buffer A, 500 mM KCl, 1 M sucrose, 2 mM DTT, 7.5 mM Mg(OAc)2). The ribosomes were pelleted by centrifugation at 264,900 × g for 106 min at 4°C. The ribosomes were resuspended in subunit separation buffer (50 mM HEPES-KOH pH 7.4, 2 mM MgCl2, 500 mM KCl, 2 mM DTT), stirred gently on ice for ∼30 min, then clarified by a 1-min spin at maximum speed in a refrigerated microfuge.
Subunits were separated by treatment with 1 mM puromycin. The ribosome solution was layered onto a 5%–20% sucrose gradient and centrifuged in a Beckman SW28 rotor at 76,221 × g for 9 h at 4°C. Separated 40S and 60S subunits were collected with the use of an in-line UV detector. Subunit-containing fractions were pooled and concentrated, and the buffer was exchanged to ribosome storage buffer (1× ribo buffer A, 250 mM sucrose, 2 mM DTT). Aliquots of the purified subunits were stored at −80°C until use.
Initiation and elongation factors
Our system utilizes the initiation factors eIF1, eIF1A, eIF5, eIF5B, and eIF2. We purified these factors according to published methods (Acker et al. 2007). The EF-Tu homolog, eEF1A, was purified in its native form from the same strain we used as the source of our ribosomes. A post-ribosomal supernatant was prepared in buffer with 50 mM KCl (buffer: 20 mM Tris pH 7.5, 0.1 mM EDTA, 1 mM DTT, 25% glycerol), and bound to DE52 resin. The unbound fraction was bound to CM-Sepharose. The bound fraction from the CM-Sepharose was eluted with 300 mM KCl, diluted to 50 mM KCl, and loaded onto a pre-equilibrated Tricorn column (Source 15S, 4.6/100 PE, GE Healthcare). Bound proteins were eluted with a linear gradient to 300 mM KCl. The eEF1A-containing fractions were identified by SDS-PAGE, pooled, and dialyzed against buffer with 100 mM KCl overnight. His-tagged eEF2 and eEF3 proteins were purified according to published procedures (Jorgensen et al. 2002; Andersen et al. 2004), and we followed these protocols as written.
Model mRNA
Model mRNAs were in vitro transcribed off of DNA oligonucleotides to generate mRNAs of the form: GG(UC)8U AUG UUC AAA UAA (UC)6. A variety of different codons were placed in the second and third coding positions of the mRNA including UUC (Phe), UUG (Leu, or near-cognate Phe), AAA (Lys), AAU (Asn, or near-cognate Lys), UAA (Stop), or UAC (Tyr, or near-cognate Stop).
Preparation and charging of tRNAs
Phenylalanine-specific tRNA from S. cerevisiae was purchased either from Sigma or from Chemblock. Charging reactions contained 1× buffer 517 (30 mM HEPES-KOH pH 7.4, 30 mM KCl, 15 mM MgCl2), 4 mM ATP, 10 uM phenylalanine, 5 mM DTT, and a 1/10th volume of an S100 extract. Reactions were incubated for 15 min at 37°C, then extracted twice with acid-buffered phenol and once with chloroform. Nucleic acids were precipitated with ethanol, resuspended in 20 mM KOAc, 2 mM DTT, pH 5.2, and stored in small aliquots at −80°C. The S100 extract was prepared as follows: Yeast cells were grown and lysed essentially as for the ribosome preparation. The lysis buffer used was 10 mM potassium phosphate buffer, pH 7.2, with the Roche EDTA-free Complete Protease Inhibitor cocktail. The post-ribosomal supernatant was applied to DE52 resin, and bound proteins were eluted with 250 mM potassium phosphate, pH 6.5. The eluate was concentrated, glycerol was added to a final concentration of 5%, and DTT was added to a final concentration of 2 mM. The extract was aliquoted and stored at −80°C.
Initiator methionine tRNA was in vitro transcribed off of a plasmid and charged as described in published literature (Acker et al. 2007).
Lysine-specific tRNA was in vitro transcribed off of a plasmid prepared in our laboratory. This tRNA was charged according to the same protocol as the tRNAPhe described above.
Release factors
The coding sequence of the S. cerevisiae gene for eRF1 (SUP45) was amplified by PCR and cloned into pPROEX-HtB (Invitrogen). The resultant plasmid (pDE8) was transformed into E. coli BL21(DE3). Cultures were grown in Terrific Broth, and expression was induced with IPTG. Cells were harvested by centrifugation and resuspended in lysis buffer (20 mM HEPES-KOH pH 7.4, 0.5 M KCl, 20 mM imidazole, 2 mM β-mercaptoethanol [β-ME]). Cells were lysed with the freezer/mill as in the ribosome preparation. The powder was resuspended in lysis buffer, clarified by centrifugation at 20,000 × g, and applied to a pre-equilibrated 5 mL HisTrap FF column (GE Healthcare). Bound proteins were eluted with elution buffer (lysis buffer with 500 mM imidazole). The eluate was applied to a 6 mL Resource Q column (GE Healthcare) and the bound proteins were eluted with a linear gradient to 1 M KCl. Fractions containing eRF1 were identified by SDS-PAGE, pooled, and concentrated. The sample was then separated on an S-75 column (GE Healthcare) in gel filtration buffer (20 mM HEPES-KOH pH 7.4, 100 mM potassium acetate pH 7.6, 2 mM DTT, 10% glycerol). Fractions containing eRF1 were pooled, concentrated, aliquoted, and stored at −80°C.
The yeast eRF3 protein (amino acids 165 through 685) was cloned into the vector pMBP-HTSHP and transformed into E. coli BL21(DE3). Cells were grown in TB media and induced with IPTG. The cells were collected by centrifugation, washed, and stored as a frozen pellet. The pellet was resuspended in amylose binding buffer (30 mM HEPES-KOH pH 7.4, 300 mM KCl, 5% glycerol, 0.1 mM GTP, 2 mM MgCl2, 2 mM β-ME) and lysed by passing twice through a French press. The lysate was clarified and applied to an amylose column. The bound protein was eluted with amylose elution buffer (same as binding buffer, but with 20 mM maltose). The eluted protein was concentrated and exchanged into cleavage buffer (30 mM HEPES KOH pH 7.4, 300 mM KCl, 20% glycerol, 0.1 mM GTP, 20 mM EDTA, 2 mM DTT), and the MBP tag was removed with Prescission protease. After cleavage, the protease was removed with a HisTrap FF column. The unbound fractions from the HisTrap column were separated on a Sephacryl S-200 column (GE Healthcare) in gel filtration buffer (30 mM HEPES-KOH pH 7.4, 300 mM KCl, 5% glycerol, 0.1 mM GTP, 2 mM MgCl2, 2 mM DTT). Fractions containing eRF3 were pooled, concentrated, aliquoted, and stored at −80°C.
Ribosome complex assembly
We assembled our ribosome complexes and carried out all our assays in buffer E (20 mM Tris-Cl pH 7.5, 100 mM KOAc, 2.5 mM Mg(OAc)2, 0.25 mM spermidine, and 2 mM DTT) (Alkalaeva et al. 2006). All reactions take place at 26°C unless otherwise noted. Ribosome complex assembly began with the preparation of ternary complex. For ternary complex preparation, 20 pmol of charged Met-tRNAiMet, 1 mM GTP, and 40 pmol of eIF2 were combined in 10 μL and incubated for 15 min. In a second tube, the 43S components were assembled: 40 pmol of 40S subunits, 400 pmol of model mRNA, 200 pmol of eIF1, and 100 pmol of eIF1A, in a total volume of 10 μL. The 43S components were added to the ternary complex and incubated for 5 min to generate 43S complex. In a fresh tube, the 80S components were assembled: 50 pmol 60S subunits, 100 pmol eIF5, 80 pmol eIF5B, and 1 mM GTP, in 10 μL. The 80S components were added to the 43S complex and incubated for 1 min to generate 80S initiation complexes. If initiation complexes are desired, they can be pelleted at this point, as will be described later.
For elongation, an eEF1A ternary complex was prepared by combining 25 pmol Phe-tRNAPhe, 50 pmol eEF1A, and 1 mM GTP in 15 μL, and preincubating for 15 min. This ternary complex was then added to the 80S initiation complex and allowed to react for 5 min. If Phe- on Leu-miscoding is desired, the magnesium concentration of the reaction was adjusted to 10 mM. Translocation was accomplished by adding GTP (1 mM final), ATP (1 mM final), 40 pmol eEF2, and 40 pmol eEF3, and incubating for an additional 5 min.
The stalled elongation complex thus generated was layered onto pelleting buffer (Buffer E with 35% sucrose). The reactions were centrifuged at 263,970 × g for 1 h at 4°C in a Beckman MLA-130 rotor. The ribosome pellets were resuspended in 50 μL buffer E, and then stored in single-use aliquots at −80°C.
Tripeptide synthesis assay
Lys- ternary complex was assembled by combining 5 pmol of Lys-tRNALys, 1 mM GTP, and 10 pmol of eEF1A in a total volume of 15 μL, and incubating for 15 min. An aliquot of dipeptide complex was thawed and mixed with the ternary complex to start the reaction. Aliquots of the reaction mixture were withdrawn and quenched with KOH. Samples were spotted onto TLC-cellulose plates, dried, and separated by electrophoresis in pyridine acetate buffer, pH 2.8 (200 mL glacial acetic acid and 6 mL pyridine per L of buffer) at 1200 V for 20 min.
Release assays
Release factor ternary complex was assembled by combining 10 pmol eRF1, 60 pmol of eRF3ΔN165, and 1 mM GTP in a volume of 15 μL for 5 min. An aliquot of pretermination complex was thawed and added to the RF ternary complex to start the reaction. Reaction aliquots were removed and quenched with formic acid. The quenched samples were stored on ice, and are spotted and run on electrophoretic TLCs as above.
Release factor binding assays
Dipeptide pretermination complexes were assembled as described above, except that the Phe-tRNAPhe used is labeled with proflavin at positions 16 and 17. Labeling was done according to published protocols (Wintermeyer and Zachau 1979), and the labeled tRNA was charged as described above. For the titrations, ∼10 nM complex, 0.5 mM GTP, and 1 μM eRF3ΔN165 were combined in a cuvette, and the fluorescent signal from the proflavin was measured. Increasing amounts of eRF1•eRF3ΔN165•GTP complex were added, and the fluorescence was measured. The change in fluorescence was plotted as a function of eRF1 concentration and fit to a hyperbola to calculate Kd.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank J. Lorsch (Johns Hopkins School of Medicine) for providing the yeast strain we use as the source of our ribosomes, the constructs for preparing initiation factors, the initiator methionine tRNA construct, and much invaluable assistance. T. Kinzy (UMDNJ-RWJMS) provided the constructs for purification of eEF2 and eEF3, and the protocol for purification of eEF1A. We also thank H. Zaher for many useful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2623711.
REFERENCES
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR 2007. Reconstitution of yeast translation initiation. Methods Enzymol 430: 111–145 [DOI] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Alksne LE, Anthony RA, Liebman SW, Warner JR 1993. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci 90: 9538–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CF, Anand M, Boesen T, Van LB, Kinzy TG, Andersen GR 2004. Purification and crystallization of the yeast translation elongation factor eEF3. Acta Crystallogr D Biol Crystallogr 60: 1304–1307 [DOI] [PubMed] [Google Scholar]
- Brown CM, McCaughan KK, Tate WP 1993. Two regions of the Escherichia coli 16S ribosomal RNA are important for decoding stop signals in polypeptide chain termination. Nucleic Acids Res 21: 2109–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348 [DOI] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R 2007. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Fan-Minogue H, Bedwell DM 2008. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA 14: 148–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Larsen N, Woese CR 1994. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev 58: 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol 164: 419–425 [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Carr-Schmid A, Ortiz PA, Kinzy TG, Andersen GR 2002. Purification and crystallization of the yeast elongation factor eEF2. Acta Crystallogr D Biol Crystallogr 58: 712–715 [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ 2010. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO, Liu R 1995. The accuracy center of a eukaryotic ribosome. Biochem Cell Biol 73: 1141–1149 [DOI] [PubMed] [Google Scholar]
- Martin R, Mogg AE, Heywood LA, Nitschke L, Burke JF 1989. Aminoglycoside suppression at UAG, UAA and UGA codons in Escherichia coli and human tissue culture cells. Mol Gen Genet 217: 411–418 [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB 2007. Origins and principles of translational control. In Translational control in biology and medicine (ed. Mathews MB et al. ), pp. 1–40 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Moazed D, Noller HF 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327: 389–394 [DOI] [PubMed] [Google Scholar]
- Parker J 1989. Errors and alternatives in reading the universal genetic code. Microbiol Rev 53: 273–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant EP, Nguyen P, Russ JR, Pittman YR, Nguyen T, Quesinberry JT, Kinzy TG, Dinman JD 2007. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS ONE 2: e517 doi: 10.1371/journal.pone.0000517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwalska M, Rospert S 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol Cell Biol 24: 9186–9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala T, Andersson D, Ehrenberg M, Kurland CG 1984. Hyper-accurate ribosomes inhibit growth. EMBO J 3: 2575–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol 348: 801–815 [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Herbert P, Lewendon A, Shaw WV, Tuite MF 1998. Missense translation errors in Saccharomyces cerevisiae. J Mol Biol 282: 13–24 [DOI] [PubMed] [Google Scholar]
- Thompson RC, Karim AM 1982. The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP [γS]. Proc Natl Acad Sci 79: 4922–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W, Zachau HG 1979. Fluorescent derivatives of yeast tRNAPhe. Eur J Biochem 98: 465–475 [DOI] [PubMed] [Google Scholar]
- Wu C, Amrani N, Jacobson A, Sachs MS 2007. The use of fungal in vitro systems for studying translational regulation. Methods Enzymol 429: 203–225 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Cochella L, Brunelle JL, He S, Green R 2006. Two distinct conformations of the conserved RNA-rich decoding center of the small ribosomal subunit are recognized by tRNAs and release factors. Cold Spring Harb Symp Quant Biol 71: 545–549 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009a. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009b. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2010. Kinetic basis for global loss of fidelity arising from mismatches in the P-site codon:anticodon helix. RNA 16: 1980–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]