Abstract
Decapping is an essential step in multiple pathways of mRNA degradation. Previously, we synthesized mRNAs containing caps that were resistant to decapping, both to dissect the various pathways for mRNA degradation and to stabilize mRNA for more sustained protein expression. mRNAs containing an α-β CH2 group are resistant to in vitro cleavage by the decapping enzyme hDcp2 but poorly translated. mRNAs containing an S substitution at the β-phosphate are well translated but only partially resistant to hDcp2. We now describe seven new cap analogs substituted at the β-phosphate with BH3 or Se, or substituted at either the α-β or β-γ O with NH. The analogs differ in affinity for eIF4E and efficiency of in vitro incorporation into mRNA by T7 RNA polymerase. Luciferase mRNAs capped with these analogs differ in resistance to hDcp2 hydrolysis in vitro, translational efficiency in rabbit reticulocyte lysate and in HeLa cells, and stability in HeLa cells. Whereas mRNAs capped with m27,2′-OGppSpG were previously found to have the most favorable properties of translational efficiency and stability in mammalian cells, mRNAs capped with m7GppBH3pm7G are translated with the same efficiency but are more stable. Interestingly, some mRNAs exhibit a lag of up to 60 min before undergoing first-order decay (t1/2 ≅ 25 min). Only mRNAs that are efficiently capped, resistant to decapping in vitro, and actively translated have long lag phases.
Keywords: boranophosphate cap analogs, phosphoroselenoate cap analogs, imidodiphosphate cap analogs, translational efficiency, mRNA stability, in vitro transcription
INTRODUCTION
mRNA turnover is a highly regulated determinant in the overall control of eukaryotic gene expression (Parker and Song 2004). The presence of a 5′-terminal cap and a 3′-terminal poly(A) tract influences both the stability and translational efficiency of mRNA. After removal of the poly(A) tract (deadenylation), mRNA is degraded by two general pathways, decapping followed by 5′→3′ hydrolysis by the exonuclease Xrn1, and 3′→5′ hydrolysis by the exosome (Mukherjee et al. 2002; Wang et al. 2002). In addition to these two canonical pathways, mRNAs containing premature termination codons are degraded by the nonsense-mediated mRNA decay pathway (Maquat and Gong 2009). Such nonsense-containing transcripts are degraded either by deadenylation-independent decapping mediated by the protein Upf1 (Johansson et al. 2007) or by accelerated deadenylation and 3′→5′ exonucleolytic digestion (Mitchell and Tollervey 2003). Another specialized decay pathway exists for mRNAs containing AU-rich sequence elements (AREs) in their 3′-UTRs, which encode proteins regulating either cell growth or the response to external factors such as microorganisms or inflammatory stimuli (Barreau et al. 2006). Degradation of mRNAs can also be initiated by sequence-specific endonucleases (Dodson and Shapiro 2002) or miRNAs (Franks and Lykke-Andersen 2008). Histone mRNAs, which contain a conserved 3′-terminal stem–loop (SL) rather than a poly(A) tract, are degraded by another specialized pathway that is initiated upon inhibition of DNA replication or at the end of S phase (Marzluff et al. 2008). Instead of deadenylation, the initial step in histone mRNA decay is the addition of 3′-terminal uridine residues (Mullen and Marzluff 2008).
Decapping of mRNA is an essential step in all of these pathways. This involves removal of the 7-methylguanosine-containing cap, which both shuts down translation and constitutes the first irreversible step in mRNA degradation. There are two types of decapping enzymes in eukaryotic cells, DcpS and the Dcp1-Dcp2 complex. DcpS is a scavenger decapping enzyme and acts on the short capped oligonucleotides remaining after 3′→5′ hydrolysis by the exosome, cleaving the cap between the β- and γ-phosphates to produce m7GMP (Liu et al. 2002). Dcp1 and Dcp2 are the central components of an RNA-dependent decapping complex. Dcp2 belongs to the Nudix family of hydrolases (Piccirillo et al. 2003; She et al. 2006). In Saccharomyces cerevisiae, Dcp1 and Dcp2 directly interact to form a stable holoenzyme in which Dcp2 is the catalytic subunit and Dcp1 plays a stimulatory role (Steiger et al. 2003). Evidence has also been presented for an interaction between human Dcp1 and Dcp2 (Lykke-Andersen 2002; van Dijk et al. 2002). The proteins Edc1, Edc2, Edc3, Dhh1, and the Lsm1–7 protein complex stimulate decapping by Dcp2 in S. cerevisiae and mammals (Coller and Parker 2004). Edc4 (also known as Hedls and Ge-1) also acts as a positive effector of Dcp2 in mammals (Fenger-Gron et al. 2005). Decapping by Dcp2 is negatively regulated by eukaryotic initiation factor 4E (eIF4E), the translational cap-binding protein, as well as by the 3′-terminal poly(A) tract (Li et al. 2008). Biochemical studies of the human decapping protein (hDcp2) indicate that it requires both an m7G-containing cap and an RNA body of at least 25 nt for full enzymatic activity and cleaves between the α- and β-phosphate moieties to produce m7GDP (Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002; Piccirillo et al. 2003).
In previous studies, we sought to develop modified cap structures that, when incorporated into mRNAs, would be resistant to hydrolysis by hDcp2 and thereby would stabilize mRNA against 5′→3′ degradation (Grudzien et al. 2006; Grudzien-Nogalska et al. 2007). Such modified cap analogs could potentially serve two purposes: to facilitate dissection of the various steps of mRNA degradation and to boost production of proteins encoded by exogenously introduced mRNAs. The new cap analogs had substitutions in the triphosphate chain intended to prevent hydrolysis by decapping enzymes, but they also had substitutions at either the C2′ or C3′ positions of m7Guo (for example, compounds 2 and 3 in Fig. 1, respectively), to ensure they are incorporated into RNA by T7 RNA polymerase exclusively in the correct orientation (Stepinski et al. 2001; Jemielity et al. 2003). We found that m27,3′-OGppCH2pG (4), which has a methylene group replacing the α-β bridging O, was resistant to hDcp2 hydrolysis in vitro and increased mRNA stability in cultured cells (Grudzien et al. 2006). However, the affinity of m27,3′-OGppCH2pG (4) for eIF4E was only 60% that of the parent compound, m27,3′-OGpppG (3) (Kalek et al. 2005; Grudzien et al. 2006). We therefore designed a new series of cap analogs that contained a phosphorothioate moiety at either the α, β, or γ position of the triphosphate chain (Grudzien-Nogalska et al. 2007). Each analog contains a stereogenic P-center and is therefore obtained as a mixture of two diastereomers that can be resolved by reverse-phase HPLC (termed D1 and D2 based on their elution order). Luciferase mRNA containing m27,2′-OGppSpG, D2 (5) was more stable and was translated more efficiently in cultured HC11 cells than mRNA containing the parent compound m27,2′-OGpppG (2) (Grudzien-Nogalska et al. 2007).
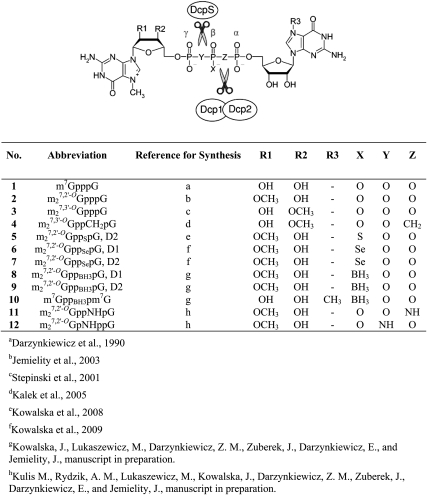
FIGURE 1.
Structures of cap analogs used in this study. D1 and D2 refer to the two diastereoisomers produced by substitution of a non-bridging oxygen atom on the β-phosphate. The cleavage sites for two decapping enzymes, DcpS and Dcp1/Dcp2, are indicated.
In the current study, we tested the resistance of mRNA capped with m27,2′-OGppSpG, D2 (5) to hydrolysis by hDcp2 under more rigorous conditions and found that it was only partially resistant. We therefore attempted to develop new cap analogs that conferred to mRNA both the favorable stability of m27,3′-OGppCH2pG (4) and the favorable translation of m27,2′-OGppSpG, D2 (5). Rather than the S substitution for a β non-bridging O of m27,2′-OGppSpG (5), we made substitutions with BH3 (m27,2′-OGppBH3pG, D1 [8] and m27,2′-OGppBH3pG, D2 [9]) and Se (m27,2′-OGppSepG, D1 [6] and m27,2′-OGppSepG, D2 [7]). Rather than the CH2 substitution for the α-β bridging O of m27,3′-OGppCH2pG (4), we made a substitution with NH (m27,2′-OGppNHpG [11]). Two related compounds, m7GppBH3pm7G (10) and m27,2′-OGpNHppG (12), were also synthesized. The seven new analogs were tested for in vitro and in vivo properties. Compared with the most favorable cap analog developed to date, m27,2′-OGppSpG, D2 (5), capping efficiency was better for two analogs (compounds 11 and 12), the same for one (10), and worse for four (6–9). mRNAs capped with the seven new analogs differed in resistance to in vitro digestion with hDcp2, but importantly, three of them (9, 10, and 11), were more resistant than m27,2′-OGppSpG, D2 (5). mRNAs capped with the new analogs had translational efficiencies in HeLa cells that, compared with m27,2′-OGppSpG, D2 (5)-capped mRNA, were either the same (7 and 10) or lower (6, 8, 9, 11, and 12). Similarly, translation in rabbit reticulocyte lysate was either the same (9, 10) or worse (6, 7, 8, 11, and 12). Finally, the stabilities of differently capped mRNAs in HeLa cells were either greater (compound 10), the same (8 and 9), or less (6, 7, and 11) than mRNA capped with m27,2′-OGppSpG, D2 (5).
RESULTS
mRNA capped by m27,2′-OGppSpG, D2 (5) is only partially resistant to hDcp2
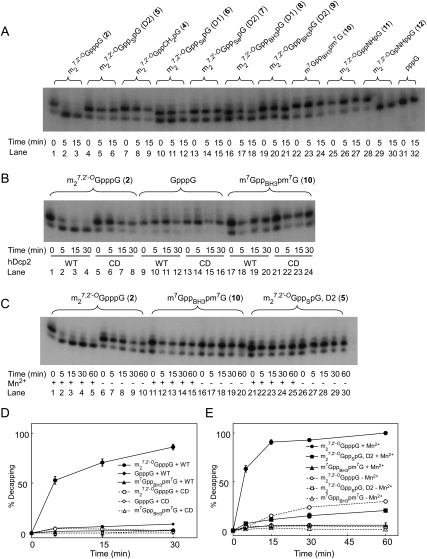
mRNA containing m27,2′-OGppSpG, D2 (5) is resistant to in vitro decapping by hDcp2 compared to the parent compound m27,2′-OGpppG (2) (Grudzien-Nogalska et al. 2007), but when we tested it under more rigorous conditions, we found it could still be partially decapped. Decapping was measured by synthesizing RNAs with T7 polymerase in the presence of a cap dinucleotide and [α-32P]GTP, incubating with recombinant hDcp2, and separating the products on an RNA sequencing gel (Fig. 2A). Capped RNAs (m27,2′-OGpppGp*GpNp…, where p* represents 32P) migrate slower than decapped RNAs (pGp*GpNp…), which migrate the same as uncapped RNAs (ppp*Gp*GpNp…) (lane 31). Bands detected by autoradiography were cut out and quantified by Cerenkov radiation. After 15 min, RNA capped with m27,2′-OGpppG (2) was completely decapped (lanes 1–3), whereas RNA capped with m27,2′-OGppSpG, D2 (5) was decapped 12 ± 3% (Fig. 2A, lanes 4–6; Table 1), indicating partial but not complete resistance to decapping. To confirm this result, capped RNAs were incubated with hDcp2 for 60 min, resulting in 20 ± 1% decapping of RNA containing m27,2′-OGppSpG, D2 (5) (Fig. 2C, lanes 21–25; Fig. 2E, squares).
FIGURE 2.
In vitro hydrolysis of capped oligonucleotides by hDcp2 analyzed on RNA sequencing gels. Oligonucleotides were synthesized from Nco1-cut pluc-A60 template with T7 polymerase in the presence of [α-32P]GTP and various cap dinucleotides. After treatment with hDcp2 for the indicated times, samples were loaded on a 10% RNA sequencing gel as described in Materials and Methods. (A) Ten RNA transcripts synthesized in the presence of either no cap dinucleotide (lanes 31,32) or the indicated cap dinucleotide were treated with 0.6 μg of hDcp2 for the indicated times and analyzed by electrophoresis and autoradiography. Samples in lanes labeled 0 min did not receive hDcp2. The quantification of decapping at 5 min and 15 min is given in Table 1. (B) Oligonucleotides synthesized in the presence of the indicated cap dinucleotides were subjected to digestion with 0.4 μg of either wild-type (WT) or catalytic dead (CD) hDcp2 for the indicated times and subjected to electrophoresis as in A. (C) Oligonucleotides synthesized in the presence of the indicated cap dinucleotides were subjected to digestion with 1 μg of WT hDcp2 for the indicated times, with or without 0.5 mM Mn2+ in the reaction mixture as indicated, and analyzed by electrophoresis and autoradiography. (D) Quantification of gel in B. (E) Quantification of gel in C. Decapping was calculated as the % loss in the upper band, normalized by the radioactivity in the upper plus lower bands. The data represent the means and SEM of three experiments. Error bars smaller than the symbols are not shown.
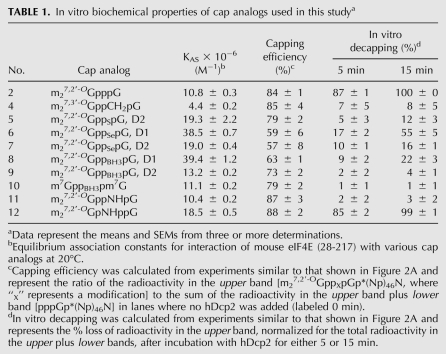
TABLE 1.
In vitro biochemical properties of cap analogs used in this studya
Development of novel borano-, seleno-, and imido-substituted cap analogs
hDcp2 is thought to cleave the cap structure between the α- and β-phosphate moieties, based on the observations that m7GDP is released (Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002) and that cleavage is blocked or retarded by substitution of the α-β bridging O with CH2 (Grudzien et al. 2006) or the β non-bridging O with S (Grudzien-Nogalska et al. 2007). However, as noted above, mRNAs capped with m27,3′-OGppCH2pG (4) are poorly translated (Grudzien et al. 2006), and mRNAs capped with m27,2′-OGppSpG, D2 (5) are not completely resistant to hDcp2 hydrolysis (Fig. 2). We therefore sought new cap analogs that would confer both of these favorable properties to mRNA: high translational efficiency and high stability. Our previous studies showed that modifications at a non-bridging O on the β-phosphate (Grudzien-Nogalska et al. 2007) or bridging O between α- and β-phosphates (Grudzien et al. 2006) rendered mRNA with the most resistance to hDcp2. Accordingly, we focused on cap analogs with substitutions at these positions (Fig. 1, compounds 6–11). The m27,2′-OGppSepG diastereoisomers (6 and 7) have a Se-for-O substitution, forming a phosphoroselenoate group at the β position. The m27,2′-OGppBH3pG diastereoisomers (8 and 9) have a BH3-for-O substitution, forming a boranophosphate group at the β position. m7GppBH3pm7G (10) is cap analog containing two identical nucleoside moieties and a boranophosphate group at the β position, so it is not synthesized as two diastereomers. m27,2′-OGppNHpG (11) has an NH-for-O substitution at the α-β bridge, forming an imidodiphosphate group. An analog with the NH group replacing the β-γ bridging O, m27,2′-OGpNHppG (12), was also included for comparison.
Fluorescence quenching measurements indicated that all seven new cap analogs have either the same or higher affinity for murine eIF4E compared with the parent compound, m27,2′-OGpppG (2) (Table 1, compounds 6–12). Previous studies (Kowalska et al. 2009) had shown that several phosphorothioate isomers of the cap dinucleotide had the highest affinity for eIF4E of all triphosphate analogs tested to date. Two of the new analogs, m27,2′-OGppSepG, D1 (6) and m27,2′-OGppBH3pG, D1 (8), also had very high affinities for eIF4E (Table 1), comparable to that of the homologous phosphorothioate m27,2′-OGppSpG, D1 (Kowalska et al. 2008). For all three substitutions for the β non-bridging O (S, Se, and BH3), the D1 diastereomer had nearly twice the affinity for eIF4E than the D2 diastereomer. An analog with a CH2 substitution for the α-β bridging O, m27,3′-OGppCH2pG (4), binds eIF4E with lower affinity than the parent compound (Table 1; Kalek et al. 2005), but substitution of the same O with NH produces an analog, m27,2′-OGppNHpG (11), with an affinity for eIF4E that is comparable to the parent compound (Table 1).
The new cap analogs differ in their ability to serve as substrates for T7 RNA polymerase
Capping efficiency reflects the degree to which a dinucleotide cap analog will be recognized as GTP by T7 RNA polymerase during in vitro transcription. We measured capping efficiency by separation on RNA sequencing gels and autoradiography (Fig. 2A). Lanes labeled 0 min (lanes 1,4,7, etc.) were not incubated with hDcp2. The ratio of the radioactivity in these lanes of the upper band [m27,2′-OGppXpGp*(Np)46N, where “X” represents a modification] to the sum of the radioactivity in the upper plus lower band [pppGp*(Np)46N] represents capping efficiency. This is summarized for all new analogs in Table 1. Under the conditions used here (10:1 ratio of cap analog to GTP in the T7 polymerase reaction), we observed 84 ± 1% capping for the parent compound, m27,2′-OGpppG (2). Higher capping efficiency can be obtained by using higher ratios of cap analog to GTP. The capping efficiency for m27,2′-OGppNHpG (11) and m27,2′-OGpNHppG (12) was slightly better than for the parent compound, followed in decreasing order by m27,3′-OGppCH2pG (4), m27,2′-OGppSpG, D2 (5), m7GppBH3pm7G (10), m27,2′-OGppBH3pG, D2 (9), m27,2′-OGppBH3pG, D1 (8), m27,2′-OGppSepG, D1 (6), and m27,2′-OGppSepG, D2 (7) (Fig. 2A; Table 1).
RNAs capped with BH3- and NH-containing analogs are resistant to decapping by hDcp2 in vitro
Transcripts synthesized in the presence of nine different cap analogs plus one uncapped transcript were incubated with hDcp2 for various times and subjected to PAGE (Fig. 2A). Transcripts capped with m27,3′-OGppCH2pG (4) were completely resistant to hDcp2 (Fig. 2A, lanes 7–9), in agreement with our previous study in which SAX chromatography was used to measure decapping (Grudzien et al. 2006). Transcripts capped with m27,2′-OGppSepG, D1 (6) (lanes 10–12), m27,2′-OGppSepG, D2 (7) (lanes 13–15), and m27,2′-OGppBH3pG, D1 (8) (lanes 16–18) were intermediate in their susceptibility to hDcp2. By contrast, transcripts capped with m27,2′-OGppBH3pG, D2 (9) (lanes 19–21), m7GppBH3pm7G (10) (lanes 22–24), and m27,2′-OGppNHpG (11) (lanes 25–27) were highly resistant to hDcp2. The average results for three experiments are presented in Table 1.
To confirm that the decapping activity observed in Figure 2A was due to hDcp2 and not to a contaminating bacterial pyrophosphatase, we used a variant of hDcp2 in which Glu-147 and Glu-148 are both changed to Gln, producing a catalytically dead (CD) enzyme (Wang et al. 2002). Under conditions where RNA containing m27,2′-OGpppG (2) was completely decapped by wild-type (WT) hDcp2 (Fig. 2B, lanes 1–4; Fig. 2D, closed circles), there was no decapping by CD hDcp2 (Fig. 2B, lanes 5–8; Fig. 2D, open circles). RNA with an unmethylated cap was inefficiently cleaved by WT hDcp2 (Fig. 2B, lanes 9–12; Fig. 2D, filled diamonds; 9.0 ± 0.5% by 60 min), in agreement with published findings (van Dijk et al. 2002; Wang et al. 2002), but was not cleaved at all by CD hDcp2 (Fig. 2B, lanes 13–16; Fig. 2D, open diamonds). RNA capped with m7GppBH3pm7G (10) was not detectably decapped by either enzyme preparation (Fig. 2B, lanes 17–24; Fig. 2D, triangles).
The original in vitro decapping assay conditions reported for hDcp2 utilized Mg2+ as the sole divalent cation (Lykke-Andersen, 2002; van Dijk et al. 2002; Wang et al. 2002), but subsequently it was shown that both hDcp2 (Piccirillo et al. 2003) and yeast Dcp2p (Steiger et al. 2003) are considerably more active if Mn2+ is added, although there is no evidence that the enzyme utilizes Mn2+ in vivo. Previous studies from our own laboratory used 2 mM Mg2+ as the sole divalent cation to demonstrate cleavage by hDcp2 of RNA capped with m27,3′-OGpppG (3) and resistance of RNA capped with m27,3′-OGppCH2pG (4) (Grudzien et al. 2006), but subsequently we used 2 mM Mg2+ plus 0.5 mM Mn2+ to show resistance to of mRNA capped with m27,2-OGppSpG, D2 (5) (Grudzien-Nogalska et al. 2007). To ensure that the resistance of mRNA capped with m27,2′-OGppSpG, D2 (5) and m7GppBH3pm7G (10) to hDcp2 is not a Mn2+-induced artifact, we carried out the decapping assay under both conditions: with Mg2+ as the sole divalent cation and with 2 mM Mg2+ plus 0.5 mM Mn2+ (Fig. 2C,E). The results confirmed that hDcp2 is much more active against RNA capped with m27,2′-OGpppG (2) in the presence of Mn2+ (Fig. 2E, filled versus open circles). Importantly, the same order of resistance to decapping is seen under both conditions. For instance, at the 30-min time point, the % decapping in Mg2+ plus Mn2+ is 92.8 ± 0.3 for RNA capped with m27,2′-OGpppG (2), 15.3 ± 2.6 for m27,2′-OGppSpG, D2 (5), and 5.5 ± 1.9 for m7GppBH3pm7G (10), whereas with Mg2+ alone, it is 24.0 ± 1.7 for m27,2′-OGpppG (2), 4.9 ± 1.9 for m27,2′-OGppSpG, D2 (5), and 1.5 ± 0.3 for m7GppBH3pm7G (10). Thus, we conclude that the resistance to in vitro decapping of these phosphate chain-modified cap structures is not a Mn2+-induced artifact.
Cap analogs substituted with BH3 stabilize mRNAs in HeLa cells
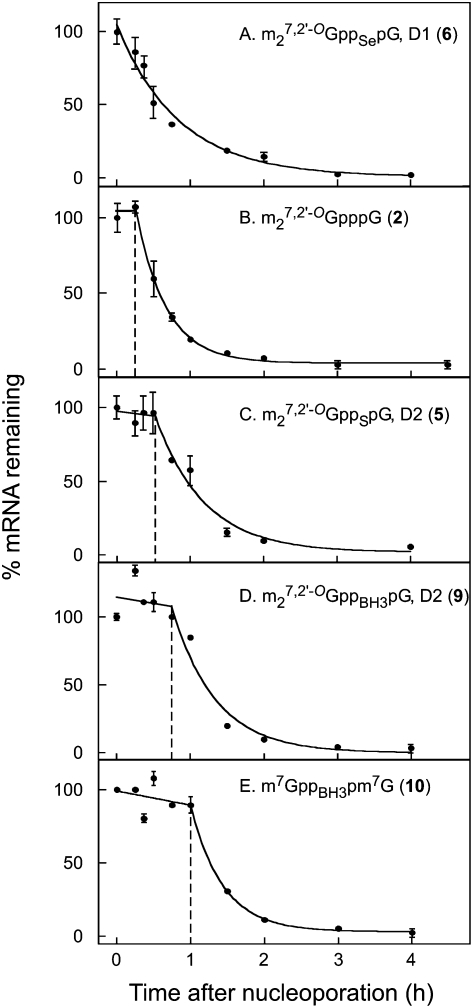
We next tested how the new cap analogs would affect mRNA stability when incorporated into RNAs and delivered into mammalian cells. Luciferase mRNAs were synthesized in vitro with various cap analogs at the 5′-termini and 60-nt poly(A) tracts at the 3′-termini. The mRNAs were introduced into HeLa cells by nucleoporation, the cells were incubated for various times up to 4 h, and total RNA was recovered. Luciferase mRNA levels were measured by qRT-PCR (Fig. 3). Interestingly, we observed a two-phase decay pattern for all mRNAs except those capped with m27,2′-OGppSepG, D1 (6) and m27,2′-OGppSepG, D2 (7); there was a lag phase of various durations lasting as long as 60 min, followed by first-order decay with approximately the same t1/2 for all mRNAs. This type of experiment was conducted three or more times for each of the seven new cap analogs, using as controls m27,2′-OGpppG (2), m27,2′-OGppSpG, D2 (5), and m27,3′-OGppCH2pG (4) (Table 2). mRNAs containing different cap analogs had similar t1/2 values for the first-order decay phase (21–32 min) but varied significantly in the length of lag phase (0–51 min).
FIGURE 3.
Decay of luciferase mRNA terminated by various cap analogs in HeLa cells. Luciferase mRNAs were synthesized in the presence of the indicated cap analogs and delivered into HeLa cells by nucleoporation as described in Materials and Methods. Cells were lysed at the indicated times and luciferase mRNA was measured by qRT-PCR. Data in panels A–E are plotted as a percentage of the luciferase mRNA present immediately after nucleoporation. In panels B–E, there is a lag before the initiation of rapid decay. The data for the post-lag period were fit to single-exponential function and the t½ of calculated as described in Materials and Methods. Vertical dashed lines mark the boundary between lag phase and first-order decay phase. The data for each transcript represent a single experiment. The error bars represent duplicate luciferase mRNA determinations. The values for replicate experiments are given in Table 2.
TABLE 2.
In vivo biochemical properties of cap analogs used in this studya
mRNAs capped with Se- and BH3-containing analogs are efficiently translated in HeLa cells and rabbit reticulocyte lysate
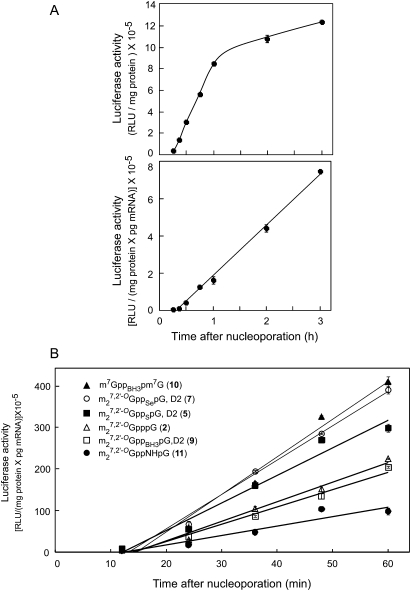
Since mRNAs capped with several of the new analogs (m27,2′-OGppBH3pG, D2 [9], m7GppBH3pm7G [10], and m27,2′-OGppNHpG [11]) are more resistant to hDcp2 hydrolysis in vitro than m27,2′-OGppSpG, D2 (5), we determined their translational properties. Translational efficiency in cultured mammalian cells can be determined by introducing luciferase mRNA by nucleoporation, removing aliquots of the cells after various times, and measuring the accumulation of luciferase (Grudzien et al. 2006; Grudzien-Nogalska et al. 2007). The concentration of luciferase mRNA is measured in the same cells by qRT-PCR, and the rate of luciferase accumulation per unit of mRNA represents translational efficiency. Figure 4A shows that luciferase accumulation begins after a ∼15 min lag, which is required for synthesis of the enzyme, and continues linearly for ∼1 h, after which it slows dramatically (upper panel). However, when the luciferase value is normalized for the amount of luciferase mRNA still remaining at each time point, the rate of luciferase accumulation is linear for at least 3 h (lower panel). Thus, the translational efficiency of luciferase mRNA remains constant over this period even though the amount of luciferase mRNA decreases.
FIGURE 4.
Translational efficiencies in HeLa cells of mRNAs containing various caps. (A) Upper panel: Luciferase activity was measured at the indicated times after introduction of luciferase mRNA capped with m27,2′-OGpppG (2) into HeLa cells by nucleoporation. Lower panel: Luciferase activity values in the upper panel were divided by the amount of luciferase mRNA at each time point as measured by qRT-PCR. (B) Luciferase activity for mRNAs synthesized in the presence of the indicated cap analogs was measured at the indicated times after nucleoporation. Data were normalized for the amount of luciferase mRNA as in A. The data for each transcript represent a single experiment. The error bars represent duplicate luciferase activity determinations. The values for replicate experiments are given in Table 2.
We chose to measure translational efficiency of mRNAs capped with the new analogs over the first 60 min after nucleoporation to minimize errors caused by mRNA decay (Fig. 4B). This type of experiment was performed three or more times for each of the seven new analogs plus three controls (Table 2). mRNAs capped with two of the new cap analogs, m27,2′-OGppSepG, D2 (7) and m7GppBH3pm7G (10), were translated 1.4- and 1.7-fold more efficiently than mRNA capped with the parent compound, m27,2′-OGpppG (2). mRNAs capped with m27,2′-OGppBH3pG, D2 (9) were translated with similar efficiency as the parent mRNA, whereas four others (m27,2′-OGppSepG, D1 [6], m27,2′-OGppBH3pG, D1 [8], m27,2′-OGppNHpG [11], and m27,2′-OGpNHppG [12]) were translated less efficiently.
Measuring translational efficiency in mammalian cells has the advantage of being more physiologically relevant than in vitro translation systems but the disadvantage that the amount of mRNA introduced by nucleoporation must be independently measured, thereby introducing experimental error. When mRNA is introduced by nucleoporation, there is also the possibility that some of it may be present in a form or location that is inaccessible to the translational machinery, although we previously showed that the majority of exogenously introduced mRNA is in polysomes (Grudzien et al. 2006; Grudzien-Nogalska et al. 2007). Both of these potential problems, as well as experimental error arising from differential degradation rates, are avoided by using an in vitro translation system. We therefore measured the translational efficiency of mRNAs capped with the seven new analogs in a rabbit reticulocyte lysate system (Table 2). Although there were quantitative differences between in vitro and in vivo translational efficiencies, most of the analogs had qualitatively similar effects in the two systems. For instance, mRNAs capped with m27,2′-OGppSepG, D2 (7) and m7GppBH3pm7G (10) were more efficiently translated than the control mRNA in both systems. Similarly, mRNAs capped with m27,2′-OGppNHpG (11) and m27,2′-OGpNHppG (12) were less efficiently translated than the control mRNA in both systems. However, mRNAs capped with m27,2′-OGppSepG, D1 (6) and m27,2′-OGppBH3pG, D1 (8) were translated somewhat better than the control mRNA in vitro but worse in vivo. Another trend observed in both systems is that the mRNAs capped with the D2 diastereomers were translated more efficiently than those capped with the D1 diastereomers, despite the higher affinity of the latter for eIF4E (Table 1).
DISCUSSION
Resistance of cap analogs substituted at the β-phosphate to hDcp2 hydrolysis
The new cap analogs with modifications at the β-phosphate position were designed to both provide resistance to hDcp2 and enhance translational efficiency. Nucleotide analogs with substitution of S, BH3, or Se for the non-bridging O in phosphate moieties share some chemical and biochemical properties such as polarity, net charge at physiological pH, and resistance to nucleases but also differ with respect to P-X bond length, van der Waals radius, and affinity of the X group for various divalent or other metal cations. S and Se, due to their larger atomic size, are more polarizable and prefer interactions with softer metal ions such as Zn2+, Mn2+, and Cd2+ compared to Mg2+ and Ca2+, which are preferred by O. The BH3 group is a very poor nucleophile because it lacks a lone electron pair and therefore does not accept H-bonds or coordinate metal ions (Li et al. 2007). It has also been reported that oligonucleotides containing boranophosphate modifications can be more resistant to enzymatic cleavage than their corresponding phosphorothioates (Sergueev and Shaw 1998). Enzymatic resistance of nucleotides and nucleic acids containing phosphorothioate, phosphoroselenoate, and boranophosphate modifications are thought to result from changes in affinities to metal ions and phosphate group geometry.
Like other Nudix-domain hydrolases, the catalytic center of Dcp2 contains three conserved Glu residues that coordinate a divalent cation responsible for activation of a water molecule to attack the phosphate moiety (She et al. 2006). The resistance of mRNA capped with m27,2′-OGppSpG, D2 (5) to hDcp2 may be due to poor interaction of S with Mg2+ or Mn2+ in the enzyme's active site or to disruption of enzyme-substrate complex geometry because of the longer P-S bond and larger S atom. The even higher resistance of BH3-containing analogs may be explained by the poor ability of BH3 to bind metal ions or accept H-bonds compared to S and Se. It is surprising that m7GppBH3pm7G (10) is less susceptible to hDcp2 than either m27,2′-OGppBH3pG, D1 (8) or m27,2′-OGppBH3pG, D2 (9). m7GppBH3pm7G (10) is not diastereomeric, but after incorporation into mRNA, it exists as a mixture of diastereomers analogous to the D1 and D2 diastereomers of m27,2′-OGppBH3pG (8 and 9, respectively). (This is because the first phosphodiester bond catalyzed by T7 RNA polymerase could be formed with the 3′-OH of either m7Guo moiety of m7GppBH3pm7G [10].) Consequently, the enzymatic resistance of mRNA prepared with m7GppBH3pm7G would be expected to be intermediate between that observed for m27,2′-OGppBH3pG, D1 (8) and m27,2′-OGppBH3pG, D2 (9). The fact that it is greater suggests that the second m7Guo moiety of m7GppBH3pm7G (10) makes an additional contribution to enzymatic resistance.
Resistance of cap analogs substituted at the α-β bridge to hDcp2 hydrolysis
The imidodiphosphate (pNHp) and methylenebisphosphonate (pCH2p) bridging modifications are both known for their resistance to enzymatic cleavage, but unlike the non-bridging modifications discussed above, their affinities for Mg2+ are higher than those of the parent compounds (Yount et al. 1971). The pNHp moiety is more similar to pyrophosphate than pCH2p in terms of bond length, geometry, and acid-base properties (Yount et al. 1971). Unlike the CH2 group, the NH group has a lone electron pair and can therefore participate in H-bonding, which is particularly important for binding to eIF4E (Tomoo et al. 2003). This is consistent with the fact that NH-containing analogs bind eIF4E with affinities similar to those of the parent compounds (Table 1), whereas the CH2-containing analogs bind eIF4E weaker (Kalek et al. 2005). The mechanism of resistance of NH- and CH2-containing analogs is different from that of non-bridging β-phosphate-substituted analogs and arises from the high chemical stability of P-C-P and P-N-P bonds compared to P-O-P. Thus, we observed complete resistance to decapping with both of these analogs (Table 1).
Translational properties of mRNAs containing the new cap analogs
For a cap analog to be incorporated into an mRNA that has both high translational efficiency and high resistance to decapping, the analog must have favorable interactions within the active sites of three structurally unrelated proteins: T7 RNA polymerase (Steitz 2009), eIF4E (Marcotrigiano et al. 1997; Matsuo et al. 1997; Tomoo et al. 2003), and hDcp2 (Deshmukh et al. 2008). Our results suggest that these interactions differ among the seven new cap analogs. mRNAs capped with compounds having BH3 in the β position (m27,2′-OGppBH3pG, D2 [9] and m7GppBH3pm7G [10]) are highly resistant to decapping, but the capping efficiency, stability, and translational efficiency of the latter is higher. m27,2′-OGppNHpG (11)-capped RNAs are also completely resistant to in vitro hydrolysis by hDcp2 but are translated less efficiently than the control mRNA both in vitro and in vivo. mRNA capped with m27,2′-OGppSepG, D2 (7) is translated more efficiently than the control mRNA despite the fact that its % capping is only 57%, suggesting that it would have an even higher translational efficiency if the % capping could be increased. Surprisingly, even though m27,2′-OGppNHpG (11) is similar to the parent m27,2′-OGpppG (2) with respect to bond length, geometry, and acid-base properties, it does not enhance translational efficiency or increase the length of the lag phase, even though it is completely resistant to hDcp2 in vitro.
Relationship of the lag in mRNA degradation to translation and decapping
Some insight into the differences among these mRNAs comes from correlations among resistance to hDcp2 in vitro, translational efficiency, and length of the lag phase. Transcripts capped with m27,2′-OGppSpG, D2 (5), m27,2′-OGppBH3pG, D2 (9), and m7GppBH3pm7G (10) are resistant to hDcp2, are translated with the same or higher efficiency than the control mRNA, and exhibit a long lag phase. On the other hand, RNAs capped with m27,3′-OGppCH2pG (4) and m27,2′-OGppNHpG (11) are resistant to hDcp2, are translated poorly, and exhibit a short lag phase. Thus, resistance to hDcp2 hydrolysis alone cannot account for a long lag phase and efficient translation. We propose that the lag is partly a function of mRNA recognition by eIF4E and recruitment to polysomes, which are determinants of the rate of both deadenylation and decapping. During translation, the cap is protected from decapping by association with eIF4E (Schwartz and Parker 2000). PABP binds to poly(A) and protects mRNA against exonucleolytic degradation (Sachs 1993; Coller et al. 1998; Wang et al. 1999). PABP also binds to the N terminus of eIF4G (Tarun and Sachs 1996) and stabilizes the eIF4G-eIF4E complex (Amrani et al. 2008). We speculate that mRNAs that are poorly recognized by the translational machinery, such as those capped with m27,3′-OGppCH2pG (4) or m27,2′-OGppNHpG (11), enter the 3′→5′ degradation pathway immediately, exhibiting a short lag, even if the cap is resistant to hDcp2. mRNAs that are well recognized by the translational machinery, such as those capped with m27,2′-OGpppG (2), are protected from decapping because Dcp2 and eIF4E compete for binding to the cap; these mRNAs exhibit an intermediate lag phase, even though the cap is susceptible to cleavage by hDcp2. When there is both efficient recognition by the translational machinery and resistance to decapping, such as for mRNAs capped with m27,2′-OGppSpG, D2 (5), m27,2′-OGppBH3pG, D2 (9), or m7GppBH3pm7G (10), the lag is the longest, presumably because both deadenylation and decapping are retarded. The absence of a lag for mRNAs capped with m27,2′-OGppSepG, D2 (7) is likely due to the fact that this analog is poorly recognized by T7 polymerase, resulting in the lowest capping efficiency of all the new analogs; nearly half of the mRNA in this preparation is uncapped (Table 1).
Utility of cleavage-resistant cap analogs
The new cap analogs differ from each other and from previously described analogs in numerous properties that make them potentially useful for a variety of applications. For example, structural studies of the catalytic site of hDcp2 and related enzymes could be carried out with a series of analogs that are completely resistant to cleavage but differ in molecular properties (m27,3′-OGppCH2pG [4], m27,2′-OGppBH3pG, D2 [9], and m27,2′-OGppNHpG [11]). Some have very high affinity for eIF4E and may point the way to new anticancer drugs based on inhibition of cap-dependent translation (m27,2′-OGppSepG, D1 [6] and m27,2′-OGppBH3pG, D1 [8]). Some confer high translational efficiency in vivo, in vitro, or both and should be useful for enhanced protein production. Efficiently translated mRNAs that differ in decapping rate (e.g., mRNAs capped with m27,2′-OGpppG [2], m27,2′-OGppSpG, D2 [5], m27,2′-OGppSepG, D2 [7], and m7GppBH3pm7G [10]) should be useful for the study of mRNA turnover. Long-lived and efficiently translated mRNAs also have a promising role is in cancer immunotherapy, where an in vitro–synthesized mRNA encoding a tumor antigen is used to elicit an immune response in dendritic cells (Weide et al. 2008). Vaccination with such mRNAs has already entered human clinical testing. Recently, capping with the D1 diastereomer of m27,2′-OGppSpG was shown to enhance RNA stability and translational efficiency in immature dendritic cells (Kuhn et al. 2010). In vivo delivery of an antigen encoded by this mRNA led to increased protein expression and enhanced priming and expansion of naïve antigen–specific-T–cells in mice. Further advances in the development of mRNAs that are both stable and translationally efficient should aid this powerful approach to induce therapeutic immune responses.
MATERIALS AND METHODS
Materials
All common reagents were of analytical grade. Oligodeoxynucleotides were synthesized by MWG/Operon (Huntsville, AL). Literature citations for synthesis of all cap analogs are given in Figure 1. The structures and homogeneities of all compounds were confirmed by mass spectrometry, 1H NMR, 31P NMR and reverse-phase HPLC. The concentrations of cap analog solutions were determined by UV absorbance at pH 7.0 using the extinction coefficient ɛ255nm = 22.6 × 103 M−1cm−1.
In vitro synthesis of RNAs
The DNA template for T7 RNA polymerase used to make capped RNAs for in vitro decapping assays was the plasmid pluc-A60 (Grudzien et al. 2006) digested with NcoI, which yields a capped RNA corresponding to the first 48 nt of firefly luciferase mRNA. Capped oligoribonucleotides were synthesized in the presence of various cap analogs and 10 μCi/μL [α-32P]GTP (ICN) in a 50-μL reaction for 1 h as previously described (Jemielity et al. 2003) except that the T7 RNA polymerase was prepared in our laboratory (Davanloo et al. 1984). Reaction mixtures were extracted with phenol and chloroform, and RNAs were separated from unincorporated nucleotides with NucAway spin columns (Ambion). The concentrations of RNAs were determined by measuring Cerenkov radiation in a scintillation counter (Beckman) and calculating from the specific radioactivity of the precursor GTP. Full-length luciferase mRNA capped with various analogs was synthesized as described above except that pluc-A60 was digested with HpaI, the reaction mixture was 200 μL, no radioactivity was included, and the incubation time was 2 h. Reaction mixtures were treated with 10 units of RQ1 RNase-Free DNase (Promega) at 37°C for 20 min. The RNAs were purified with an E.Z.N.A. Total RNA Miniprep Kit (Omega Bio-Tek). RNA concentration was determined by UV absorbance at 260 nm, and RNA integrity was verified by electrophoresis on 1.2% agarose gels containing formaldehyde.
Decapping assays
GST-hDcp2 and a variant of GST-hDcp2 in which Glu-147 and Glu-148 is replaced with Gln to produce a catalytically inactive enzyme were expressed in BL21(DE3)pLysS Escherichia coli cells from plasmids pGEX-hDcp2 and pGEX hDcp2Q147/8, respectively (Wang et al. 2002), which were generously donated by Megerdich Kiledjian, Rutgers University. The enzymes were expressed (Wang et al. 2002) and purified (Zhao et al. 2003) as previously described. Capped 32P-labled oligonucleotides were subjected to digestion with either wild-type or catalytically inactive GST-hDcp2 at 37°C for various times. The conditions were the same as described previously (Grudzien-Nogalska et al. 2007) except that spermidine was omitted. In Figure 2C, Mn2+ was omitted from some reactions as indicated. Reactions were stopped by adding two volumes of Precipitation/Inactivation buffer (Ambion), and RNAs were precipitated at −20°C for 20 min and collected by centrifugation at 13,000 × g at 4°C for 20 min. Precipitated samples were resuspended in 5 μL of Sequencing Gel Loading Buffer (Ambion) and denatured at 95°C for 5 min. RNA sequencing gels (10% polyacrylamide) were run at 45–70 W for ∼3 h on a Base Runner Nucleic Acid Sequencer apparatus (International Biotechnologies) (Slatko and Albright 1992). Gels were fixed in 5% acetic acid, 5% methanol for 10–15 min, dried onto Whatman 3MM filter paper (Fisher Scientific), and exposed to Blue X-ray film (Kodak). Radioactivity in individual bands was quantified by two methods: analyzing scanned film using ImageQuant TL program (GE Health Care, version 7.0), and cutting them out and determining Cenrenkov radiation.
Cell culture and nucleoporation of mRNA
HeLa cells were grown in DMEM high glucose medium (Hyclone) containing 10% fetal bovine serum (Atlanta Biologicals) and 1× penicillin-streptomycin antibiotics (Mediatech). Cells were seeded 1 d prior to nucleoporation and detached from plates by treatment with 0.05% trypsin and 2 mM EDTA when they reached ∼60% confluency. Cells were counted, incubated in fresh media at 37°C for 30 min, and subjected to nucleoporation with a Nucleofector II (Lonza), following the manufacture's protocol. One microgram of RNA was introduced into 106 cells in Nucleoporation Solution R by using program I-13.
Measurement of translational efficiency and mRNA decay in HeLa cells
For assay of in vivo translational efficiency, aliquots of 0.5 × 106 cells were shaken in 1.5-mL Eppendorf tubes at 37°C for various times after nucleoporation. Total protein was extracted and luciferase activity was measured as described previously (Grudzien-Nogalska et al. 2007). For measurement of mRNA stability over periods <1 h, cells were shaken in Eppendorf tubes as described above. For periods >1 h, cells were plated onto 35-mm cell culture dishes and incubated at 37°C in 5% CO2. The extraction of total RNA and measurement of luciferase mRNA levels by quantitative RT-PCR (qRT-PCR) were performed as described previously (Grudzien-Nogalska et al. 2007). The amount of luciferase mRNA at different time points was expressed as a percentage of the mRNA at time zero. Data were plotted versus time of incubation after nucleoporation and fitted to a single-exponential function:
where k, ΔA, and A∞ are the observed first-order rate constant for mRNA decay, amplitude, and final value of mRNA, respectively. KaleidaGraph (Synergy Software, version 3.06) was used for nonlinear least-squares fitting of decay data and determination of standard errors for parameters obtained from the fits. The t1/2 of luciferase mRNAs was calculated using the equation t1/2 = ln2/k. When a lag in decay of mRNA was observed, only the data points occurring after the lag were fit to Equation 1.
Other methods
Mouse eIF4E (residues 28–217) was expressed in E. coli (Zuberek et al. 2003), reconstituted from inclusion bodies, and purified by ion-exchange chromatography on a HiTrap SP column (Marcotrigiano et al. 1997). Fluorescence time-synchronized titrations were performed as previously described (Niedzwiecka et al. 2002). In vitro translational efficiency was measured in a rabbit reticulocyte lysate system (Jemielity et al. 2003).
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (R01GM20818 to R.E.R.), the Polish Ministry of Science and Higher Education (N N204 089438 to J.J., N N301 096339 to E.D.), and the Howard Hughes Medical Institute (55005604 to E.D.). The authors are grateful to Megerditch Kiledjian (Rutgers University) for the pGEX-hDcp2 and pGEX hDcp2Q147/8 plasmids, Eric First (LSUHSC-S) for assistance with RNA sequencing gels, and the Research Core Facility of the Louisiana State University Health Sciences Center, Shreveport, for instrumentation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2430711.
REFERENCES
- Amrani N, Ghosh S, Mangus DA, Jacobson A 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453: 1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB 2006. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Coller JM, Gray NK, Wickens MP 1998. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev 12: 3226–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz E, Stepinski J, Tahara SM, Stolarski R, Ekiel I, Haber D, Neuvonen K, Lehikoinen P, Labadi I, Lönnberg H 1990. Synthesis, conformation and hydrolytic stability of 1,P3-dinucleoside triphosphates related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleoside monophosphate analogs. Nucleosides Nucleotides 9: 599–618 [Google Scholar]
- Davanloo P, Rosenberg AH, Dunn JJ, Studier FW 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci 81: 2035–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD 2008. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol Cell 29: 324–336 [DOI] [PubMed] [Google Scholar]
- Dodson RE, Shapiro DJ 2002. Regulation of pathways of mRNA destabilization and stabilization. Prog Nucleic Acid Res Mol Biol 72: 129–164 [DOI] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE 2006. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem 281: 1857–1867 [DOI] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Jemielity J, Kowalska J, Darzynkiewicz E, Rhoads RE 2007. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA 13: 1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity J, Fowler T, Zuberek J, Stepinski J, Lewdorowicz M, Niedzwiecka A, Stolarski R, Darzynkiewicz E, Rhoads RE 2003. Novel “anti-reverse” cap analogues with superior translational properties. RNA 9: 1108–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJO, He F, Spatrick P, Li C, Jacobson A 2007. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci 104: 20872–20877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalek M, Jemielity J, Grudzien E, Zuberek J, Bojarska E, Cohen LS, Stepinski J, Stolarski R, Davis RE, Rhoads RE, et al. 2005. Synthesis and biochemical properties of novel mRNA 5′ cap analogs resistant to enzymatic hydrolysis. Nucleosides Nucleotides Nucleic Acids 24: 615–621 [DOI] [PubMed] [Google Scholar]
- Kowalska J, Lewdorowicz M, Zuberek J, Grudzien-Nogalska E, Bojarska E, Stepinski J, Rhoads RE, Darzynkiewicz E, Davis RE, Jemielity J 2008. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA 14: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska J, Lukaszewicz M, Zuberek J, Darzynkiewicz E, Jemielity J 2009. Phosphoroselenoate dinucleotides for modification of mRNA 5′ end. ChemBioChem 10: 2469–2473 [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, Darzynkiewicz E, Huber C, Tureci O, Sahin U 2010. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther 17: 961–971 [DOI] [PubMed] [Google Scholar]
- Li P, Sergueeva ZA, Dobrikov M, Shaw BR 2007. Nucleoside and oligonucleoside boranophosphates: chemistry and properties. Chem Rev 107: 4746–4796 [DOI] [PubMed] [Google Scholar]
- Li Y, Song M-G, Kiledjian M 2008. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol 28: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rodgers ND, Jiao X, Kiledjian M 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 21: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Gong C 2009. Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochem Soc Trans 37: 1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras A-C, Sonenberg N, Burley SK 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras A-C, Sonenberg N, Wagner G 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 4: 717–724 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol Cell 11: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras A-C, Mak P, Darzynkiewicz E, Sonenberg N, et al. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319: 615–635 [DOI] [PubMed] [Google Scholar]
- Parker R, Song H 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Piccirillo C, Khanna R, Kiledjian M 2003. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9: 1138–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB 1993. Messenger RNA degradation in eukaryotes. Cell 74: 413–421 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergueev DS, Shaw BR 1998. H-Phosphonate approach for solid-phase synthesis of oligodeoxyribonucleoside boranophosphates and their characterization. J Am Chem Soc 120: 9417–9427 [Google Scholar]
- She M, Decker CJ, Chen N, Tumati S, Parker R, Song H 2006. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol 13: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko BE, Albright LM 1992. Denaturing gel electrophoresis for sequencing. In Current protocols in molecular biology, pp. 7.6.1–7.6.13 John Wiley & Sons, Inc, NJ: [DOI] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA 2009. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr Opin Struct Biol 19: 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE 2001. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogues 7-methyl(3′-O-methyl)GpppG and 7-methyl(3′-deoxy)GpppG. RNA 7: 1486–1495 [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15: 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, et al. 2003. Structural feature of human factor 4E, studied by X-ray crystal analysis and molecular dynamics simulations. J Mol Biol 328: 365–383 [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19: 4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide B, Garbe C, Rammensee HG, Pascolo S 2008. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol Lett 115: 33–42 [DOI] [PubMed] [Google Scholar]
- Yount RG, Babcock D, Ballantyne W, Ojala D 1971. Adenylyl imidodiphosphate, an adenosine triphosphate analog containing a P-N-P linkage. Biochemistry 10: 2484–2499 [DOI] [PubMed] [Google Scholar]
- Zhao X, Lamphear BJ, Xiong D, Knowlton K, Rhoads RE 2003. Protection of cap-dependent protein synthesis in vivo and in vitro with an eIF4G-1 variant highly resistant to cleavage by Coxsackievirus 2A protease. J Biol Chem 278: 4449–4457 [DOI] [PubMed] [Google Scholar]
- Zuberek J, Wyslouch-Cieszynska A, Niedzwiecka A, Dadlez M, Stepinski J, Augustyniak W, Gingras A-C, Zhang Z, Burley SK, Sonenberg N, et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA cap analogs by electrostatic repusion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]