Abstract
Translational control of gene expression is essential for development in organisms that rely on maternal mRNAs. In Drosophila, translation of maternal nanos (nos) mRNA must be restricted to the posterior of the early embryo for proper patterning of the anterior-posterior axis. Spatial control of nos translation is coordinated through the localization of a small subset of nos mRNA to the posterior pole late in oogenesis, activation of this localized mRNA, and repression of the remaining unlocalized nos mRNA throughout the bulk cytoplasm. Translational repression is mediated by the interaction of a cis-acting element in the nos 3′ untranslated region with two proteins, Glorund (Glo) and Smaug (Smg), that function in the oocyte and embryo, respectively. The mechanism of Glo-dependent repression is unknown. Previous work suggests that Smg represses translation initiation but this model is not easily reconciled with evidence for polysome association of repressed nos mRNA. Using an in vitro translation system, we have decoupled translational repression of nos imposed during oogenesis from repression during embryogenesis. Our results suggest that both Glo and Smg regulate translation initiation, but by different mechanisms. Furthermore, we show that, during late oogenesis, nos translation is also repressed post-initiation and provide evidence that Glo mediates this event. This post-initiation block is maintained into embryogenesis during the transition to Smg-dependent regulation. We propose that the use of multiple modes of repression ensures inactivation of nos RNA that is translated at earlier stages of oogenesis and maintenance of this inactivate state throughout late oogenesis into embryogenesis.
Keywords: Drosophila, Glorund, Smaug, development, nanos
INTRODUCTION
Translational control of gene expression is widespread among eukaryotes and is essential for development in organisms like Drosophila, Caenorhabditis, and Xenopus, where zygotic gene expression is delayed after fertilization and the earliest events of embryogenesis such as cell fate decisions and axial patterning must be controlled by proteins synthesized from maternal mRNAs. Since these mRNAs are present in the egg at fertilization, subsequent spatial and temporal restrictions on their expression must be imposed post-transcriptionally (Evans and Hunter 2005; Tadros and Lipshitz 2005). Even after the transition to zygotic gene expression, transcriptional regulation is not sufficient to coordinate developmental events, as evidenced by requirements for translational regulation in developmental timing, germline development, sex determination, and neuronal morphogenesis (Thompson et al. 2007).
Translational control may be global, involving the entire complement of cellular mRNAs, or highly selective, targeting only one or a few specific mRNAs. While global regulatory mechanisms have long been studied, more recent work has begun to investigate the mechanisms that impose selective control of particular mRNAs during development (Sonenberg and Hinnebusch 2007). Studies of mRNAs whose translation is repressed at specific developmental stages or in particular spatial domains have shown that these mRNAs often contain cis-regulatory elements within their 3′ untranslated regions (3′ UTRs) (Kuersten and Goodwin 2003). In some cases, proteins that bind to these elements have been identified (Thompson et al. 2007), but how proteins bound to the 3′ UTR of a message abrogate translation is less well understood. The Drosophila nanos (nos) mRNA, whose translation must be spatially controlled for proper embryonic development, has provided a useful model for dissecting such 3′ UTR-dependent regulatory mechanisms.
Nos protein is required in the posterior of the early embryo to direct the formation of abdominal segments during embryonic development but must be prevented from accumulating in the anterior of the embryo, where it will suppress head and thorax development (Wang and Lehmann 1991; Gavis and Lehmann 1992). This problem is solved through a coupling of nos mRNA localization with translational regulation that restricts the synthesis of Nos to the posterior. As a maternal mRNA, nos is transcribed and translated in the ovarian nurse cells that are connected to the oocyte and support its growth and development (Wang et al. 1994). During the latter period of oogenesis, the nurse cells initiate apoptosis and “dump” their contents, including nos, into the oocyte (Becalska and Gavis 2009). A small fraction (4%) of nos then becomes localized to the specialized germ plasm at the oocyte posterior, providing a concentrated local source for production of Nos protein (Bergsten and Gavis 1999; Forrest and Gavis 2003; Forrest et al. 2004). The remainder of nos that enters the oocyte is translationally silenced (Forrest et al. 2004). This translational repression of unlocalized nos, coupled with the selective translation of nos localized to the germ plasm, persists in the early embryo, limiting the accumulation of Nos to the posterior region (Gavis and Lehmann 1994).
Translational repression of unlocalized nos mRNA is conferred by a translational control element (TCE) in the nos 3′ UTR (Dahanukar and Wharton 1996; Gavis et al. 1996; Smibert et al. 1996). The TCE comprises two stem–loops, designated II and III, that exhibit developmentally distinct activities through their interactions with repressor proteins (Crucs et al. 2000; Forrest et al. 2004). Stem–loop II mediates repression strictly during embryogenesis, through its interaction with Smaug (Smg), a SAM-domain protein whose own synthesis is repressed during oogenesis (Dahanukar et al. 1999; Smibert et al. 1999; Forrest et al. 2004). Smg binds to a motif in stem–loop II designated as the Smg recognition element (SRE), and mutation of either the SRE or Smg itself results in ectopic nos activity in the early embryo (Smibert et al. 1996; Dahanukar et al. 1999). Stem–loop III is bound by the hnRNP F/H protein Glorund (Glo) and TCE mutations that disrupt Glo binding or mutation of Glo abrogate repression of nos during oogenesis (Crucs et al. 2000; Kalifa et al. 2006).
The mechanism by which the binding of Glo to the TCE represses translation has not yet been investigated. In contrast, previous work has shown that Smg interacts with the eIF4E-binding protein Cup, suggesting a possible mechanism for repression whereby Smg bound to the TCE recruits Cup, which in turn binds to eIF4E and blocks the interaction of eIF4E with eIF4G (Nelson et al. 2004). However, a role for Smg in regulating translation initiation of nos is not easily reconciled with results from sucrose density gradient sedimentation experiments showing that nos mRNA in the early embryo is associated with polysomes, even under conditions when it is completely unlocalized and Nos protein is undetectable (Clark et al. 2000). Because nos mRNA is translated in the ovarian nurse cells before being transferred to the oocyte (Wang et al. 1994; Forrest et al. 2004), a mechanism that acts post-initiation would be well suited to repression of nos, permitting rapid inactivation of polysomal nos mRNA as it enters the oocyte. However, whether TCE-mediated repression operates at the initiation phase, at a post-initiation step or both, and whether Glo and Smg contribute to one or both of these modes of regulation remain to be determined.
To investigate mechanisms of TCE function, we have taken advantage of an in vitro translation system based on ovary and embryo extracts that are capable of TCE-dependent translational repression. This system has enabled us to decouple regulation enacted during embryogenesis from regulation imposed previously during oogenesis, which have not been separable in vivo. Our results reconcile seemingly disparate previous findings by showing that there are two modes of repression established late in oogenesis, one that is initiation-dependent and another that operates post-initiation. Although the post-initiation block may be maintained into embryogenesis, only Smg-dependent repression at translation initiation can be imposed after fertilization. We show that repression of nos translation initiation at late stages of oogenesis, but not during embryogenesis, depends on the presence of a poly(A) tail. In addition, our results support a role for Glo in mediating repression of nos translation post-initiation. These findings suggest that different modes of regulation adapted to the differing physiologies of oogenesis and embryogenesis are coupled to ensure that regulation is robust to developmental transitions.
RESULTS
Reconstitution of TCE-mediated repression in Drosophila ovary and embryo extracts
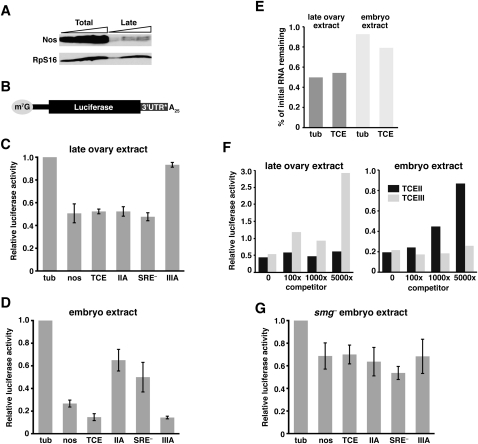
We previously developed an in vitro system that recapitulates TCE-mediated repression in embryo extracts (Clark et al. 2000). However, attempts to generate a comparable ovary extract failed to obtain TCE-dependent regulation. nos is actively translated in ovarian nurse cells and early oocytes, and is only repressed in late-stage oocytes, which are no longer supported by nurse cells (Forrest et al. 2004). Thus, factors required for repression may be diluted or inhibited when extracts are prepared from ovaries that contain the full complement of developmental stages. To overcome this problem, we prepared extract from ovaries enriched for late-stage oocytes (see Materials and Methods) as confirmed both by visual inspection of the tissue and by immunoblotting with anti-Nos antibody. Previous analysis showed that Nos protein synthesized during early and mid-oogenesis is degraded by late oogenesis and that Nos levels are low in late oocytes, where the majority of nos mRNA is repressed (Forrest et al. 2004). In comparison to the total ovary extract which contains abundant Nos protein, only low levels of Nos are detected in the late-stage extract, validating the enrichment method (Fig. 1A). The late-stage extract translates luciferase reporter mRNAs with similar efficiency to that of the total extract and translation is dependent on both m7G cap and poly(A) tail (see below and data not shown).
FIGURE 1.
In vitro translation assay for TCE-mediated repression in late ovary and embryo extracts. (A) Immunoblot of total ovary and late ovary extracts. Lanes contain increasing amounts of each extract, in twofold increments. Nos and the RpS16 loading control were detected simultaneously. (B) Architecture of the reporter RNAs used in C-G, with the m7GpppG cap, nos 5′ UTR (thinner black bar), firefly luciferase coding region, and 25-nt poly(A) tail in common. 3′ UTR* denotes sequences tested in C, D, and G: α-tubulin 3′ UTR (tub), nos 3′ UTR (nos), 3xTCE (TCE), 3xTCEIIA (IIA), 3xTCE[SRE–] (SRE–), or 3xTCEIIIA (IIIA). (C,D) Luciferase assays. The indicated reporter RNAs were translated in late ovary extract (C) or embryo extract (D) together with an internal control Renilla luciferase RNA. For each reaction, firefly luciferase activity of the reporter was normalized to Renilla luciferase activity. Relative luciferase activity was calculated by dividing the normalized firefly luciferase value for the indicated reporter by the normalized value for the luc-tub3′UTR reporter RNA (tub = 1.0). The graph reports the mean and standard deviation from at least three independent experiments for each reporter. (E) Quantitation of luc-tub3′UTR and luc-3xTCE RNA stability. RNA purified from in vitro translation reactions after 5 min and either 120 min (ovary) or 90 min (embryo) of incubation was analyzed by Northern blotting. The ratio of the final level to the initial level for each reporter, as determined by quantitation of the Northern blots, is plotted. (F) Competition experiment. luc-tub3′UTR and luc-3xTCE RNAs were translated in either ovary or embryo extract in the presence of 0-, 100-, 1000-, or 5000-fold molar excess of TCE stem–loop II or TCE stem–loop III RNA. Relative luciferase activity was determined as described in C and D. Similar results were obtained in multiple independent experiments. (G) Luciferase assays as described for C and D using smg mutant embryo extract.
To test the TCE dependence of the late-stage ovary extract (referred to hereafter simply as late ovary extract), the extract was programmed with capped and polyadenylated luciferase reporter RNAs bearing either nos 3′ UTR sequences or α-tubulin (tub) 3′ UTR sequences (Fig. 1B). We have previously established that the tub 3′ UTR serves as an unregulated control 3′ UTR both in vivo and in vitro (Gavis and Lehmann 1994; Gavis et al. 1996; Clark et al. 2000). All reactions also contained a Renilla luciferase standard RNA to control for experimental variability. Luciferase activity was measured using a dual luciferase assay and firefly luciferase activity was normalized to Renilla activity. In the late ovary extract, the luc-nos3′UTR and luc-3xTCE reporter RNAs produced two- to threefold less luciferase activity than the control luc-tub3′UTR RNA (Figs. 1C, 2B). This repression of the two TCE-containing RNAs in the late ovary extract is less robust than the four- to sixfold repression of the same RNAs observed in embryo extract (Fig. 1D; see also Clark et al. 2000) but is reproducible among independent preparations of extract. Northern blot analysis confirmed that the decreased production of luciferase by the luc-nos3′UTR and luc-3xTCE reporters is not due to differential RNA stability but reflects differences in translational efficiency (Fig. 1E; Clark et al. 2000). As further confirmation of these results, repression of luc-3xTCE RNA in ovary extract is diminished by the addition of excess TCE stem–loop III RNA, but not by excess stem–loop II RNA whereas repression in embryo extract is effectively competed by addition of excess stem–loop II RNA but not by excess stem–loop III (Fig. 1F).
FIGURE 2.
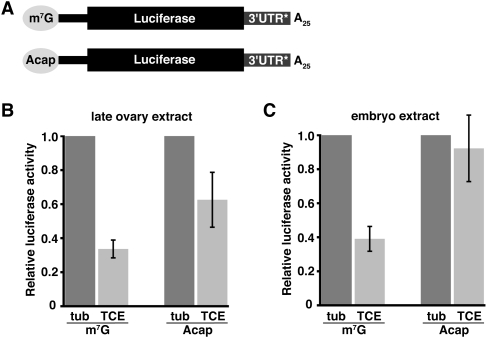
Cap dependence of TCE-mediated repression. (A) Reporter RNAs used in B and C are similar to those in Figure 1 and differ from each other by the presence of an m7GpppG cap (m7G) or an ApppG cap (Acap). 3′UTR* denotes tub or 3xTCE (TCE) sequences. (B,C) Luciferase assays of the m7G and Acap luc-tub3′UTR (tub) and luc-3xTCE (TCE) reporter RNAs in late ovary or embryo extract. Relative luciferase activity was determined as described in Figure 1, except that the normalized luciferase activity of each luc-3xTCE RNA was divided by the normalized activity of its cognate luc-tub3′UTR RNA (for each, tub = 1.0).
Analysis of nos transgenes bearing wild-type and mutant TCE sequences established that TCE stem–loop III mediates repression primarily during oogenesis whereas TCE stem–loop II acts strictly in the embryo (Forrest et al. 2004). Moreover, TCE stem–loop III function during oogenesis requires binding by Glo, whereas TCE stem–loop II function in the embryo requires binding by Smg (Dahanukar et al. 1999; Smibert et al. 1999; Kalifa et al. 2006). We therefore tested whether the temporal specificity exhibited by stem–loops II and III in vivo is reflected in vitro. Luc-3xTCEmut reporters were constructed bearing stem–loop II or III mutations shown to disrupt TCE function in vivo and were analyzed for their behavior in ovary and embryo extracts. Quantitation using the dual luciferase assay showed that repression in the late ovary extract is nearly abolished by the IIIA mutation, which disrupts formation of TCE stem–loop III and the Glo binding site (Crucs et al. 2000; Kalifa et al. 2006), but is not affected by mutations that disrupt formation of stem–loop II (IIA) (Crucs et al. 2000) or the Smg-binding site (SRE–) (Fig. 1C; Smibert et al. 1996). In contrast, the IIA and SRE– mutations compromise TCE-mediated repression in the embryo extract (Fig. 1D), consistent with previous results (Forrest et al. 2004). Similarly, repression of luc-3xTCE RNA is compromised in extract prepared from smg mutant embryos and TCE mutations have no additional effect in this extract (Fig. 1G). We have not been able to carry out the reciprocal experiment with late ovary extract lacking Glo protein because glo mutants exhibit pleiotropic oogenesis defects and do not produce late oocytes in sufficient quantity to prepare translation extract. However, the nearly complete abrogation of repression by the IIIA mutation strongly implicates Glo as the repressor. Taken together, these results demonstrate that temporally distinct modes of TCE function in vivo are preserved in vitro.
The TCE targets translation initiation in ovary and embryo extract
The interaction of Smg with the eIF4E-binding protein Cup suggests that TCE-mediated repression of nos translation in the embryo occurs at the initiation step (Nelson et al. 2004). To test whether TCE-mediated regulation is cap-dependent, we transcribed luc-3xTCE and luc-tub3′UTR RNAs in the presence of the cap analog, ApppG, which does not bind to eIF4E (Fig. 2A). Consistent with previous results (Clark et al. 2000), reporters capped with the analog are translated five- to sixfold less efficiently than their m7GpppG capped cognates in both ovary and embryo extracts, but this level of translation is still within the linear range of the assay (data not shown). Repression of ApppG versus m7GpppG capped luc-3xTCE RNAs was therefore compared relative to the corresponding luc-tub3′UTR control RNAs. In the late ovary extract, luc-3xTCE RNA bearing the ApppG cap is still repressed relative to ApppG capped luc-tub3′UTR RNA, but less well than m7GpppG capped luc-3xTCE RNA (1.6-fold compared to 3.5-fold; Fig. 2B). In contrast, the presence of the ApppG cap completely abolishes repression of luc-3xTCE RNA in the embryo extract (Fig. 2C). This cap dependence of TCE-mediated repression in embryo extract suggests that, in the early embryo, the TCE functions primarily by blocking initiation. In contrast, TCE-mediated repression in the ovary may not depend entirely on cap-dependent initiation.
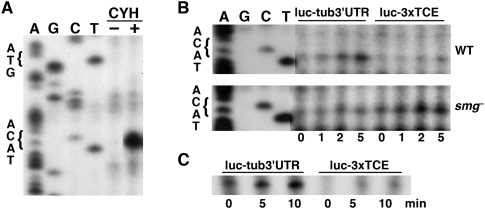
To monitor initiation more directly, we used a toeprinting assay, which maps ribosomes and other complexes on RNA by their ability to block reverse transcriptase (RT). m7GpppG capped, polyadenylated luc-3xTCE and luc-tub3′UTR reporter RNAs were translated in ovary or embryo extract and subjected to reverse transcription with a radiolabeled primer complementary to a sequence 100 nucleotides (nt) downstream from the AUG initiation codon. When the translation reaction is carried out in the presence of the elongation inhibitor cycloheximide, 80S ribosomes cannot proceed beyond the initiation codon, resulting in an RT block at the leading edge of the ribosome, ∼17 nt downstream from the AUG (Sachs et al. 2002). The occupancy of the AUG by ribosomes can therefore be monitored by the appearance of the predicted RT product on a denaturing polyacrylamide gel.
Addition of cycloheximide to embryo extract programmed with luc-tub3′UTR RNA resulted in accumulation of the expected 83-nt RT product (Fig. 3A,B). The AUG toeprint is cycloheximide-dependent, indicating that it reflects arrested ribosomes (Fig. 3A). Compared to luc-tub3′UTR RNA, toeprinting of ribosomes at the AUG of luc-3xTCE in embryo extract showed reduced ribosome occupancy (Fig. 3B). Moreover, this decreased accumulation of 80S ribosomes at the AUG is reversed in smg mutant extract, where repression of luc-3xTCE is compromised. A similar toeprinting analysis performed using the late ovary extract also showed that AUG occupancy of luc-3xTCE RNA is reduced compared to that of luc-tub3′UTR RNA (Fig. 3C). Together, these results are consistent with a function for the TCE in repressing translation initiation during both embryogenesis and oogenesis.
FIGURE 3.
The TCE decreases AUG occupancy. (A) Toeprinting experiment to monitor AUG occupancy on luc-tub3′UTR RNA in the presence (+) or absence (–) of cycloheximide (CYH). Dideoxysequencing of the reporter DNA template (four lanes on left using lighter exposures) shows the position of the initiation codon. The toeprint occurs 17 nt downstream from the A of the AUG as expected (Sachs et al. 2002). (B,C) Time-course experiments monitoring AUG toeprints on luc-tub3′UTR and luc-3xTCE reporter RNAs. (B) Reporter RNAs were translated in wild-type (WT) or smg mutant (smg–) embryo extract and aliquots were removed at the indicated time points after addition of cycloheximide (at t = 0 min) for reverse transcription. (C) Toeprinting experiment performed using late ovary extract.
A poly(A) tail is required for TCE-mediated repression during oogenesis
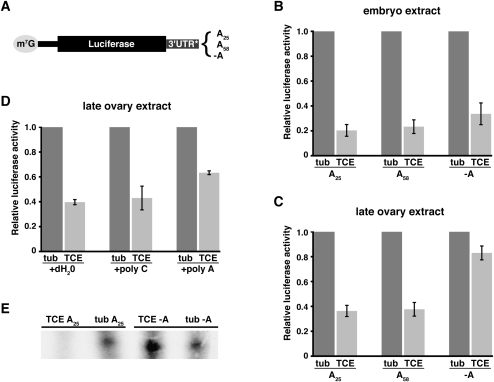
Translational control of maternal mRNAs is often associated with changes in polyadenylation status, with deadenylation being characteristic for translational repression (Tadros and Lipshitz 2005). To investigate whether TCE-mediated repression in vitro involves a poly(A)-dependent mechanism, we assayed the effect on translational repression of either removing the poly(A) tail or increasing its length. In addition to our standard luc-3xTCE and luc-tub3′UTR RNAs with 25-residue poly(A) tails, we synthesized capped luc-3xTCE and luc-tub3′UTR RNAs without poly(A) tails or with 58-residue poly(A) tails (Fig. 4A). Translation of these RNAs in ovary and embryo extract was then monitored using the dual luciferase assay. Because the absolute translational efficiency of RNA in the extract is dependent on poly(A) tail length (Clark et al. 2000 and data not shown), the effect of poly(A) length on repression was evaluated by determining the ratio of luciferase activity produced by luc-3xTCE, luc-3xTCE(A58), or luc-3xTCE(-A) RNAs to luciferase activity produced by luc-tub3′UTR RNA with the corresponding poly(A) tail.
FIGURE 4.
Poly(A) tail dependence of TCE-mediated repression during oogenesis. (A) Reporter RNAs used for the experiments in B–E with differences in poly(A) tail length as shown. 3′UTR* denotes tub or 3xTCE (TCE) sequences. (B–D) Luciferase assays, with relative luciferase activity determined as described for Figure 2. (B,C) Reporter RNAs shown in A were translated in late ovary or embryo extract. (D) Translation reactions containing luc-tub3′UTR (tub) and luc3xTCE (TCE) RNAs with standard 25A tails were challenged by addition of a 1000-fold molar excess of poly(C) or poly(A), or an equivalent volume of dH2O. (E) Toeprinting showing AUG occupancy for the indicated reporter RNAs in late ovary extract.
In the embryo extract, there is no significant difference in repression of luc-3xTCE, luc-3xTCE(A58), and luc-3xTCE(-A) RNAs (Fig. 4B). While increasing poly(A) tail length also has no effect on repression in the late ovary extract, elimination of the poly(A) tail severely compromises repression (Fig. 4C). A difference in poly(A) tail dependence between the two extracts could result if poly(A) addition occurs in the embryo but not in the late ovary extract. To rule out this possibility, we repeated the in vitro translation assays in the presence of the adenosine analog cordycepin, which blocks poly(A) addition by poly(A) polymerase. Addition of cordycepin did not alter the poly(A) tail independence of translational repression in the embryo extract, nor did it affect the poly(A) tail dependence in the late ovary extract (data not shown).
Our results therefore suggest that the presence of a poly(A) tail is necessary for translational repression of nos during oogenesis, but that the length of the tail is not critical. This poly(A) tail dependence could be explained if one or more trans-acting factors that bind to the poly(A) tail act in concert with TCE stem–loop III and possibly Glo to establish repression. We tested this possibility by using poly(A) RNA to competitively inhibit binding of such factors to RNA poly(A) tails in the late ovary extract. Excess poly(A) or poly(C) RNA was added to translation reactions containing luc-3xTCE or luc-tub3′UTR RNA and translation of the reporter was measured by the dual luciferase assay. Whereas addition of poly(C) had no effect on translational repression, addition of excess poly(A) compromised translational repression (Fig. 4D). Similarly to the behavior of luc-3xTCE(-A) RNA, repression of luc-3xTCE is not completely abrogated. These results suggest that factors binding to the poly(A) tail contribute to repression during oogenesis but do not account for the entirety of repression.
To determine whether the poly(A) tail is required for TCE-mediated repression of translation initiation, we performed a toeprinting experiment with luc-3xTCE(-A) and luc-tub3′UTR(-A) reporters in the late ovary extract. In contrast to our standard A25 reporters where occupancy of the AUG by ribosomes is decreased by the TCE, AUG occupancy on the luc-3xTCE(-A) reporter is similar to occupancy on the luc-tub3′UTR(-A) reporter (Fig. 4E). Thus, elimination of the poly(A) tail mimics elimination of the TCE, suggesting that the poly(A) tail is necessary for initiation-based repression in the ovary.
Repression of translation at a post-initiation step during oogenesis
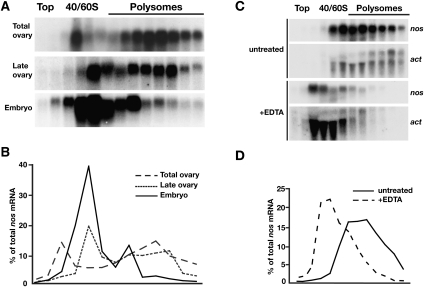
In vivo, translationally repressed nos mRNA in the early embryo is polysome-associated, indicating that nos repression by the TCE includes a component that operates at a late step in the translation cycle. The finding that ApppG capped luc-3xTCE RNA is fully derepressed in embryo extract but remains partially repressed in the late ovary extract suggests that, in addition to poly(A) tail-dependent repression of initiation, a second mechanism that acts post-initiation may be first deployed during oogenesis. To determine whether this is the case, we performed a temporal analysis of the polysome profile of nos mRNA by sucrose gradient sedimentation of total ovary, late ovary, and embryo extracts. The distribution of nos in each gradient was determined by Northern blotting. Consistent with the robust accumulation of Nos protein through mid-oogenesis (Fig. 1A), nos mRNA is detected primarily in the polysomal fractions of total ovary extract (Fig. 5A,B). In the late ovary extract, the distribution of nos shifts toward the smaller polysomes and nonpolysomal fractions. However, a substantial amount of nos remains in the polysomal fractions (Fig. 5A,B), even though Nos protein levels are greatly reduced in late-stage oocytes (Fig. 1A). Comparison of late ovary extract with embryo extract shows a further reduction in the amount of nos co-sedimenting with polysomes (Fig. 5A,B). In all cases, when extracts are treated with EDTA to destabilize polysomes, nos sediments more slowly, indicating the presence of nos in the heavier fractions is due to polysome association (Fig. 5C,D and data not shown). These results, taken together with data from the in vitro translation assays, suggest that TCE-mediated repression of nos targets both cap-dependent initiation and a post-initiation step during oogenesis. In the early embryo, the post-initiation repression can be maintained but repression becomes primarily cap-dependent.
FIGURE 5.
Temporal analysis of nos polysome association. (A) Total ovary, late ovary, and embryo extracts were fractionated on 20%–50% sucrose density gradients and RNA isolated from gradient fractions was analyzed by Northern blotting with probes for nos mRNA. (B) Quantitation of nos mRNA distribution from Northern blots in A as a % of total nos radioactivity. (C) Fractionation of late ovary extract on 20%–50% sucrose density gradients with or without addition of EDTA to disrupt polysomes. Polysomes are generally less well preserved in this experiment than in the experiment shown in A due to the lowered Mg2+ concentration in the extract necessary for polysome disruption (see Materials and Methods; Clark et al. 2000). RNA isolated from gradient fractions was analyzed by Northern blotting with probes for nos and actin mRNAs (actin migrates as two species). (D) Quantitation of nos mRNA distribution in C as a % of total nos radioactivity.
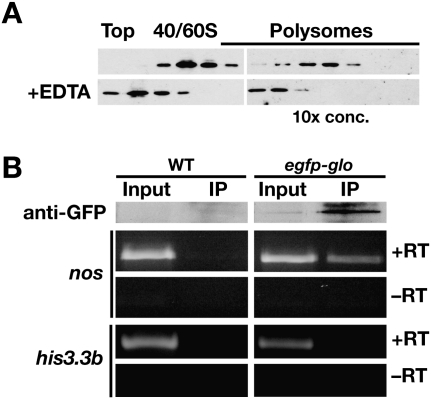
Given that Glo is the only known regulator of nos translation in the ovary, we investigated whether Glo might mediate repression of nos post-initiation. We have not been able to investigate the effect of glo mutation on the polysomal association of nos mRNA due to the pleoitropic role of glo in oogenesis described above. However, immunoblot analysis of sucrose gradient fractions showed that Glo cosediments with the polysomes and this distribution is EDTA-sensitive (Fig. 6A). To determine whether polysomal Glo is bound to nos mRNA, we performed co-immunoprecipitation experiments from late ovary polysomes. Polysomes isolated from wild-type ovaries or ovaries expressing a functional EGFP–Glo fusion protein (Kalifa et al. 2009) were concentrated by centrifugation through a sucrose cushion and solubilized, and then Glo RNP complexes were immunoprecipitated with anti-GFP antibodies. RT-PCR analysis detected nos mRNA specifically in RNA isolated from the EGFP–Glo immunoprecipitate whereas a control mRNA was not detected in either immunoprecipitate (Fig. 6B). These results indicate that Glo interacts with translationally repressed, polysomal nos mRNA, implicating Glo in post-initiation control of nos translation.
FIGURE 6.
Glo is associated with polysomal nos mRNA in vivo. (A) Immunoblot analysis of Glo protein in fractions from the late ovary gradients shown in Figure 5C. Polysomal fractions were TCA-precipitated prior to immunoblotting, and ∼10-fold more total protein was loaded per lane. The distribution of Glo shifts to lighter fractions when polysomes are disrupted by EDTA treatment. (B) Co-immunoprecipitation of Glo and nos mRNA. A polyclonal anti-GFP antibody was used for immunoprecipitation from pelleted polysomes from wild-type (WT) or EGFP–Glo expressing late ovaries. Top panels: immunoblot analysis with a monoclonal anti-GFP antibody confirms specific purification of EGFP–Glo. Input sample is 1/10 volume equivalent of the immunoprecipitate sample (IP). Bottom panels: RT-PCR analysis of RNA isolated from immunoprecipitates detects nos but not the control his3.3b RNA. Reactions were performed with (+RT) or without (−RT) reverse transcriptase.
DISCUSSION
In the early embryo, silencing of unlocalized nos together with translation of posteriorly localized nos mRNA provides the spatial control of Nos that is essential for embryonic patterning. Control of nos translation is first exerted during oogenesis, through the interaction of Glo with TCE stem–loop III, and is perpetuated in the early embryo, through the interaction of Smg with TCE stem–loop II (Forrest et al. 2004). By using in vitro translation systems derived from ovary and embryos, respectively, we have separated the mechanisms that establish nos repression during oogenesis from those that maintain repression during embryogenesis. We show that translation is repressed at initiation during embryogenesis by Smg and during oogenesis, most likely by Glo, but by different means. Moreover, we provide evidence that an additional cap-independent component of TCE-mediated repression is imposed during oogenesis and maintained in the early embryo. The association of Glo with polysomal nos mRNA implicates Glo in this post-initiation mechanism.
Translation initiation is the most frequent target of translational control mechanisms (Sonenberg and Hinnebusch 2009). One common mechanism of regulating cap-dependent translation involves eIF4E-binding proteins that block the interaction of eIF4E with eIF4G, thereby inhibiting recruitment of the small ribosomal subunit to the message. In an extension of this paradigm, eIF4E binding proteins may be tethered to specific mRNAs through their interactions with mRNA-binding proteins, allowing for selective control of these messages. A role for Smg in repressing translation initiation is predicted from its interaction with the eIF4E-binding protein Cup but has not previously been demonstrated. Cup is required for repression of a luc-3xTCE reporter RNA by Smg in an RNA injection assay and binding of Cup to eIF4E prevents interaction of eIF4G in vitro (Nelson et al. 2004). Although we could not test the involvement of Cup using our assay, because cup mutants produce few and fragile embryos, our data confirm that binding of Smg to the TCE results in the inhibition of translation initiation. The poly(A) independence of TCE-mediated repression in the embryo is also consistent with a model whereby Smg serves as an adaptor that tethers Cup to nos RNA. Similarly to nos, translation of another posteriorly localized maternal mRNA, oskar (osk), is regulated during oogenesis by an RNA-binding protein—Bruno (Bru) in this case—that interacts with Cup to inhibit recruitment of the small ribosomal subunit (Chekulaeva et al. 2006).
TCE-mediated repression of translation initiation during oogenesis depends on the presence of a poly(A) tail, indicating that it must occur by a different mechanism. Consistent with this result, nos mRNA isolated from late-stage oocytes, where the majority of nos is repressed, has a longer poly(A) tail than nos mRNA isolated from early and mid-stage egg chambers, where nos is highly translated (Benoit et al. 2008). In addition, TCE-mediated repression remains intact in late ovary extracts prepared from cupΔ212 mutants, in which the eIF4E binding motif is disrupted (Nakamura et al. 2004), suggesting that Cup is not essential for repression of nos during oogenesis (S Andrews and ER Gavis, unpubl.). Pleiotropic requirements for glo during oogenesis and nonspecific effects caused during attempts at immunodepletion of Glo have precluded a direct analysis of Glo function in our in vitro system. However, the fact that the TCEIIIA mutation, which disrupts the Glo binding site (Kalifa et al. 2006), completely eliminates repression during oogenesis suggests that Glo mediates this function. Moreover, the requirement for a poly(A) tail, together with poly(A) competition experiments, suggests a mechanism whereby interaction of poly(A) tail and 3′ UTR binding factors represses initiation. The poly(A) tail can impact translation initiation through the interaction of PABP with eIF4G. Simultaneous binding of eIF4G to PABP and eIF4E circularizes the message, forming a closed loop that stimulates translation initiation by promoting ribosome recruitment and possibly recycling of ribosomes to the 5′ end of the message after termination (Mangus et al. 2003; Kahvejian et al. 2005). Although we have not detected a specific interaction of Glo with PABP, eIF4G, or eIF4E (K Hughes and ER Gavis, unpubl.), the interaction of Glo with auxiliary factors bound to the poly(A) tail could block the PABP–eIF4G interaction or alter the ability of this complex to interact with eIF4E, thereby breaking the stimulatory loop. Intriguingly, a similar requirement for both the cap and poly(A) tail has been reported for microRNA-dependent repression in transfected HeLa cells (Humphreys et al. 2005).
Smg-dependent repression of translation initiation has been difficult to reconcile with the presence of translationally repressed polysomal nos mRNA in the embryo. Results from our in vitro translation assays and temporal analysis of polysomes now shed light on this apparent paradox. Whereas TCE-dependent repression in the embryo extract is entirely cap-dependent, repression of the ApppG capped luc-3xTCE reporter RNA that cannot bind eIF4E is only partially compromised in the late ovary extract. This result suggests that there is a cap-independent component to regulation in the late ovary extract that is not recapitulated in the embryo extract. Although we cannot rule out the possibility that factors required for this post-initiation mechanism are differentially stable between the two extracts, a likely explanation lies in the ovarian history of endogenous embryonic nos mRNA that is not recapitulated when embryo extract is programmed with exogenous mRNA. We therefore propose that repression at a post-initiation step can only be established during oogenesis, but once established, can be maintained in the embryo.
Whereas blocking translation at the initiation step may be cost-effective for a cell, preserving energy resources and ribosomes to be used for other purposes, it may not provide a sufficiently rapid on/off switch required by mRNAs like nos. During oogenesis, a post-initiation block would allow rapid inactivation of polysomal nos RNA upon transfer from the nurse cells into the oocyte, while non-polysomal nos mRNA would be repressed at initiation. Our results suggest that Glo mediates both levels of control. In the early embryo, the polysomal association of nos is reduced, suggesting that, while the post-initiation mechanism is maintained into embryogenesis, it begins to break down and is replaced by Smg-dependent repression. The combination of post-initiation and initiation-based repression, the latter reinforced by the production of Smg in the embryo, ensures that Nos is produced only in the germ plasm, where nos mRNA can be efficiently translated. Whether this localized nos mRNA is protected from repressors or actively derepressed remains to be determined.
MATERIALS AND METHODS
Generation of reporter and competitor RNAs
The luc-3xTCE, luc-3xTCE(A58), luc-tub3′UTR, and luc-tub3′UTR(A58) reporter plasmids are described in Clark et al. (2000). The luc-3xTCEIIA, luc-3xTCEIIIA, and luc-3xTCE[SRE–] reporter plasmids are described in Forrest et al. (2004). The luc-nos3′UTR plasmid was generated from luc-tub3′UTR by replacing the tubulin 3′ UTR sequences with a fragment encompassing nucleotides 6–842 of the nos 3′ UTR.
For each construct, 1 μg of linearized plasmid DNA was transcribed using the mMessage mMachine SP6 kit (Ambion). Linearization with NsiI results in transcripts containing 25- or 58-nt poly(A) tails. Linearization of plasmids with SacI, which cuts 5′ to the encoded poly(A) sequence, results in production of transcripts lacking a poly(A) tail. To produce ApppG capped RNAs, the Ambion 2x NTP/Cap mixture was substituted by a mixture containing 8 mM ApppG and 10 mM each of ATP, CTP, UTP, and GTP. RNA for competition assays was generated from linearized plasmids containing individual TCE stem–loop II and TCE stem–loop III sequences (Kalifa et al. 2006) using the MEGAscript T7 High Yield Transcription Kit (Ambion). In all cases, transcription reactions were treated with 1 μL TURBO DNase (Ambion) for 15 min at 37°C and RNA was extracted with phenol:chloroform and precipitated with isopropanol. Prior to use, the TCE stem–loop III competitor RNA was heated to 65°C for 5 min, then slowly cooled to room temperature (Kalifa et al. 2006).
In vitro translation assay
The y w67c23 strain (Lindsley and Zimm 1992) was used for preparation of wild-type ovary and embryo translation extracts. smg mutant extracts were prepared from ovaries or embryos produced by smg1/Df(ScfR6) females (Dahanukar et al. 1999). Embryo extract was prepared as described previously (Clark et al. 2000). For preparation of late ovary extract, females were fed for 3 d in bottles of corn meal agar food supplemented with yeast paste, and then transferred to bottles containing only a wet Kimwipe for 20 h to promote accumulation of late-stage oocytes. Ovaries were visibly enriched for late-stage oocytes and enrichment was confirmed by anti-Nos immunoblotting. Ovaries were dissected on ice in PBS and washed twice in 12 volumes of a 1:1 mix of PBS:Buffer A (10 mM HEPES pH 7.5, 5 mM DTT, 0.5 mM PMSF), followed by two washes in 12 volumes of Buffer A. The liquid was removed and ovaries were homogenized and centrifuged at 16,000g for 10 min at 4°C. The supernatant was supplemented with 1/9 volume Buffer O (100 mM HEPES pH 7.5, 1 M potassium acetate, and 50 mM DTT), then centrifuged at 16,000g for 10 min at 4°C. The supernatant was again collected and mixed with 1/50 volume RNase inhibitor, aliquoted, frozen in liquid nitrogen, and stored at −80°C for no more than 48 h before use.
In vitro translation reactions (20 μL) contained 50% extract, 1x translation mix (25 mM HEPES pH 7.5, 1.5 mM MgOAc [embryo extract] or 3 mM MgOAc [late ovary extract], 0.2 mM spermidine, 2.5 mM DTT, 25 μM amino acids, 1.2 mM ATP, and 0.3 mM GTP), 15 mM creatine phosphate, 0.1 mg/mL creatine phosphokinase, 0.1 nM Renilla luciferase RNA, and 1 nM firefly luciferase reporter RNA. For each experiment, all components except for the firefly reporter RNAs were pre-mixed and aliquoted. Reactions were allowed to proceed for 90 min for embryo extract or 120 min for late ovary extract, the earliest time point at which maximum repression is achieved for each extract. For competition assays, reactions contained 100-, 1000-, or 5000-fold molar excess of competitor RNA. Cordecypin was used at a final concentration of 200 μM. Poly(A) and poly(C) 15-nt oligomers were added to achieve a final concentration of 10 μM. Luciferase activity was assayed using the Dual-Luciferase Assay system (Promega) and luminescence was monitored with a Glomax 20/20 Luminometer (Promega). For each experiment, firefly luciferase was normalized to Renilla luciferase to control for differences among samples. Each experiment was performed three times, in some cases using different preparations of extract.
To monitor RNA stability, RNA was extracted from translation reactions frozen in liquid N2 at t = 0, t = 90 min (embryo) or t = 120 min (ovary) using acid phenol:chloroform (Tri-Reagent, Sigma) and analyzed by Northern blotting. Blots were quantitated by phosphorimaging (Molecular Dynamics).
Toeprinting assay
Toeprinting assays were carried out as previously described (Hartz et al. 1988) except that translation reactions were pre-incubated on ice for either 30 min (late ovary extract) or 15 min (embryo extract) to ensure that repression was established prior to addition of 10 μg/mL cycloheximide. At each time point after cycloheximide addition, reactions were placed on ice and the reporter RNA was reverse-transcribed from a 32P-end-labeled primer (5′-GCAATTGTTCCAGGAACC-3′) that anneals 100 nt downstream from the AUG start codon in firefly luciferase. Dideoxy sequencing reactions were performed using the same 32P-end-labeled primer and the USB Sequenase 2.0 kit (Amersham).
Immunoblotting
Immunoblotting of ovary extracts was performed according to Forrest et al. (2004) except that nitrocellulose membrane was used and the membrane was blocked overnight at 4°C in TBST (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Tween-20) with 5% nonfat dry milk. Antibody incubations and washes were performed using this solution, except for a final wash in TBST. Polyclonal anti-Nos antibody (1:1000; gift of A. Nakamura) and polyclonal anti-RpS16 antibody (1.2 μg/mL; Abcam ab26159a) were applied to the blot for 2 h at room temperature and detected using HRP-conjugated secondary antibodies and ECL (Amersham).
Polysome fractionation
Sucrose density gradient centrifugation was performed as described in Clark et al. (2000) with the following modifications. Approximately 500 μL ovaries were homogenized on ice in 0.5 M NaCl, 25 mM MgOAc, 50 mM Tris-HCl pH 7.5, 1 mg/mL heparin, 50 units/mL RNase Inhibitor (NEB), 0.5 mg/mL cycloheximide and 1× complete EDTA-free protease inhibitor cocktail (Roche). Either 42 A260 units (total or late ovary extract) or 35 A260 units (embryo extract) were layered onto 11 mL 20%–50% sucrose gradients. EDTA disruption was also performed as described (Clark et al. 2000). For RNA isolation, gradient fractions were supplemented to 10 mM EDTA/1% SDS and treated with 150 μg/μL proteinase K for 30 min at 37°C. RNA was extracted with Tri-Reagent LS (Sigma), ethanol-precipitated, resuspended in DEPC-treated H2O, and analyzed by Northern blotting. Blots were quantitated by phosphorimaging (Molecular Dynamics). For immunoblotting with anti-Glo antibody (1:500 5B6; Kalifa et al. 2006), gradient fractions were either supplemented with SDS-PAGE sample buffer or TCA-precipitated and resuspended in sample buffer.
Polysome concentration and RNA co-immunoprecipitation
Approximately 250 μg ovaries were homogenized on ice in 0.25 M NaCl, 5 mM MgOAc, 25 mM Tris-HCl pH 7.5, 1 mg/mL heparin, 50 units/mL RNase Inhibitor (NEB), 0.5 mg/mL cycloheximide, and 1× complete EDTA-free protease inhibitor cocktail (Roche). Homogenates were cleared by centrifugation for 10 min at 13,000 RPM at 4°C and 45 A260 units of cleared extract were layered onto a 4 mL, 30% sucrose cushion containing 0.25 M NaCl, 5 mM MgOAc, 50 mM Tris pH 7.5. Cushions were centrifuged in a SW 50.1 rotor for 2.5 h at 33,000 RPM at 4°C. The pelleted material was washed twice in RNA co-immunoprecipitation buffer (RCB: 25 mM HEPES pH 7.4, 150 mM NaCl, 2.5 mM MgCl, 0.5mM EDTA, 0.01% Triton X-100, 50 units/mL RNase Inhibitor, 1x complete EDTA-free protease inhibitor cocktail), then homogenized in 500 μL RCB buffer. EGFP–Glo was immunoprecipitated by incubation for 1 h at room temperature with 50 μL Protein G Dynabeads (Invitrogen) bound with anti-GFP antibody (Abcam 290). Immunoprecipitates were washed six times in RCB. Half of each immunoprecipitate was resuspended in SDS-PAGE sample buffer for immunoblotting with anti-GFP antibody (1:1000 JL-8, Clonetech). The remainder was resuspended in RCB and treated with four units RQ1 RNase-free DNase (Promega) for 15 min at room temperature, then extracted with acid phenol:chloroform (Tri-Reagent, Sigma) and ethanol-precipitated. RNA was reverse-transcribed using SuperScript II Reverse Transcriptase (Invitrogen) and oligo d(T)15 for 2 h at 42°C and cDNA was amplified for 33 cycles with primers for nos (Jain and Gavis 2008) or for his3.3B (Becalska et al. 2011).
ACKNOWLEDGMENTS
We thank S. Chatterjee for technical assistance and A. Nakamura for generously providing anti-Nos antibody. I.E.C. was supported by an NIH Postdoctoral Training Grant (CA09528) and an NIH National Research Service Award (F32 GM18770-01A1). This work was supported by NIH grant GM061107 to E.R.G.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2478611.
REFERENCES
- Becalska AN, Gavis ER 2009. Lighting up mRNA localization in Drosophila oogenesis. Development 136: 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER 2011. Aubergine is a component of a nanos mRNA localization complex. Dev Biol 349: 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten SE, Gavis ER 1999. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development 126: 659–669 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533 [DOI] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER 2000. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational mechanism. Curr Biol 10: 1311–1314 [DOI] [PubMed] [Google Scholar]
- Crucs S, Chatterjee S, Gavis ER 2000. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol Cell 5: 457–467 [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Wharton RP 1996. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes & Dev 10: 2610–2620 [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP 1999. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell 4: 209–218 [DOI] [PubMed] [Google Scholar]
- Evans TC, Hunter CP 2005. Translational control of maternal RNAs. WormBook 10: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER 2003. Live imaging of endogenous mRNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13: 1159–1168 [DOI] [PubMed] [Google Scholar]
- Forrest KM, Clark IE, Jain RA, Gavis ER 2004. Temporal complexity within a translational control element in the nanos mRNA. Development 131: 5849–5857 [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R 1992. Localization of nanos RNA controls embryonic polarity. Cell 71: 301–313 [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R 1994. Translational regulation of nanos by RNA localization. Nature 369: 315–318 [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lunsford L, Bergsten SE, Lehmann R 1996. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122: 2791–2800 [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol 164: 419–425 [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T 2005. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci 102: 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Gavis ER 2008. The Drosophila hnRNP M homolog, Rumpelstiltskin, regulates nanos mRNA localization. Development 135: 973–982 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER 2006. Glorund, an hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell 10: 291–301 [DOI] [PubMed] [Google Scholar]
- Kalifa Y, Armenti ST, Gavis ER 2009. Glorund interactions in the regulation of gurken and oskar mRNAs. Dev Biol 326: 68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB 2003. The power of the 3′UTR: Translational control and development. Nat Rev Genet 4: 626–635 [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG 1992. The genome of Drosophila melanogaster. Academic Press, San Diego [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi: 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA 2004. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 23: 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MS, Wang Z, Gaba A, Fang P, Belk J, Ganesan R, Amrani N, Jacobson A 2002. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 26: 105–114 [DOI] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM 1996. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes & Dev 10: 2600–2609 [DOI] [PubMed] [Google Scholar]
- Smibert CA, Lie YS, Shillingaw W, Henzel WJ, Macdonald PM 1999. Smaug, a novel and conserved protein contributes to repression of nanos mRNA translation in vitro. RNA 5: 1535–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2007. New modes of translational control in development, behavior, and disease. Mol Cell 28: 721–729 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD 2005. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev Dyn 232: 593–608 [DOI] [PubMed] [Google Scholar]
- Thompson B, Wickens M, Kimble J 2007. Translational control in development. In Translational control in biology and medicine (ed. Mathews MB et al. ), pp. 507–544 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wang C, Lehmann R 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66: 637–647 [DOI] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R 1994. Genetics of nanos localization in Drosophila. Dev Dyn 199: 103–115 [DOI] [PubMed] [Google Scholar]