Abstract
MicroRNAs (miRNAs) are small non-coding RNAs involved in fine-tuning of gene regulation. Antisense oligonucleotides (ONs) are promising tools as anti-miRNA (anti-miR) agents toward therapeutic applications and to uncover miRNA function. Such anti-miR ONs include 2′-O-methyl (OMe), cationic peptide nucleic acids like K-PNA-K3, and locked nucleic acid (LNA)-based anti-miRs such as LNA/DNA or LNA/OMe. Northern blotting is a widely used and robust technique to detect miRNAs. However, miRNA quantification in the presence of anti-miR ONs has proved to be challenging, due to detection artifacts, which has led to poor understanding of miRNA fate upon anti-miR binding. Here we show that anti-miR ON bound to miR-122 can prevent the miRNA from being properly precipitated into the purified RNA fraction using the standard RNA extraction protocol (TRI-Reagent), yielding an RNA extract that does not reflect the real cellular levels of the miRNA. An increase in the numbers of equivalents of isopropanol during the precipitation step leads to full recovery of the targeted miRNA back into the purified RNA extract. Following our improved protocol, we demonstrate by Northern blotting, in conjunction with a PNA decoy strategy and use of high denaturing PAGE, that high-affinity anti-miRs (K-PNA-K3, LNA/DNA, and LNA/OMe) sequester miR-122 without causing miRNA degradation, while miR-122 targeting with a lower-affinity anti-miR (OMe) seems to promote degradation of the miRNA. The technical issues explored in this work will have relevance for other hybridization-based techniques for miRNA quantification in the presence of anti-miR ONs.
Keywords: microRNA, miR-122, oligonucleotide, Northern blot, RNA extraction
INTRODUCTION
MicroRNAs (miRNAs) are an important class of small non-coding RNAs that regulate gene expression post-transcriptionally. They are transcribed as stem–loop-containing precursors and are processed into shorter duplex RNAs by a series of enzyme complexes, first in the nucleus and then following transport into the cytosol, in order to become fully active (Siomi and Siomi 2010). The final complex, miRISC, directs one of the two RNA strands (the guide strand) to bind to the 3′-UTR of target mRNAs resulting in repression of gene expression. Most miRNAs are able to target a number of mRNAs (Lim et al. 2005; Friedman et al. 2009), and their expression and activity are often associated with complex cellular pathways, for example, cell growth and apoptosis (Cheng et al. 2005), metabolism (Esau et al. 2006), viral infection (Skalsky and Cullen 2010), and cancer (Lu et al. 2005).
Anti-miRNA oligonucleotides (ONs) capable of forming complementary base pairs with the guide strand of miRNAs have proved to be of great value in recent years as tools to understand miRNA action and as potential therapeutics (Krützfeldt et al. 2005; Esau et al. 2006; Kloosterman et al. 2007; Vermeulen et al. 2007; Elmén et al. 2008a; Lu et al. 2009; Fabani et al. 2010; Lanford et al. 2010; Robertson et al. 2010). Several types of ON analogs have been proposed as anti-miRNA (anti-miR) agents in order to enhance steric-block potency, biostability, and binding affinity for their RNA targets. Some of these include charge neutral ONs, such as peptide nucleic acids (PNA) (Fabani and Gait 2008; Oh et al. 2009; Fabani et al. 2010) and phosphorodiamidate morpholino oligonucleotides (PMO) (Flynt et al. 2007; Kloosterman et al. 2007; Eberhart et al. 2008); 2′-modified ONs such as 2′-O-methyl (OMe) (Hutvágner et al. 2004; Meister et al. 2004; Cheng et al. 2005), which also form the basis of antagomiRs (Krützfeldt et al. 2005, 2007) 2′-O-methoxyethyl (MOE) (Davis et al. 2006; Esau et al. 2006) and 2′-fluoro/2′-methoxyethyl mixmers (2′F/MOE) (Davis et al. 2009); and the particularly strong RNA binding analog locked nucleic acid (LNA) in mixmers with DNA (Ørom et al. 2006; Elmén et al. 2008a; Lanford et al. 2010), OMe (Fabani and Gait 2008), or MOE (Davis et al. 2006, 2009).
Anti-miR ONs inhibit their miRNA target by a steric blocking mechanism. However, the fate of the miRNA following anti-miR ON binding is still unclear (Horwich and Zamore 2008). Some evidence suggests that upon binding of an LNA/DNA anti-miR, the miRNA becomes complexed and sequestered in cells, but not degraded (Elmén et al. 2008a,b). In contrast, there are also data suggesting anti-miR-mediated miRNA degradation for antagomiRs, MOE, and PNA anti-miRs (Krützfeldt et al. 2005; Esau et al. 2006; Fabani and Gait 2008; Ameres et al. 2010). More recently, a report suggested that the miRNA fate upon anti-miR binding might be chemistry-dependent. While miRNA inhibition by a 2′F/MOE anti-miR ON would not degrade the miRNA, treatment with MOE anti-miR ONs would induce miRNA degradation (Davis et al. 2009).
Many techniques have been proposed for detection of miRNAs. Most of these techniques are dependent on the hybridization properties between the target miRNA and a complementary nucleic acid strand probe (Cissell and Deo 2009). Among these methods, Northern blotting has proved to be a robust and widely used technique that also allows detection of not only mature miRNAs but also their precursors (Lee and Ambros 2001; Lee et al. 2002; Obernosterer et al. 2006) and that does not require specialized equipment. It is useful to quantify miRNA levels, to determine their size and to validate predicted miRNAs (Várallyay et al. 2007). The sensitivity, safety, and speed of this assay have been improved significantly by use of LNA-modified detection probes (Várallyay et al. 2008), by non-radioactive labeling of the detection probe (Kim et al. 2010), and by EDC-mediated cross-linking of the RNA to appropriate membranes (Pall and Hamilton 2008). Thus, it is not surprising that Northern blotting is also the most frequently used method to monitor miRNA abundance after treatment with anti-miR ONs (Chan et al. 2005; Davis et al. 2006; Esau et al. 2006; Krützfeldt et al. 2007; Elmén et al. 2008b; Fabani and Gait 2008; Fabani et al. 2010; Lanford et al. 2010).
Despite the advantages of Northern blotting over other techniques to detect miRNAs, miRNA detection by Northern blot in the presence of anti-miR ONs can give misleading results, which partly explains the lack of consensus on the mode of action of anti-miR ONs. It has been shown that anti-miR ONs can interfere with miRNA detection at two different steps. First, during the Northern blot hybridization step, the probe for miRNA detection may not be able to displace the anti-miR ON already bound to the miRNA of interest, even in samples analyzed in denaturing PAGE conditions (Chan et al. 2005; Davis et al. 2009). In this case, uncomplexed miRNA would be detected, while an miRNA:anti-miR duplex would not be detected, thus leading to the interpretation that there is a (partial) knockdown of the miRNA. Second, the miRNA:anti-miR duplexes might not be recovered in the purified RNA fraction during the commonly used guanidinium thiocyanate-phenol-choloroform RNA extraction protocol (Chomczynski and Sacchi 2006) (commercially known as TRI-Reagent or TRIzol), yielding an RNA extract that does not represent the physiological levels of the particular miRNA (Fabani and Gait 2008; Lu et al. 2009; Davis et al. 2009). Therefore, in light of these technical issues, the inability to detect an miRNA by Northern blot in the presence of anti-miRs is not sufficient evidence for anti-miR-mediated miRNA degradation.
There have been suggestions on how to overcome some of these problems. It has been shown that miRNA masking by the anti-miR ON can be relieved, at least for lower-affinity analogs, by increasing the denaturing conditions of the electrophoresis separation (Krützfeldt et al. 2005). Alternatively, a competitor PNA ON, having the same sequence as the miRNA of interest and that binds to the anti-miR ON, can be introduced in the RNA samples prior to PAGE separation, allowing for miRNA release and its detection by Northern blot (Davis et al. 2009). However, there are currently no reports on how to recover miRNA:anti-miR duplexes that have been lost during the RNA extraction procedure.
MicroRNA-122 (miR-122), a liver-specific miRNA associated with lipid metabolism (Esau et al. 2006) and virus infection (Triboulet et al. 2007; Chang et al. 2008), has become one of the main miRNA models for anti-miR-mediated inhibition of miRNAs, mainly due to the huge interest in it as a target for therapeutics (Jopling et al. 2005; Krützfeldt et al. 2005, 2007; Esau et al. 2006; Elmén et al. 2008b; Fabani and Gait 2008; Davis et al. 2009; Lanford et al. 2010).
Here we show in detail how anti-miRs of four different chemistries—2′-O-methyl (OMe), peptide nucleic acid (PNA), and LNA/OMe and LNA/DNA mixmers targeting miR-122—interfere at the two aforementioned steps of the miRNA detection process by Northern blotting. Furthermore, we demonstrate complete recovery of miRNA:anti-miR duplexes back into the RNA extract, even for high-affinity anti-miR ONs, by modification of the widespread TRI-Reagent-based RNA extraction procedure. Finally, by use of the new RNA extraction procedure together with a decoy PNA ON strategy (Davis et al. 2009) and polyacrylamide gel electrophoresis (PAGE) separation of RNA samples under highly denaturing conditions (Krützfeldt et al. 2005), we were able to shed light on the cellular outcome for miRNAs upon anti-miR targeting of different chemistries.
RESULTS
OMe, LNA/OMe, and LNA/DNA anti-miR interfere with miR-122 detection by Northern blotting
We first wished to verify whether anti-miR ONs interfere with miRNA detection. We addressed this issue by detection of miR-122 by Northern blotting in the presence of anti-miR-122 ONs of four different chemistries: K-PNA-K3 (23-mer + 4 Lys) (Fabani and Gait 2008), OMe (31-mer) (Jopling et al. 2005), LNA/OMe (23-mer) (Fabani and Gait 2008), and a commercial 23-mer LNA/DNA knockdown probe (Exiqon). PNA, OMe, and LNA/OMe anti-miR ONs have been validated previously as effective miR-122 inhibitors through additional assays (Jopling et al. 2005; Fabani and Gait 2008). For miR-122 detection, RNA was extracted using TRI-Reagent (Sigma), which is also available (TRIzol) from multiple suppliers, following the manufacturer's protocol that involves the use of the standard 0.5 volumes of isopropanol for each volume of TRI-Reagent for RNA precipitation. RNA separations were carried out by use of very highly denaturing PAGE in the presence of 8 M urea and 20% formamide (Krützfeldt et al. 2005). Additionally, 8 M urea and 20% formamide were included in the loading buffer, and the sample was preheated before loading. These highly stringent PAGE conditions were used to maximize the likelihood of the miRNA being single-stranded and thus most easily detectable. Finally, LNA probes were used for miRNA detection by Northern blotting because of their superior sensitivity compared to standard DNA probes (Várallyay et al. 2008) and their ability to detect miRNAs in the presence of anti-miRs (Horwich and Zamore 2008).
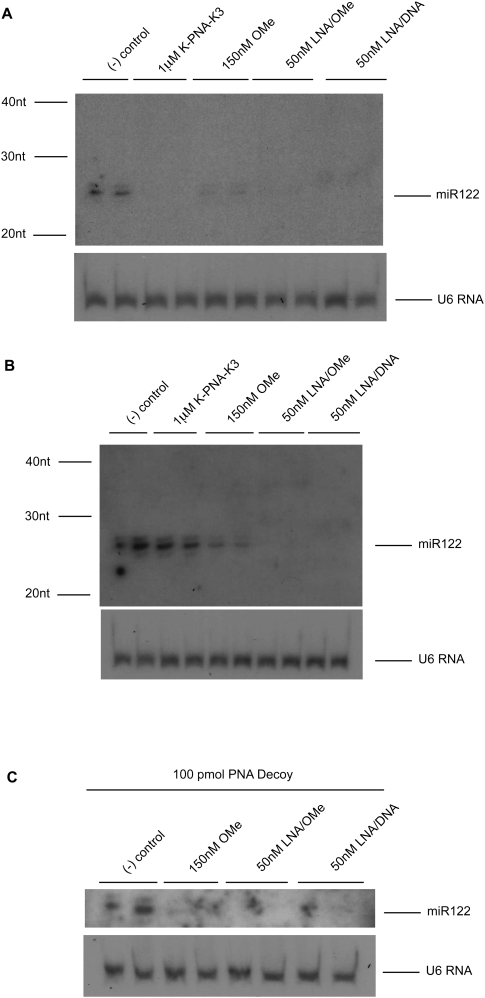
We treated Huh7 cells that express endogenous miR-122 with the anti-miRs and measured miR-122 levels by Northern blot analysis. K-PNA-K3 anti-miR was added to the cells without the use of a transfection agent as described previously (Fabani and Gait 2008), while the other three anti-miRs were lipofected at the indicated concentrations. As expected from previous studies, treatment with any of the anti-miRs led to a reduction in miR-122 signal (Fig. 1A). OMe anti-miR treatment resulted in partial miR-122 loss of signal, while for K-PNA-K3 and LNA/OMe anti-miRs, the loss was much stronger. MiR-122 detection following LNA/DNA anti-miR treatment resulted in a slight band shift consistent with miR122:LNA/DNA anti-miR duplex formation as previously reported (Elmén et al. 2008a,b). However, the strength of the miR-122:LNA/DNA anti-miR duplex signal was much weaker as compared to untreated cells (negative control) (Fig. 1A). Note that in our conditions, an approximate loss of 95% of miR-122 signal was achieved in the low nanomolar range for lipofected LNA/DNA and LNA/OMe anti-miRs, in the low hundreds nanomolar range for lipofected OMe anti-miR and in the low micromolar range for free delivered K-PNA-K3 anti-miR. Also note that pre-miR122 bands could be detected and they remained unchanged in the presence of any of the anti-miRs tested (data not shown).
FIGURE 1.
Anti-miRs can interfere with miR-122 detection by Northern blot. (A) Northern blot of total purified RNA from Huh7 cells treated with miR-122 anti-miRs K-PNA-K3, OMe, LNA/OMe, or LNA/DNA. Total purified RNA from untreated cells was used as negative controls. (B) Northern blot of total purified RNA after addition of miR-122 anti-miRs at equivalent amounts to mimic transfections in A to Huh7 cell lysates. Total purified RNA from Huh7 cells lysates without addition of anti-miR was used as negative control. (C) Northern blot of total RNA from Huh7 cell lysates treated as in B with OMe, LNA/OMe, or LNA/DNA anti-miR and further addition of 100 pmol of decoy PNA ON to the purified RNA. RNA from untreated lysates but with 100 pmol of decoy PNA ON addition was used as negative control. U6 RNA was used as loading control for all gels.
To explore this loss of signal, we added to untreated Huh7 cell lysates in TRI-Reagent equivalent amounts of each anti-miR to mimic the cell treatments carried out in the previous experiment prior to RNA extraction and Northern blotting, an approach that has been commonly used previously (Krützfeldt et al. 2005, 2007; Esau et al. 2006; Davis et al. 2006, 2009; Fabani and Gait 2008; Lu et al. 2009). As shown in Figure 1B, the presence of a band corresponding to miR-122 in the sample to which K-PNA-K3 anti-miR had been added suggests that this anti-miR does not interfere with miR-122 detection by Northern blotting under these conditions. In contrast, partial interference was seen with OMe anti-miR, and a complete loss of miR-122 signal was seen in cell lysates containing LNA/OMe or LNA/DNA anti-miR, showing in these cases major interference with miR-122 detection. To test whether the lack of miR-122 signal was due to complex formation with the corresponding anti-miR, hindering the miRNA from the labeled miRNA detection probe, we repeated the previous experiment, but before PAGE separation we added an excess of a competitor (decoy) PNA ON having the same sequence as miR-122, as previously described by Davis et al. (2009) (Fig. 1C). No further recovery of miR-122 signal was achieved under these conditions, suggesting that even in the presence of excess decoy, miR-122 is not fully detected in the presence of these anti-miRs. These results show that OMe, LNA/OMe, and LNA/DNA anti-miR ONs interfere with the Northern blot detection of the targeted miRNA and, further, that it is not possible to conclude using published protocols for this assay whether or not miR-122 is degraded or sequestered upon anti-miR treatment.
K-PNA-K3 anti-miR sequesters miR-122 without causing miR-122 degradation
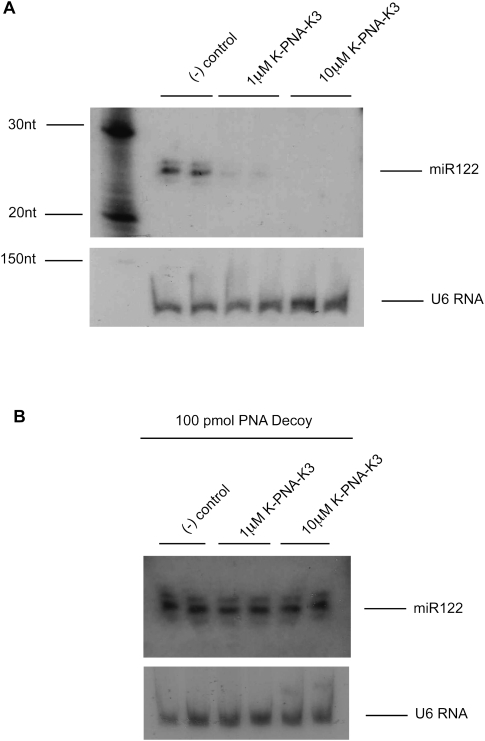
Since K-PNA-K3 anti-miR was the only backbone chemistry that did not seem to interfere with miR-122 detection when Huh7 cell lysates were supplemented with this anti-miR, it appeared likely that the reduction in the miR-122 signal strength seen in Huh7 cells treated with this anti-miR represented a genuine miRNA knockdown (Fig. 1A,B), as we suggested previously (Fabani and Gait 2008). To further examine this observation, we treated Huh7 cells with K-PNA-K3 anti-miR at two different concentrations (1 and 10 μM). As expected, a dose-dependent loss of miR-122 signal was seen (Fig. 2A). We then investigated whether the observed decrease in miR-122 signal was due to target degradation by supplementing the RNA samples with decoy PNA ON prior to high denaturing PAGE. To our surprise, the recovery of miR-122 signal was complete, demonstrating that K-PNA-K3 anti-miR sequesters miR-122 in Huh7 cells and does not cause its degradation, even at high anti-miR concentrations (Fig. 2B). Thus, the loss of signal in Figure 2A cannot be attributed to degradation and must instead be due to undetected K-PNA-K3:miR-122 complex formation, resulting because bound K-PNA-K3 anti-miR is not displaced by the miRNA detection probe under these conditions. Note that when K-PNA-K3 anti-miR was added to untreated Huh7 cell lysates in TRI-Reagent at equivalent amounts to mimic the concentrations used in transfections in Figure 2A, we found it to interfere with miR-122 detection only at the highest concentration tested, 10 μM (Supplemental Fig. 1). This result and the dose-dependent effect seen in Figure 2A suggest that the level of interference by anti-miR ONs in detection of targeted miRNAs by Northern blotting is also dependent on the concentration of anti-miR used. A similar anti-miR dose-dependent effect on miR-122 detection by Northern blotting was seen for the other anti-miRs tested in cell lysates that received addition of each anti-miR to mimic transfections in the nanomolar range (data not shown).
FIGURE 2.
miR-122 fate upon targeting with K-PNA-K3 anti-miR. (A) Northern blot of total purified RNA from Huh7 cells treated with K-PNA-K3 anti-miR at 1 μM and 10 μM. RNA from untreated cells was used as negative control. (B) Northern blot of total purified RNA from Huh7 cells treated with K-PNA-K3 anti-miR as in A and further addition of 100 pmol of decoy PNA ON to the purified RNA. RNA from untreated cells but with 100 pmol of decoy PNA ON addition was used as negative control. U6 RNA was used as loading control for all gels.
miR-122:anti-miR duplex loss and recovery during RNA extraction
The experiments so far indicated that all anti-miR ONs tested interfere with miRNA detection by Northern blotting. However, in contrast to our observation for K-PNA-K3 anti-miR, miR-122 signal could not be fully recovered when OMe, LNA/OMe, or LNA/DNA anti-miRs were added to untreated Huh7 cell lysates, even in the presence of decoy PNA ON (Fig. 1B,C). This might indicate that either the decoy PNA ON is not able to displace the anti-miR from the miRNA in the conditions tested, or that miRNA:anti-miR complexes are lost during RNA purification, prior to Northern blotting, as previously suggested (Fabani and Gait 2008; Davis et al. 2009; Lu et al. 2009). To explore the second possibility, we added anti-miRs to untreated Huh7 cell lysates and supplemented the lysates with small amounts of 32P-labeled single-stranded miR-122 mimic (32P-miR122). Use of 32P-miR122 allows for easy identification of miR-122 throughout the RNA extraction procedure and eliminates the need for a radiolabeled anti-miR-122 detection probe for Northern blotting. Furthermore, 32P-miR122:anti-miR duplexes should be detectable as well as free 32P-miR122. Figure 3A (upper panel) confirms that, under our conditions, 32P-miR122 is partially or completely lost from the purified RNA fraction when RNA was extracted in the presence of OMe, LNA/OMe, or LNA/DNA anti-miRs. As expected, 32P-miR122 is totally recovered in the purified RNA fraction in the presence of equivalent amounts of K-PNA-K3 anti-miR to mimic 1 μM transfections, a concentration at which K-PNA-K3 did not interfere with miR-122 detection in lysates of untreated cells (Fig. 1B). Careful analysis of the individual fractions obtained during the RNA extraction procedure showed that most of the missing 32P-miR122 remained in the supernatant of the RNA precipitation step with isopropanol (Fig. 3A, lower panel). Note that under our conditions 32P-miR122 was not differentially distributed during the initial phase separation of the RNA extraction protocol in the presence or absence of an LNA-based anti-miR (Supplemental Fig. 2), in contrast to a previous suggestion (Davis et al. 2009).
FIGURE 3.
miR-122 loss and recovery during RNA extraction. Denaturing PAGE of cell lysates supplemented with 32P-miR122 and further addition of K-PNA-K3, OMe, LNA/OMe, or LNA/DNA anti-miRs before RNA extraction, at equivalent amounts to mimic transfections in Figure 1A. Negative controls correspond to cell lysates spiked with 32P-miR122 only. (A) Purified RNA (upper panel) and the remaining isopropanol supernatant (lower panel) of samples after RNA extraction with 1 equivalent (v/v) isopropanol. (B) Purified RNA (upper panel) and the remaining isopropanol supernatant (lower panel) from cell lysates treated as in A with LNA/OMe anti-miR after RNA extraction with 1, 2, or 3 equivalents (v/v) isopropanol. (C) Purified RNA from samples treated as in A after RNA extraction with 3 equivalents (v/v) isopropanol.
All available kits based on the guanidinium thiocyanate–phenol–chloroform RNA-extraction method (Chomczynski and Sacchi 2006) (TRIzol, Tri-Reagent, etc.) stipulate the use of 0.5 volumes of isopropanol for each volume of TRI-Reagent for RNA precipitation. This corresponds approximately to 1 equivalent (v/v) of isopropanol relative to the volume of the aqueous RNA-containing phase after the first phase separation step of the protocol. We thought that RNA precipitation could be improved by increasing the amount of isopropanol at the RNA precipitation step. Indeed, by adjusting the volume of isopropanol from 1 to 3 equivalents (v/v) for RNA precipitation, we observed increasing recovery of 32P-miR122 in the purified RNA phase (Fig. 3B, upper panel) and decreasing abundance of 32P-miR122 in the remaining isopropanol supernatant (Fig. 3B, lower panel; Supplemental Fig. 3) in the presence of LNA/OMe anti-miR, as analyzed by denaturing PAGE. Similarly, 32P-miR122 could also be completely recovered by precipitating the RNA with 3 equivalents (v/v) of isopropanol in the presence of OMe and LNA/DNA anti-miRs (Fig. 3C). The superior binding strength of LNA-based anti-miRs is evidenced by the presence of lower-mobility complexes formed between LNA/DNA or LNA/OMe anti-miRs with 32P-miR122, even under highly denaturing PAGE conditions (8 M urea/20% formamide). The same conditions seem to be sufficient for complex dissociation between OMe anti-miR and 32P-miR122. In the absence of 20% formamide, the OMe anti-miR showed complex formation (Supplemental Fig. 4), as previously shown for antagomiRs (Krützfeldt et al. 2005). Interestingly, under denaturing conditions, duplexes did not migrate in the gels at the expected molecular size. However, in vitro binding assays between 32P-miR122 and each anti-miR under these highly denaturing conditions confirmed complex formation (Supplemental Fig. 5). Note that in Figure 3B, partial melting of the 32P-miR122:LNA/OMe anti-miR duplex is seen in the isopropnaol supernatant phase; this is due to the NAP-10 purification (desalting) procedure.
The fate of miR-122 upon anti-miR targeting is dependent on the anti-miR chemistry
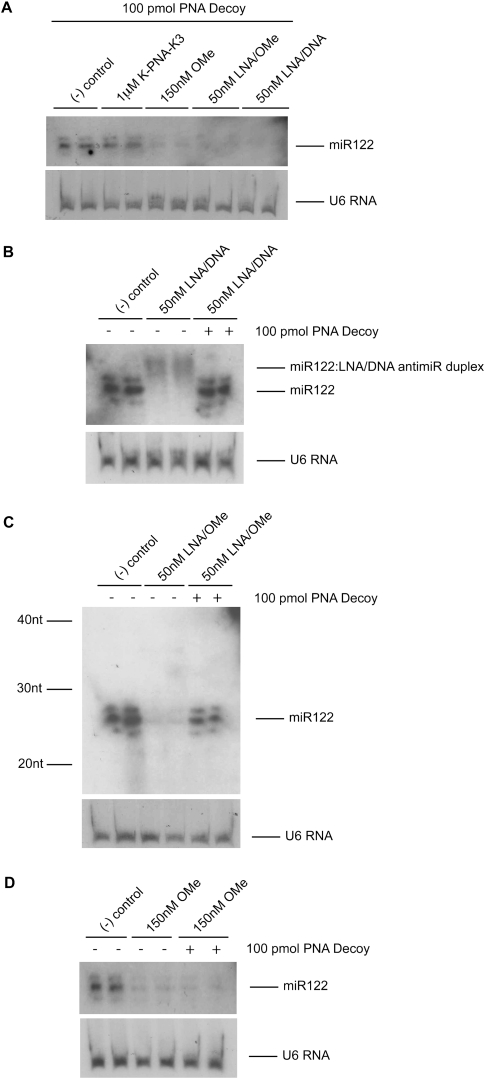
Having optimized the RNA extraction procedure in order to recover miRNA:anti-miR duplexes back into the RNA extract (Fig. 3), we set out to determine the cellular outcome for miR-122 upon targeting with LNA/DNA, LNA/OMe, or OMe anti-miRs. First, total RNA was obtained from Huh7 cells transfected with each of these anti-miR oligonucleotides by standard purification (1 equivalent of isopropanol compared to the aqueous phase for the RNA precipitation step) that had been supplemented with PNA decoy and analyzed by Northern blotting. Figure 4A shows that under these standard purification conditions, miR-122 signal was only recovered through decoy use in the presence of K-PNA-K3 anti-miR, but not in the presence of any of the other three anti-miRs of different chemistry, and confirms the lack of degradation of the miR-122 target in the presence of K-PNA-K3 seen in Figure 2B.
FIGURE 4.
miRNA fate upon targeting with LNA/DNA, LNA/OMe, or OMe anti-miR. (A) Northern blot of total purified RNA extracted with 1 equivalent (v/v) isopropanol from Huh7 cells treated as in Figure 1A and further addition of decoy PNA ON to the RNA samples before PAGE separation. RNA from untreated Huh7 cells but in the presence of decoy PNA ON was used as negative control. (B–D) Northern blots of total purified RNA extracted with 3 equivalents (v/v) isopropanol from HuH7 cells treated as in A with either LNA/DNA anti-miR (B), LNA/OMe anti-miR (C), or OMe anti-miR (D) in the presence or absence of decoy PNA ON. RNA from untreated Huh7 cells was used as negative control. U6 RNA was used as loading control for all gels.
As suggested in Figure 3, OMe, LNA/OMe, and LNA/DNA anti-miR chemistries prevent the miR-122:anti-miR duplexes from being properly recovered in the RNA extract following the standard RNA extraction procedure. Therefore, we decided to test our improved RNA extraction protocol (3 equivalents [v/v] isopropanol for RNA precipitation step) in Huh7 cells treated with these three anti-miR chemistries and by analyzing by Northern blotting as before. Figure 4B shows that for cells treated with LNA/DNA anti-miR, the procedure allows for partial detection of miR-122 in the form of a low-mobility complex. MiR-122 is released by treatment of this complex with decoy PNA ON, showing that this high-affinity anti-miR sequesters miR-122 without inducing target degradation in cells, just as for K-PNA-K3 anti-miR. Furthermore, when LNA/DNA anti-miR was used in mice, the miR-122 signal was recovered in Northern blotting under the improved RNA extraction conditions, and once again decoy use resulted in recovery of the complete miR-122 signal in the case of K-PNA-K3 (Supplemental Fig. 6).
Treatment of Huh7 cells with the very-high-affinity LNA/OMe anti-miR showed a similar result, but the miR-122:anti-miR duplex was not detected, and the recovery of the miR-122 signal upon addition of decoy PNA ON was not complete (Fig. 4C). The most likely explanation is that the less strongly binding decoy PNA ON is not able to completely displace the highly stable miR-122:LNA/OMe anti-miR duplex. Addition of a greater excess of decoy PNA ON could not be used to enhance the recovery of the miR-122 signal because the PNA decoy detection competes too efficiently under these conditions for the radioactive detection probe and masks the ability to detect the miR-122 (data not shown). Also, cell treatment with a higher amount (150 nM) of LNA/OMe anti-miR did not result in a lower miR-122 signal than in the case of use of 50 nM anti-miR (data not shown), thus ruling out dose dependence of the signal detection. Our results are best consistent with the view that this high-affinity anti-miR sequesters miR-122 without causing significant miRNA degradation.
In contrast, for the OMe anti-miR, miR-122 signal was not recovered following use of the improved method for miRNA recovery and detection by Northern blotting in the presence of decoy PNA ON (Fig. 4D). The lack of miR-122 signal in the presence of OMe anti-miR under these conditions is very unlikely to be due to an artifact. We have shown that in the presence of these amounts of OMe anti-miR, miR-122 would have been detected if present in the RNA extract when RNA precipitation is carried out with 3 equivalents (v/v) of isopropanol (Fig. 3C). Furthermore, the miR-122:OMe anti-miR duplex is clearly fully disrupted under our highly denaturing conditions (Supplemental Figs. 4, 5), and this is independent of whether the decoy PNA ON is present or not, ruling out the possibility that decoy PNA ON is unable to displace the OMe anti-miR bound to miR-122. Thus, uniquely among the four anti-miR chemistries tested, OMe anti-miR usage in Huh7 cells is consistent with degradation of the miR-122 target.
DISCUSSION
Northern blotting has become a primary method for detection, identification, and validation of novel miRNAs, and it has been extensively used to demonstrate the effect of anti-miR ONs on miRNA expression (Chan et al. 2005; Davis et al. 2006; Esau et al. 2006; Krützfeldt et al. 2007; Elmén et al. 2008b; Fabani and Gait 2008; Fabani et al. 2010; Lanford et al. 2010). Of the four chemistries we investigated, only LNA/DNA has advanced to clinical trials as reported by Santaris Pharma in 2010. OMe has been the most widely used anti-miR chemistry (Chang et al. 2004; Hutvágner et al. 2004; Meister et al. 2004; Jopling et al. 2005; Horwich and Zamore 2008), which also forms the basis of antagomiRs (Krützfeldt et al. 2005, 2007). PNAs containing cationic amino acids have been shown to inhibit miRNAs in cells (Fabani and Gait 2008; Oh et al. 2009, 2010; Fabani et al. 2010) and in vivo (Fabani et al. 2010). We reported high-affinity LNA/OMe anti-miRs for miR-122 inhibition in cell culture, which proved to be more potent than the OMe anti-miR, (Fabani and Gait 2008) and for other steric-blocking applications (Arzumanov et al. 2001; Brown et al. 2005).
Problems with the Northern blotting technique have hitherto served to obscure the fate of miRNAs targeted by anti-miRs. We have now demonstrated, through use of a PNA decoy approach (Davis et al. 2009), that K-PNA-K3 anti-miR acts by sequestering the miRNA without causing its degradation in cell culture (Fig. 2) or in vivo (Supplemental Fig. 6). For OMe, LNA/OMe, and LNA/DNA anti-miRs, neither analysis under highly denaturing conditions nor addition of a PNA decoy was sufficient to reveal the effect of these anti-miRs on miR-122 levels (Figs. 1C, 4A). Loss of miR-122 in the purified RNA samples treated with these anti-miRs (Fig. 3A, upper panel) has been reported previously for high-affinity LNA-containing anti-miRs by us and others (Fabani and Gait 2008; Davis et al. 2009; Lu et al. 2009). We did not find anti-miRs to partition radiolabeled miR-122 (32P-miR-122) into the organic phase during the RNA extraction (TRI-Reagent) protocol as measured by scintillation counting (Supplemental Fig. 2), in contrast to similar experiments reported for LNA/MOE anti-miRs (Davis et al. 2009). Instead, the LNA-containing anti-miRs, and OMe anti-miR (partially), caused retention of targeted 32P-miR122 in the isopropanol supernatant during precipitation when the standard RNA extraction procedure was followed (Fig. 3A). However, miR-122 could be recovered fully by use of 3 equivalents (v/v) of isopropanol instead of the standard single equivalent (Fig. 3B,C; Supplemental Fig. 3).
By use of these improved techniques, it now becomes clear that LNA/DNA and LNA/OMe anti-miRs, just like K-PNA-K3, also inhibit miR-122 in cells by complex formation without causing significant miRNA degradation (Fig. 4B,C), and this applies also for LNA/DNA anti-miRs in vivo (Supplemental Fig. 6). Our results are consistent with the conclusions of Elmén et al. (2008a,b) that LNA/DNA anti-miRs act to complex miR-122 without degradation, but note that we and others (Chan et al. 2005; Naguibneva et al. 2006; Laneve et al. 2007; Inomata et al. 2009) have been unable to clearly visualize such complexes by denaturing PAGE under standard RNA extraction conditions. With our higher isopropanol precipitation conditions during extraction and under denaturing PAGE, miR-122 can be detected by Northern blotting partially as a complex with the LNA/DNA anti-miR (Figs. 3C, 4B; Supplemental Fig. 6b). Addition of decoy PNA ON resulted in full recovery of the miR-122 signal as single-stranded miRNA, thus confirming the miR-122 sequestration during cell treatment rather than degradation (Fig. 4B). Our study is the first to report that high-affinity LNA/OMe anti-miR also does not give rise to significant miRNA degradation (Fig. 4C). Davis et al. (2009) reported that the high-affinity 2′F/MOE anti-miR also sequesters miR-122 without causing miRNA degradation, but we have been unable to investigate MOE-containing anti-miR ONs, because such ONs are not commercially available. We therefore conclude that miRNA sequestration without significant miRNA degradation is a general feature of high-affinity anti-miRs.
The OMe anti-miR was the only one of the four tested that appears to induce miR-122 degradation upon targeting (Fig. 4D). This 31-mer OMe ON is the same as that shown to be functionally active as an anti-miR122 in Huh7 cells (Jopling et al. 2005). Several other studies that showed functional inhibition of miRNA activities in a variety of cell types by OMe ONs also showed loss of miRNA signal on Northern blotting, suggesting that these miRNAs were degraded by the OMe anti-miR in cells (Hutvágner et al. 2004; Chan et al. 2005; Krichevsky et al. 2006; Yang et al. 2008). However, in all these cases, the standard RNA extraction conditions were used, and in some cases, only lower-affinity DNA probes were used for miRNA detection, which may have compromised the Northern blotting assays. Horwich and Zamore (2008) reported that in Drosophila cells OMe anti-miRs had no significant effect on endogenous miR-277 levels in Northern blotting, which contrasts with all other studies in mammalian cells. Krützfeldt et al. (2005, 2007) reported that in mice, inhibition of miR-122 by antagomiRs (cholesterol-conjugated OMe ONs partially modified with a phosphorothioate backbone) led to target miRNA degradation, but that the equivalent OMe anti-miR (partially or fully modified with phosphorothioate backbone but not cholesterol-conjugated) resulted in detection of the miR-122 RNA in Northern blotting in denaturing PAGE, suggesting miR-122 complex formation (Krützfeldt et al. 2005). Cholesterol-unconjugated all PO OMe ONs were not reported in this study. Very recently, Ameres et al. (2010) showed that in HeLa cells, antagomiRs targeted to miR-16 or miR-21 lead to miRNA degradation through tailing (addition of predominately adenosines and uridines at the 3′ end of the miRNA) and trimming (usually 3′ to 5′) of the targeted miRNA. Ameres's work suggests an interesting mechanism for anti-miR-mediated degradation of miRNAs; however, under our conditions, we failed to detect trimming or tailing of miR-122 by Northern blotting after cell treatment with anti-miRs.
Under our improved isopropanol precipitation conditions of RNA and use of a decoy PNA ON, we observed clear loss of the miRNA target in Northern blotting and therefore, in agreement with the large majority of studies using OMe ONs as anti-miRs, we conclude that degradation remains the most probable outcome of OMe targeting. Note also that Esau et al. (2006) reported miR-122 degradation upon MOE/PS anti-miR targeting in mice under standard RNA extraction conditions, and Davis et al. (2009), using the PNA decoy approach, recently showed clear evidence for miR-122 degradation in mice treated with MOE/PS anti-miRs, even though MOE/PS ONs were, in principle, able to be complexed fully by the PNA decoy.
While high-affinity anti-miR chemistries (K-PNA-K3, LNA/DNA, LNA/OMe) in all cases maintain complex formation with their target miRNAs without evidence for significant miRNA degradation, the overwhelming balance of experimentation across the available literature is consistent with miRNA degradation being observed only when lower-affinity anti-miRs are used. It seems unlikely that this degradation is because of cellular instability of the anti-miR, because miRNA degradation is clearly seen when, for example, highly stable MOE/PS ONs are used (Esau et al. 2006; Davis et al. 2009). Instead, it seems likely that miRNA duplexes formed with lower-affinity anti-miRs are more prone to being broken in cells during inhibition experiments, which are often carried out over extended periods and subsequently become vulnerable to degradation by nucleases. This would imply possible displacement of the miRNA from miRISC. In contrast, stronger binding PNA and LNA anti-miRs are highly stable inside cells and form very strong complexes with miRNAs. Irrespective of whether they are displaced from miRISC, such strong complexes would protect the miRNA from nuclease degradation. Verification of this hypothesis would ideally require a fully independent method of miRNA level measurement within cells, perhaps through use of exogenously introduced and quantifiable miRNA mimics as well as anti-miR complements in otherwise miRNA-depleted cells, experiments beyond the scope of this study.
Our improvements to the technical aspects of Northern blotting in the presence of anti-miRs will have relevance also to other hybridization-based miRNA detection procedures such as in situ hybridization (Kloosterman et al. 2006, 2007; Elmén et al. 2008b; Jørgensen et al. 2010), microarrays, BRET-based, bioluminescence and electrochemical assays, and real-time quantitative PCR (RT-qPCR) (for an overview on these techniques, see Cissell and Deo 2009), which are subject to misleading interpretation due to anti-miR interference, for example, shown in RT-qPCR (Davis et al. 2009; Lu et al. 2009). However, for each technique, further particular optimization will need to be addressed to develop a reliable method for miRNA quantification and detection in the presence of anti-miRs. We believe that our results will be of wide interest to those using such miRNA quantification and detection methods and serve also as an additional reminder that evaluation of anti-miR efficiency as potential therapeutics in cells and in vivo requires additional criteria beyond Northern blotting, such as measurement of the levels of mRNA targets of the specific miRNA inhibited by the anti-miR, as we and others have advocated previously (Esau 2008; Fabani and Gait 2008; Davis et al. 2009).
MATERIALS AND METHODS
Oligonucleotides
Synthetic miR-122 RNA ON corresponding to the human miR-122 sense strand was purchased from Dharmacon, and the sequence was obtained from the miRBase Sequence Database (Release 9.2): 5′-UGGAGUGUGACAAUGGUGUUUGU-3′.
LNA/OMe ON was synthesized as previously described (Turner et al. 2006), and the ON sequence corresponded to that previously reported (Fabani and Gait 2008): 5′-aCaAaCaCcAuuGuCaCaCuCca-3′ (LNA/OMe).
PNA ONs were synthesized on a Liberty (CEM Corporation) microwave-assisted peptide synthesizer as described previously (Fabani et al. 2010). For in vitro studies and cell work, a 23-mer, fully complementary to mature wild-type miR-122, PNA sequence was synthesized containing an N-terminal Cys and four (L)-Lys residues K-PNA-K3: Cys-(L)K-5′-ACAAACACCATTGTCACACTCCA-3′–(L)K(L)K(L)K (Fabani and Gait 2008).
The 31-mer 2′-OMe fully modified ON (Jopling et al. 2005) was purchased from Dharmacon: 5′-AGACACAAACACCAUUGUCACACUCCACAGC-3′.
For in vitro and cell assays, an miRCURY LNA/DNA knockdown probe was used and was purchased from EXIQON (5′-ACAAACACCATTGTCACACTCCA-3′; Cat No: 118019-00 hsa-miR-122a).
Decoy PNA was obtained from Panagene (Davis et al. 2009): 5′-TGGAGTGTGACAATGGTGTTTGT-3′.
Cell culture, transfections, and RNA extractions
Huh7 cells were plated in a 6-well plate format and maintained in DMEM/10% FBS with antibiotics (Full Media) for ∼20 h before transfection at 37°C/5% CO2. For PNA ON treatment, cells were carefully washed once with PBS and media was replaced by 1 mL of opti-MEM (Invitrogen) containing PNA ON at the desire concentration. Four hours later, media was replaced by Full Media. All other ONs were lipofected using Lipofectamine 2000 (Invitrogen) in serum-free media following the manufacturer's protocol. Four hours after lipofection, the media was replaced by Full Media. Cells were incubated for 20 h at 37°C/5% CO2 after transfection, washed once with PBS, and lysed using 1 mL of TRI-Reagent (Sigma). RNA was extracted following the TRI-Reagent manufacturer's protocol unless stated otherwise. For 2-equivalent and 3-equivalent isopropanol (v/v) samples, the TRI-Reagent protocol was slightly modified as follows: ∼500 μL of aqueous (RNA) phase was extracted and precipitated with 1 mL or 1.5 mL of isopropanol, respectively. The RNA pellet was then washed twice with 2 mL of 75% ethanol and dissolved in 100 μL of water. RNA quantification and quality control were measured using a NanoDrop ND-1000 spectrophotometer (only for non-radioactive RNA). Addition of more than recommended isopropanol during the RNA precipitation step induces precipitation of other contaminants to the RNA fraction; hence the need for more RNA washing steps. For samples extracted with 3 equivalents (v/v) isopropanol, RNA was re-precipitated after the extraction protocol using 2.5 volumes of ethanol 96% and 0.1 volume of 3 M NaOAc, samples were incubated for 2 h at −80°C, and the RNA was recovered by centrifugation for 30 min at 4°C at 13,200 rpm. Re-precipitated RNA was washed once with 1.5 mL of 75% ethanol, dried at room temperature, and dissolved in 100 μL of water. MiR-122 recovery in the presence of any of the anti-miRs used in this study was not compromised during the RNA re-precipitation procedure.
Isopropanol supernatants of radioactive samples were concentrated to ∼200 μL (using a Savant SpeedVac DNA 110 concentrator) where a precipitate was detected. The precipitate was dissolved in 500 μL of water and was NAP-10-purified (GE Healthcare) following the manufacturer's protocol.
Cell lysate experiments
Huh7 cells were plated in 6-well plate format and kept in Full Media for 48 h as described above. Cells were then lysed using 1 mL of TRI-Reagent (Sigma). Equivalent amounts of anti-miR ONs were added to the cell lysates to mimic transfections at the desired concentrations, and RNA was extracted.
For experiments containing 32P-miR122 ON, 250 pmol of synthetic miR-122 sense strand ON was 5′-end-radiolabeled using [γ-32P]ATP. The reaction product was NAP-10-purified (GE Healthcare), and the eluate was concentrated to half of its volume, ∼750 μL, using a Savant SpeedVac DNA 110 concentrator. Twenty microliters of this solution was added to each cell lysate.
Polyacrylamide gel electrophoresis (PAGE) and Northern blots
PAGE and Northern blotting were carried out as described previously (Fabani and Gait 2008) with minor modifications: 4 to 10 μg of total RNA from each experimental condition was dissolved in loading buffer (8 M urea; 50 mM EDTA; 20% formamide; Bromophenol Blue; Xylene Cyanol F), loaded onto the gels, and run for 2 h at 10 W at room temperature. A 32P-labeled RNA ladder (Decade-Markers; Applied Biosystems) was used to estimate band sizes. Blotted membranes were cross-linked but not baked. After overnight membrane hybridization with specific radiolabeled probes, membranes were sequentially washed with 2× SSC/0.1% SDS (30 min), 1× SSC/0.1% SDS (30 min), and 0.5× SSC/0.1% SDS (1 min) at 42°C.
For experiments containing 32P-miR122, 5 μL of extracted total RNA or purified isopropanol supernatant was mixed with 10 μL of loading buffer and loaded in a gel and run as described above. After the electrophoresis, the gels were exposed to a radiographic film for 30 min at −80°C.
PNA decoy experiments
PNA decoy experiments were carried out as described by Davis et al. (2009) with minor modifications: Gel composition and RNA loading buffer contained 20% formamide as described above. The whole gel was transferred to a membrane as described previously (Fabani and Gait 2008), and the membrane was cut prior to hybridization with miR-122 probe. The pre-hybridization buffer was not exchanged after the membrane pre-hybridization step. Hybridization of the miR-122 probe was carried out overnight, and the membrane was then washed as described above.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Donna Williams for PNA synthesis. We also thank David Loakes and Andrey Arzumanov for technical advice. We thank Andrew Newman for reading and commenting on the manuscript. A.G.T. is funded by a Cesar Milstein Scholarship from the Darwin Trust of Edinburgh, Scotland. The work was supported by the Medical Research Council (MRC Unit programme U105178803).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2533811.
REFERENCES
- Ameres SL, Horwich MD, Hung J-H, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzumanov A, Walsh AP, Rajwanshi VK, Kumar R, Wengel J, Gait MJ 2001. Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry 40: 14645–14654 [DOI] [PubMed] [Google Scholar]
- Brown D, Arzumanov A, Turner J, Stetsenko D, Lever A, Gait M 2005. Antiviral activity of steric-block oligonucleotides targeting the HIV-1 trans-activation response and packaging signal stem–loop RNAs. Nucleosides Nucleotides Nucleic Acids 24: 393–396 [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS 2005. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033 [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 1: 106–113 [DOI] [PubMed] [Google Scholar]
- Chang J, Guo J-T, Jiang D, Guo H, Taylor JM, Block TM 2008. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol 82: 8215–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP 2005. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33: 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat Protoc 1: 581–585 [DOI] [PubMed] [Google Scholar]
- Cissell KA, Deo SK 2009. Trends in microRNA detection. Anal Bioanal Chem 394: 1109–1116 [DOI] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C 2006. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 34: 2294–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C 2009. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res 37: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, Yan Y-L, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH 2008. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet 40: 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. 2008a. LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899 [DOI] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, et al. 2008b. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 36: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau CC 2008. Inhibition of microRNA with antisense oligonucleotides. Methods 44: 55–60 [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98 [DOI] [PubMed] [Google Scholar]
- Fabani MM, Gait MJ 2008. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 14: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KGC, Enright AJ, Gait MJ, Vigorito E 2010. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res 38: 4466–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG 2007. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-h, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Zamore PD 2008. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc 3: 1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD 2004. Sequence-specific inhibition of small RNA function. PLoS Biol 2: e98 doi: 10.1371/journal.pbio.0020098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata M, Tagawa H, Guo Y-M, Kameoka Y, Takahashi N, Sawada K 2009. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood 113: 396–402 [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Jørgensen S, Baker A, Møller S, Nielsen BS 2010. Robust one-day in situ hybridization protocol for detection of microRNA in paraffin samples using LNA probes. Methods 52: 375–381 [DOI] [PubMed] [Google Scholar]
- Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B 2010. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res 38: e98 doi: 10.1093/nar/gkp1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RHA 2006. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods 3: 27–29 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RHA 2007. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol 5: e203 doi: 10.1371/journal.pbio.0050203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag K-C, Isacson O, Kosik KS 2006. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24: 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with ‘antagomirs.’ Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M 2007. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res 35: 2885–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, Bozzoni I, Caffarelli E 2007. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci 104: 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee J-T, Kim S, Kim VN 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z, Yang B 2009. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res 37: e24 doi: 10.1093/nar/gkn1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Nonne N, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Pritchard LL, Harel-Bellan A 2006. An LNA-based loss-of-function assay for micro-RNAs. Biomed Pharmacother 60: 633–638 [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJF, Alenius M, Martinez J 2006. Post-transcriptional regulation of microRNA expression. RNA 12: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SY, Ju Y, Park H 2009. A highly effective and long-lasting inhibition of miRNAs with PNA-based antisense oligonucleotides. Mol Cells 28: 341–345 [DOI] [PubMed] [Google Scholar]
- Oh SY, Ju Y, Kim S, Park H 2010. PNA-based antisense oligonucleotides for microRNAs inhibition in the absence of a transfection reagent. Oligonucleotides 20: 225–230 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Kauppinen S, Lund AH 2006. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 372: 137–141 [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ 2008. Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Robertson B, Dalby AB, Karpilow J, Khvorova A, Leake D, Vermeulen A 2010. Specificity and functionality of microRNA inhibitors. Silence 1: 10 doi: 10.1186/1758-907X-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2010. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332 [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR 2010. Viruses, microRNAs, and host interactions. Annu Rev Microbiol 64: 123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin Y-L, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Turner JJ, Williams D, Owen D, Gait MJ 2006. Disulfide conjugation of peptides to oligonucleotides and their analogs. Current Protoc Nucleic Acid Chem 24: 4.28.1–4.28.21 [DOI] [PubMed] [Google Scholar]
- Várallyay E, Burgyán J, Havelda Z 2007. Detection of microRNAs by Northern blot analyses using LNA probes. Methods 43: 140–145 [DOI] [PubMed] [Google Scholar]
- Várallyay E, Burgyán J, Havelda Z 2008. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 3: 190–196 [DOI] [PubMed] [Google Scholar]
- Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S 2007. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA 13: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao J-J, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. 2008. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 68: 425–433 [DOI] [PubMed] [Google Scholar]