Abstract
Superfamily 1 (SF1) and superfamily 2 (SF2) RNA helicases are ubiquitous mRNA-protein (mRNP) remodelling enzymes that play critical roles in all aspects of RNA metabolism1, 2. The SF2 DEAD-box ATPase Dbp5/Ddx19 functions in mRNA export and is thought to remodel mRNPs at the nuclear pore complex (NPC)3–8. Dbp5 is localized to the NPC via an interaction with Nup159/Nup2143–5, 8, 9 and is locally activated there by Gle1 together with the small-molecule inositol hexakisphosphate (IP6) 10, 11. Local activation of Dbp5 at the NPC by Gle1 is essential for mRNA export in vivo11, 12; however, the mechanistic role of Dbp5 in mRNP export is poorly understood and it is not known how Gle1IP6 and Nup159 regulate the activity of Dbp5. Here we report structures of Dbp5 in complex with Gle1IP6, Nup159/Gle1IP6, and RNA. These structures reveal that IP6 functions as a small-molecule tether for the Gle1-Dbp5 interaction. Surprisingly, the Gle1IP6-Dbp5 complex is structurally similar to another DEAD-box ATPase complex essential for translation initiation, eIF4G-eIF4A, and we demonstrate that Gle1IP6 and eIF4G both activate their DEAD-box partner by stimulating RNA release. Furthermore, Gle1IP6 relieves Dbp5 auto- regulation and cooperates with Nup159 in stabilizing an open Dbp5-intermediate that precludes RNA binding. These findings explain how Gle1IP6, Nup159, and Dbp5 collaborate in mRNA export and provide a general mechanism for DEAD-box ATPase regulation by Gle1/eIF4G-like activators.
Gle1 and IP6 robustly activate the ATPase activity of Dbp510, 11, but interactions between Dbp5 and Gle1IP6 are weak and transient in vitro10, 12. However, gain-of-function mutations in Dbp5 and Gle1 (Dbp5L327V and Gle1H337R)10 allowed for the purification of a stable Dbp5-Gle1IP6 complex for protein crystallography (data not shown). Multiwavelength anomalous dispersion (MAD) phased protein crystal structures were initially obtained for Gle1IP6 and the C-terminal RecA-like domain (CTD) of Dbp5L327V, both in the presence and absence of the Gle1 H337R mutation at a resolution of 2.6 Å (Rwork/Rfree of 18.7%/21.3%) and 2.5 Å (Rwork/Rfree of 20.2%/23.8%), respectively (Supplementary Table 1). This model allowed us to solve the crystal structure of an N-terminally truncated Δ90Dbp5L327V construct containing both RecA-like domains of Dbp5 (residues 91–482) bound to ADP and Gle1H337RIP6 (residues 244–538) by molecular replacement at a resolution of 4.0 Å (Rwork/Rfree of 21.4%/23.9%) (Fig. 1 and Supplementary Table 2). Structures were also obtained for Δ90Dbp5L327 or Δ90Dbp5WT in complex with RNA and ADP BeF3 at a resolution of 1.5 Å (Rwork/Rfree of 16.5%/18.9%) and 1.4 Å (Rwork/Rfree of 16.0%/18.0%), respectively (Supplementary Table 2).
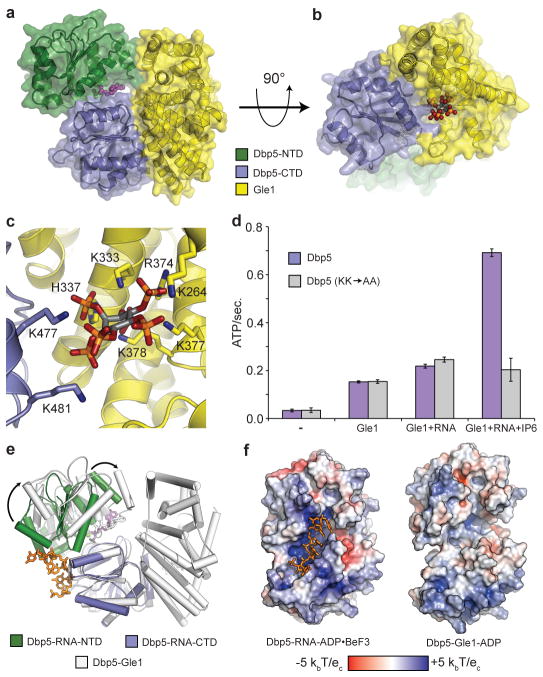
Figure 1. The Gle1IP6-Δ90Dbp5-ADP complex.
a, Structure of Dbp5 bound to ADP (magenta spheres) and Gle1 (see colour key). b, Side view showing IP6 (coloured spheres) bound at the interface between Gle1 and Dbp5. c, Detailed view of the IP6 binding interface (see colour key). IP6 is shown as grey sticks with orange phosphate and red oxygen atoms. Nitrogen atoms are in dark blue. d, IP6 binding is required for maximal Dbp5 ATPase stimulation with RNA. Error bars represent s.d. (n=3). e, Structural superposition of the RNA-Δ90Dbp5 complex with Gle1IP6-Δ90Dbp5. Arrows depict large rigid body movement in Dbp5. f, Van der Waals surface view of the RNA-Δ90Dbp5 complex and the Gle1IP6-Δ90Dbp5 complex coloured by solvent accessible electrostatics. Abbreviations: kb – Boltzmann’s constant (Joules/Kelvin), T – temperature (310 Kelvin), ec – charge of an electron (1.602 x 10−19 Coulombs).
The structures reveal that Gle1 comprises an all α-helical HEAT repeat protein that makes contact with both RecA-like domains of Dbp5 (Fig. 1a–b). The CTD interface buries 2230 Å2 of surface area, and contains a number of residues previously shown to be important for Gle1-mediated activation of Dbp512. The second interaction surface uses the N-terminal RecA-like domain (NTD) of Dbp5 and an exposed hydrophobic α-helix of Gle1, which buries an additional 930 Å2 of surface area (Fig. 1a). Interestingly, and despite the lack of obvious homology in sequence alignments, a search of the structural database using the DALI server13 showed that the Gle1-fold is similar to that of eIF4G (14% identity, 4.1Å root mean squared deviation), a cofactor for the SF2 DEAD-box ATPase eIF4A14–16, and to that of Upf2 (8% identity, 3.0 Å root mean squared deviation), which stimulates the SF1 protein Upf117–19.
A unique feature of the Gle1IP6-Dbp5 structures is the presence of IP6, which bridges the protein- protein interaction by binding in a positively charged pocket at the interface between Gle1 and the CTD of Dbp5 (Fig. 1b–c). Residues K264/K333/H337/R374/K377/K378 in Gle1 and K477/K481 in Dbp5 interact with phosphate groups in IP6 (Fig. 1c and Supplementary Fig. 1a– b). Two of these residues in Gle1 (K377 and K378) were recently identified as being involved in IP6 binding20. Gle1 residues (H337/R374/K377/K378) show charge conservation, and Dbp5 residue K477 is conserved as a hydrogen bond donor in most metazoans, suggesting an evolutionary pressure to maintain a polar residue that can interact with IP6 at these positions. (Supplementary Fig. 1c–d). To our knowledge, this represents the first structural characterization of an endogenous small molecule that localizes at the interface of two heterologous proteins to mediate a protein-protein binding interaction.
To assess the importance of IP6 binding by Dbp5, we mutated the Dbp5-IP6 contacts K477 and K481 to Ala (Dbp5KK→AA). Dbp5KK→AA exhibited no synergistic ATPase activation in the presence of IP6, RNA, and sub-saturating amounts of Gle1, but was still active as an ATPase and was stimulated by RNA and Gle1 comparable to wild-type protein in the absence of IP6 (Fig. 1d). Furthermore, Dbp5KK→AA (L327V variant) no longer formed a stable complex with Gle1H337R and IP6 (Supplementary Fig. 1e). These results demonstrate that IP6 stabilizes the Gle1-Dbp5 interaction by acting as a small-molecule tether.
A comparison of the RNA and ADP BeF3-bound Δ90Dbp5 structure with that of Gle1IP6-Dbp5 reveals that Gle1IP6 in the presence of ADP induces a large rigid body movement in Dbp5, opening the tandem RecA-like domains relative to their positions in the RNA-bound state (Fig. 1e and Supplementary Movie 1). Smaller conformational changes occur at the Gle1IP6-CTD interface, and structures obtained with wild-type vs. gain-of-function mutations show that these mutations subtly alter Gle1IP6-Dbp5 contacts, likely contributing additional binding energy to stabilize the complex (Supplementary Fig. 2). Interestingly, binding to Gle1IP6 dramatically alters the RNA binding site in Dbp5, which opens and loses its localized positive charge (Fig. 1f and Supplementary Movie 2). This suggests that Gle1IP6 binding leads to the formation of a partially open, catalytically-inactive, and ADP-bound complex that precludes RNA binding.
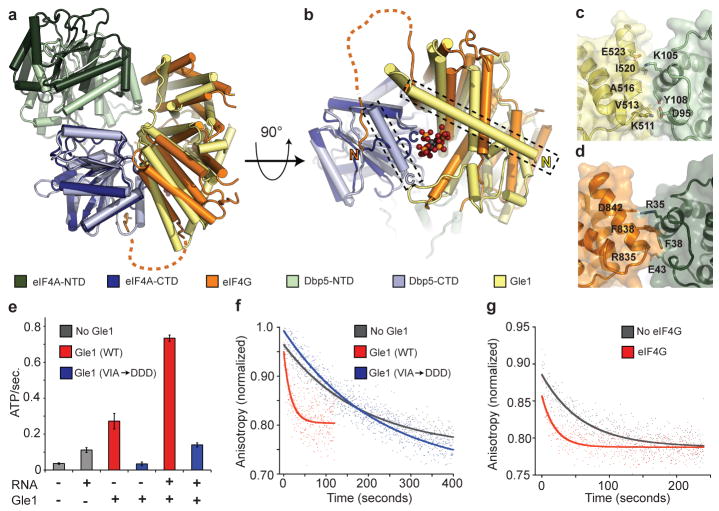
Due to the unexpected similarity between Gle1 and eIF4G, we also compared our structures to the eIF4A/4G complex. The open conformation of Dbp5 induced by Gle1IP6 is similar to that described for eIF4A in the presence of eIF4G14, 21, 22, and there is an overall similarity between the interactions of eIF4G and Gle1IP6 with the CTDs of their DEAD-box ATPase partners (Fig. 2a). However, eIF4G lacks an N-terminal helix that in Gle1 forms part of the binding pocket for IP6; instead, eIF4G forms a flexible loop that makes additional contacts with eIF4A, effectively compensating for the lack IP6 (Fig. 2b). The interface between eIF4G and the eIF4A-NTD also exhibits inter-domain differences when compared to Gle1IP6-Dbp5 (Fig. 2c–d), causing eIF4G to stabilize a more open eIF4A intermediate14, 21. Thus, the eIF4G-eIF4A complex appears more competent for ADP exchange than the Gle1IP6-Dbp5 complex. Despite these small differences, both Gle1IP6 and eIF4G stabilize their DEAD-box binding partner in an open state that is incompatible with RNA binding.
Figure 2. Comparison of Gle1IP6-Δ90Dbp5 and eIF4A-eIF4G.
a, Structural superposition of Gle1IP6-Δ90Dbp5 with eIF4G-eIF4A and AMP (PDB ID: 2VSO)21 (see colour key). b, View of the C-terminal RecA-like domain binding interface. Unique α-helices present in both Dbp5 and Gle1 form the IP6 biding pocket (boxed in figure). c, d, Residues involved in the formation of the N-terminal RecA-like domain binding interface in (c) Gle1IP6-Δ90Dbp5 and (d) eIF4A-eIF4G. e, Measured ATPase activity using wild type or mutant Gle1. Error bars represent s.d. (n=3). f, g, RNA release from Dbp5E240Q or eIF4AE172Q monitored by fluorescence polarization. Representative curves are shown.
Gle1IP6 is a potent activator of the overall ATPase activity of Dbp510, 11, but in our structures, Gle1 is not positioned to directly stimulate ATP hydrolysis (Fig. 1). We therefore hypothesized that Gle1IP6 activates Dbp5 by enhancing RNA release, a step known to be rate-limiting in the hydrolytic cycle of other DEAD-box ATPases23, 24. In such a model, product release would rely upon the ability of Gle1IP6 to induce the structural rearrangement of the two tandem RecA-like domains of Dbp5 (Fig. 1e–f). Thus, loss of contact between Gle1 and the NTD of Dbp5 should impair the ability of Gle1IP6 to activate Dbp5 without affecting the binding between these proteins, which is primarily mediated by the Gle1IP6-CTD interaction (Fig. 1a). To test this idea, residues V513, A516, and I520 on a solvent exposed helix of Gle1 (Supplementary Fig. 3a), were mutated (Gle1VAI→DDD) to disrupt the Gle1-NTD interface (Fig. 2c and Supplementary Fig. 3b). In an ATPase assay, Gle1VAI→DDD was unable to activate Dbp5 regardless of the presence of RNA and IP6 (Fig. 2e), but still formed a complex with Dbp5L327V by gel filtration (Supplementary Fig. 3c), demonstrating that these mutations did not disrupt the primary Gle1IP6-CTD association.
Next, we directly assessed the effect of Gle1IP6 on RNA release from a hydrolysis-deficient Dbp5 mutant, Dbp5E240Q, bound to both RNA and ATP (Fig. 2f and Supplementary Table 3). Dbp5E240Q released RNA very slowly with a t1/2 of 180 +/− 15 seconds (koff=0.0039 s−1) and Gle1IP6 stimulated this release ~9-fold (t1/2 of 21 +/− 6 seconds, koff=0.033 s−1). Importantly, no enhancement of RNA release was observed with the Gle1VAI→DDD mutant (166 +/− 50 seconds, koff=0.042 s−1). Since mutation of Gle1 at these residues in vivo resulted in lethality (Supplementary Fig. 3d) and Gle1IP6 did not affect the ability of Dbp5 to bind or release ADP (Supplementary Table 3), we conclude that an essential function of Gle1IP6 is to promote RNA release from Dbp5. This may explain how Gle1IP6 helps to terminate mRNA export in the cytoplasm.
Given the structural similarities between Gle1IP6-Dbp5 and eIF4G-eIF4A, we also tested eIF4G in the same RNA release assay and found that RNA release from eIF4A is similarly stimulated in the presence of its ATPase activator (eIF4AE172Q alone = 55 +/−8 seconds, koff = 0.019+/−0.03 s −1 compared to eIF4AE172Q + eIF4G = 11+/−3 seconds, koff = 0.091 +/− 0.03 s−1) (Fig. 2g). This behaviour corresponds well with the ~3X stimulation of ATPase activity reported for eIF4G21. Together with the structural similarity between Gle1, eIF4G, and other RNA helicase activators such as Upf2, these findings support a model in which Gle1/eIF4G-like proteins generally activate SF1 and SF2 ATPases by promoting RNA release and enzyme recycling.
Recent structural studies of the human Dbp5 ortholog, Ddx19, identified a N-terminal α-helix that lowers basal Dbp5/Ddx19 ATPase activity, but that is also required for RNA-stimulated ATP turnover9, 25. Our structures lack this N-terminal helix, but structural alignments show that Gle1IP6 and a C-terminal helix of Dbp5 would clash with this N-terminal domain in the RNA bound state (Supplementary Fig. 4a–b). This observation suggests that the autoinhibitory N-terminal helix must be displaced upon Gle1IP6 association, and should have little effect on ATPase activity in the presence of Gle1IP6. As seen for human Ddx1925, the yeast Δ90Dbp5 construct lacking this regulatory domain displays a basal ATPase rate ~3-fold higher than full- length Dbp5. However, Δ90Dbp5 only reaches the same maximum ATPase level as full-length Dbp5 in the presence of Gle1IP6 (Supplementary Fig. 4c). This result indicates that release of Dbp5 auto-regulation accounts for a portion of Gle1IP6-mediated ATPase stimulation.
Dbp5 is an ATP-dependent RNA binding protein that dissociates from RNA upon each hydrolysis event (Supplementary Fig. 4d), and an interaction between full-length Dbp5 and RNA can only be detected at steady-state when turnover is inhibited (Supplementary Table 3)10. Intriguingly, deletion of the N-terminal regulatory α-helix within Dbp5 allowed for the formation of a steady-state RNA-Dbp5 complex under conditions of hydrolysis (Supplementary Fig. 4e). These data imply that the N-terminal 90 residues of Dbp5 are involved in regulating Dbp5-RNA binding. Furthermore, the observation that Δ90Dbp5 binds RNA (Supplementary Fig. 4e), but is not stimulated for hydrolysis by RNA (Supplementary Fig. 4c), suggests that these 90 residues play a critical role in the coupling of RNA binding to ATPase activity. To test whether Gle1IP6 can alter steady-state RNA binding by displacing this α-helix, we added Gle1IP6 to RNA binding assays containing full length Dbp5 and ATP. We observed that addition of Gle1IP6 allowed for steady-state binding with an apparent affinity for RNA equal to that measured for Δ90Dbp5 (Supplementary Fig. 4e and Supplementary Table 3). This observation suggests that the N-terminal extension of Dbp5 regulates RNA association, and that Gle1IP6 binding can alter this activity. Overall, our results are consistent with a model in which Gle1IP6 regulates Dbp5 by: (1) altering the regulatory function of an N-terminal helix within Dbp5, and (2) promoting substrate release through separating the Dbp5 RecA-like domains. Together, these findings explain the robust activation of Dbp5 by Gle1 and the small molecule IP6.
In addition to Gle1, Dbp5 interacts with the nucleoporin Nup159 (Nup214 in vertebrates)3, 4, 8, 9,26. Nup159 and RNA bind to overlapping surfaces on Dbp5, and Nup159 inhibits RNA-dependent ATPase stimulation of Dbp5 in vitro9. To better understand how this potentially inhibitory role of Nup159 contributes to mRNA export, we solved the crystal structure of a Dbp5L327V-Gle1H337R-Nup159 complex bound to IP6 and ADP at a resolution of 2.9 Å (Rwork/Rfree of 22.9%/26.2%) (Fig. 3a and Supplementary Table 2). Within this structure, Nup159 interacts with Dbp5 in a manner similar to that of Ddx19 and Nup2149, occluding the RNA binding site, and sterically clashing with the closed, RNA- and ATP-bound state of Dbp5 (Supplementary Fig. 5). While the Gle1IP6-CTD interface remains relatively static in all of our structures, the Gle1-NTD interface displays distinct structural differences in the presence of Nup159 and now buries only 672 Å2 of solvent exposed surface area (compare Fig. 2c and 3b). Furthermore, the two RecA-like domains of Dbp5 become further separated in the Dbp5- Gle1IP6-Nup159 structure, and the Dbp5 catalytic centre becomes exposed to solvent as functionally critical arginine fingers are pulled out from the nucleotide binding site (Fig. 3c). A comparison of Dbp5 among all structures reveals that inter-domain structural changes proceed along a pair of related arcs, suggesting that Nup159 may further assist Gle1IP6 in stabilizing a post-hydrolysis state of Dbp5 to allow for efficient product release at the cytoplasmic face of the NPC (Supplementary Movie 3).
Figure 3. The Gle1IP6-Δ90Dbp5-Nup159 complex.
a, Two views of the Gle1IP6-Dbp5-Nup159 complex (see colour key). b, The N-terminal RecA-like domain binding interface between Dbp5- Gle1IP6 is altered in the presence of Nup159 (compare to Fig. 2c). Residues involved in the binding interface (sticks) are labelled. c, Superposition of the N-terminal RecA-domain among the three structural states of Dbp5. Arrow highlights the movement of the CTD and catalytic arginine finger residues (sticks) among the three states (see colour key). For clarity, the Gle1-ADP intermediate is shown slightly transparent, and both the arginine finger side chains and ADP have been removed. d, Inhibition of the RNA stimulated ATPase activity of Dbp5 by Nup159 is overcome in the presence of Gle1. Error bars represent s.d. (n=3).
To test this biochemically, we assayed Dbp5 ATPase activity and found that upon addition of Gle1IP6, Nup159 no longer inhibits ATP turnover (Fig. 3d) indicating that Gle1IP6 can overcome the inhibitory effect of Nup159. This suggests that during the hydrolysis cycle in the presence of Gle1, the association of Dbp5 with Nup159 is transient, which is in agreement with the observation that the presence of adenosine nucleotide and/or RNA weakens the Dbp5-Nup159 interaction3, 9. This provides for the possibility that Nup159 not only increases the local concentration of Dbp5 at the NPC4, 8, but also cooperates with Gle1 to promote mRNA export. Mechanistically, Gle1 and Nup159 may spatially coordinate the loading of Dbp5 onto an mRNP to target the removal of mRNA-bound export factors and contribute to the overall efficiency of mRNA export4, 8, 9. Alternatively, in a non-mutually exclusive model, the Dbp5- Gle1IP6-Nup159 structure may represent a post-mRNA export complex formed after mRNP remodelling is complete and Dbp5 is removed from the mRNP. In this model, the interactions of Gle1 and Nup159 with Dbp5 would be critical for enzyme recycling and/or preventing spurious re-binding of Dbp5 to an mRNA in the terminal stages of export.
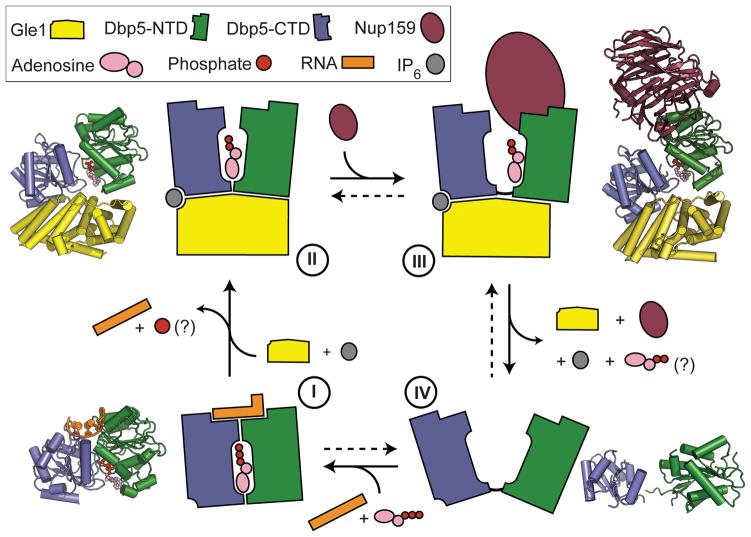
In conclusion, our study reveals a surprising structural similarity between Gle1 and other helicase regulators including eIF4G and Upf214, 19, 21. Together with our biochemical data, these data suggest a common mode of activation for DEAD-box ATPases and RNA helicases by Gle1/eIF4G-like protein activators. Critical for the activation are a primary stable interaction between the activator and the C-terminal RecA-like domain of the ATPase, and a secondary transient interaction between the activator and the N-terminal RecA-like domain that is required to trigger nucleic-acid release (Fig. 2). For Dbp5, we propose that RNA and Gle1IP6/Nup159 promote the formation of closed (ATP binding and hydrolysis) and open (substrate release) conformations of the ATPase, respectively, by influencing the position of the NTD of Dbp5 relative to the CTD (Fig. 4 and Supplementary Movie 3). Complex disassembly may be further promoted by the ~50-fold ATP-dependent lowering of Dbp5 binding affinity for Nup159 observed in the presence of ATP9, thus allowing Dbp5 to once again bind an mRNA and re-enter the ATPase cycle. For DEAD-box ATPases in general, such a toggling mechanism of the two RecA-like domains could be used to facilitate recycling of enzymes that have just melted RNA duplexes or remodelled RNA-protein complexes. Furthermore, in the presence of additional RNA-binding domains provided in cis or trans, this activity might also allow for a processive movement of this class of SF2 proteins along an RNA substrate27.
Figure 4. Model of the Dbp5 mechanochemical cycle.
In the presence ATP, Dbp5 binds RNA causing local destabilization and remodelling of duplexed RNA or RNA-protein complexes (state I). ATP hydrolysis then allows the activator (Gle1) to bind both the C-terminal and N-terminal RecA-like domains, separating the two RecA-like domains and promoting RNA release (state II). For Dbp5 it is currently not known when Pi is released following hydrolysis, but this will likely occur prior to the formation of state II. Subsequent release of the bound RNA allows Nup159 to bind Dbp5 causing the two RecA domains to further separate (state III). The formation of this state could then facilitate ADP release, prevent rebinding of the RNA, and aid in enzyme recycling (state IV). Crystal structure model in state IV is PDB ID:3FHO28. For colour coding details see key.
The unique role of IP6 as a small molecule tether within the Dbp5-Gle1 complex may allow for IP6-dependent fine-tuning of interaction strength. Functionally, this could be used to modulate RNA release/turnover and consequently regulate mRNA export under various growth conditions (e.g. in response to stress). Additional experiments will be needed to elucidate the functional importance of IP6 in vivo. However, the structural and biochemical framework presented here should guide future experiments not only to determine how Dbp5 in combination with its interaction partners confers directionality to mRNA export, but also to dissect the cellular functions of other RNA helicases and their activators.
Methods Summary
Yeast strains and plasmids are listed in Supplementary Table 4–5. RNA binding and ATPase assays were performed as described in Methods. ssRNA substrates used for binding and release assays carrying a single fluorescein conjugated UTP were made by in vitro transcription. RNA release assays were performed by monitoring the change in fluorescent polarization after rapid 1:1 mixing of DEAD-box protein-RNA binding reactions with excess polyA-RNA using a stopped flow apparatus linked to a Fluoromax-3 fluorimeter. For crystallography, protein purification was performed by affinity and ion exchange chromatography followed by affinity tag cleavage, and complex formation. Protein complexes were separated by gel filtration over a HiPrep Sephacryl S-200 column. Fractions containing the Dbp5-Gle1 or Nup159-Dbp5-Gle1 complex were used for crystal screening in a hanging drop format (200 nL) using a Mosquito robotics platform at 18°C. After 3–7 days of growth, crystals were looped and flash frozen in liquid nitrogen, and diffraction data collected on Beamline 8.3.1 at the Advanced Light Source. Detailed methods and references related to data processing, structure solution, and model building, plus all other methods used are outlined in the Methods section.
Methods
Protein purification for crystallography
Dbp5, Gle1 (a.a. 244–538), Nup159 (a.a. 2–387), eIF4A, and eIF4G (a.a. 572–952) from S. cerevisiae were expressed as previously described8, 10, 21 in BL21-CodonPlus (DE3)-RIPL cells (Strategene) using a combination of nickel affinity, ion exchange, and size exclusion chromatography. For the production of selenomethionine-labeled protein, Dbp5 and Gle1 proteins were purified as above, except that cells were grown in minimal media supplemented with selenomethionine and β-ME and DTT were replaced with TCEP throughout the purification. TEV cleavage was done at a 1:50 ratio (w/w) in 30 mM HEPES (pH 7.5), 400 mM NaCl, 1 mM DTT, 0.25 mM IP6, and 10% glycerol at 22°C for 4 hours29. Following removal of uncleaved protein, by orthogonal purification over a mixture of HIS-Select HF Nickel Affinity Gel (Sigma) and amylose resin (NEB), proteins were mixed in an equimolar ratio, diluted to 150 mM NaCl and concentrated to ~15 mg/mL at 4°C (Amicon Ultra; Millipore). The protein mixture was then further separated by gel filtration over a HiPrep Sephacryl S-200 column (GE Healthcare) at 0.4 ml/min in 10 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM DTT, 0.50 mM IP6, and 5% glycerol at 4°C. Fractions containing the desired protein complex, in an equimolar ratio (as judged by SDS-PAGE), were then pooled and concentrated to ~25 mg/mL at 4°C.
Protein crystallization
Protein solutions were stored at 4°C and used for crystallization within 1 week, during which time there was no detectable degradation of the proteins. All crystallization was performed in a hanging drop format using a Mosquito robotics platform (TTP LabTech) at 18°C.
The Dbp5-CTDL327V/IP6/Gle1H337R crystals were grown by mixing 100 nL of selenomethionine-labeled protein solution at 14 mg/mL in gel filtration buffer with 100 nL of a solution containing 30% PEG 3350, 100 mM HEPES pH 8.0, 50 mM NaOAc, and 200 mM LiSO4, and incubating for 5 days. Crystals were cryoprotected by dilution into a solution containing 24% PEG 3350, 80 mM HEPES 8.0, 160 mM LiSO4, 40 mM NaOAc, 20% glycerol, 0.25 mM IP6, 1 mM DTT and 100 mM NaCl, and flash frozen in liquid nitrogen.
The Dbp5-CTDL327V/IP6/Gle1WT crystals were grown using the same methods as for Dbp5-CTDL327V/IP6/Gle1H337R, but proteins were subjected to gel filtration individually prior to forming 1:1 complexes by direct mixing. Crystallization was performed by mixing 20 mg/mL of selenomethionine-labeled protein solution in gel filtration buffer (with NaCl increased to 150 mM) with a solution containing 30% PEG 3350, 100 mM HEPES 7.8, and 200 mM LiSO4 and incubating for 5 days. Crystals were cryoprotected by diluting into a solution containing 24% PEG 3350, 80 mM HEPES 7.8, 160 mM LiSO4, 10% glycerol, 0.4 mM IP6, 1 mM DTT and 120 mM NaCl, and flash frozen in liquid nitrogen.
The Δ90Dbp5L327V/RNA/ADP•BeF3 complex was formed by mixing a solution containing unlabeled Δ90Dbp5L327V in gel filtration buffer (with the addition of 5 mM MgCl2) with a 1.2:1 molar ratio of rU10 RNA (IDT), incubating for 15 minutes, and then mixing with ADP•BeF3 (prepared in a 1:3:15 ratio of ADP:Be:F) to a final concentration of 1 mM, followed by another 15 minutes of incubation. Crystals of the Δ90Dbp5L327V/RNA/ADP•BeF3 complex were grown by mixing 100 nL of the complex at a protein concentration of 15 mg/mL with 100 nL of a solution containing 200 mM MgNO3 and 20% PEG 3350 (Qiagen “The PEGs Suite” condition #64) and incubating for 7 days. Crystals were cryoprotected by diluting in a solution containing 18% PEG 3350, 9 mM HEPES pH 7.5, 180 mM MgNO3, 4.5% glycerol, 1 mM DTT, 90 mM NaCl, 0.5 mM IP6, 4.5 mM MgCl2 and 1 mM ADP BeFx (prepared in a 1:3:15 ratio of ADP:Be:F), and flash frozen in liquid nitrogen.
The Δ90Dbp5L327V/IP6/Gle1H337R /ADP complex was formed by mixing the unlabeled protein solution with ADP to a final concentration of 0.5 mM incubated for 15 minutes. Crystals were grown by mixing 200 nL of a solution containing 30% PEG 300 and 100 mM MES pH 6.5 with 50 nL of a 10% MPD additive solution, adding 250 nL of protein/ADP complex at 15 mg/mL protein in gel filtration buffer (with the addition of 10 mM MgCl2), and incubating for 9 days. Crystals were cryo-protected by diluting in a solution containing 27% PEG 3350, 90 mM MES pH 6.5, 9 mM HEPES pH 7.5, 5.4% MPD, 90 mM NaCl, 9 mM MgCl2, 0.9 mM DTT, 0.9 mM ADP, 0.45 mM IP6 and 4.5% glycerol and flash frozen in liquid nitrogen. The Δ90Dbp5/RNA/ADP•BeF3 crystals were grown in the same manner using wild type Δ90Dbp5.
The Nup159/Δ90 Dbp5L327V/IP6/Gle1H337R/ADP complex was formed by mixing the unlabeled protein solution with ADP to a final concentration of 1.0 mM, and incubating for 15 minutes. Crystals were grown by mixing 200 nL of a solution containing 20% PEG 3350 and 200 mM KOAc with 50 nL of a 100 mM sarcosine additive solution, adding 250 nL of protein/ADP complex at 15 mg/mL protein in gel filtration buffer (with the addition of 10 mM MgCl2), and incubating for 5 days. Crystals were cryo-protected by diluting in a solution containing 27% PEG 3350, 180 mM KOAc, 90 mM NaCl, 9 mM HEPES pH 7.5, 9 mM sarcosine, 9 mM MgCl2, 0.9 mM DTT, 0.9 mM ADP, 0.45 mM IP6 and 4.5% glycerol and flash frozen in liquid nitrogen.
Data collection, structure solution and refinement
All data were collected at beamline 8.3.1 at the Advanced Light Source at 100 Kelvin30, and processed using either HKL2000 or ELVES31, 32(see also below). Structures were solved with the AutoSol or AutoMR components of PHENIX33. Iterative rounds of manual model building in COOT34, coupled with refinement using PHENIX33 and stereochemical validation using MOLPROBITY35, resulted in final models displaying excellent geometry and Rwork/Rfree values for their respective resolutions, as well as no Ramachandran outliers. Rigid body refinement was conducted on Gle1 and Dbp5 RecA-like domains for all structures solved by molecular replacement in order to reveal any rigid-body differences among the structures. Simulated annealing omit maps were calculated to confirm the presence of all ligands (Supplementary Fig. 6). For all structures solved by molecular replacement, simulated annealing composite omit maps and prime and switch maps, both made using the AutoBuild component of PHENIX33, were calculated to help remove model bias.
Multiwavelength anomalous dispersion (MAD) data were collected at wavelengths of 0.9796, 0.9798, and 1.020 Å for the Dbp5-CTDL327V/IP6/Gle1H337R crystals in inverse beam mode and processed in HKL200031 (Supplementary Table 1). The structure was solved using PHENIX AutoSol, which found 11 selenium sites and gave figures of merit of 0.5 and 0.7 for phasing and density modification, respectively. An initial model was built using PHENIX AutoBuild33. For the final model, atomic coordinates, individual atomic displacement parameters (B-factors), translation/libration/screw (TLS) parameters, and f’ and f’’ parameters for the Se atoms were refined against the unmerged Friedel pairs (F+ and F−) of the remote dataset as implemented in PHENIX. Ramachandran statistics are 98.3% preferred, 1.7% allowed and 0% outliers as reported by Molprobity35.
A single low energy remote dataset was collected at a wavelength of 1.116 Å for the Dbp5-CTDL327V/IP6/Gle1WT crystals and processed in HKL2000 with merging of Friedel pairs. The structure was solved using PHENIX AutoMR by searching for a single copy of the fully refined Dbp5-CTDL327V/IP6/Gle1H337R structure33. For the final model, atomic coordinates, individual atomic displacement parameters (B-factors), and translation liberation screw (TLS) parameters were refined. The refinement test set was copied from the isomorphous Dbp5-CTDL327V/IP6/Gle1H337R data and extended to higher resolution in order to avoid biasing the Rfree. Ramachandran statistics are 98.0% preferred, 2.0% allowed and 0% outliers as reported by Molprobity35.
Native data were collected on the Δ90Dbp5L327V/RNA/ADP•BeF3 crystals at a wavelength of 1.116 Å and processed in HKL2000. The structure was solved using PHENIX AutoMR by searching for a single copy of a Saccharomyces cerevisiae Dbp5 homology model (SWISS-MODEL) 33, 36, created using the human Ddx19/RNA/AMPPNP structure as a template (PDB: 3G0H)25. For the final model, atomic coordinates, individual atomic displacement parameters (B-factors), TLS parameters, and occupancies for residues with alternate conformations were refined. Custom bond restraints were used for octahedral Mg2+-H2O clusters and the ADP BeF3 molecule. Ramachandran statistics are 99.2% preferred, 0.8% allowed and 0% outliers as reported by Molprobity35.
Native data were collected on the Δ90Dbp5WT/RNA/ADP•BeF3 crystals at a wavelength of 1.116 Å and processed in HKL2000. The structure was solved using PHENIX AutoMR by searching for a single copy of the fully refined Δ90Dbp5L327V/RNA/ADP•BeF3 structure and refined in the same manner33. Ramachandran statistics are 98.5% preferred, 1.5% allowed and 0% outliers as reported by Molprobity35.
Native data were collected on the Δ90 Dbp5L327V/IP6/Gle1H337R/ADP crystals at a wavelength of 1.116 Å and processed in Elves. The structure was solved using PHENIX AutoMR by searching for a single copy of the fully refined Gle1H337R/IP6/Dbp5-CL327V structure and a single copy of the Dbp5-NTD (residues 101–293) of the fully refinedΔ90Dbp5L327V/RNA/ADP•BeF3 structure33. B-factor sharpening was used throughout model building. Initial Fo–Fc difference density showed that two additional IP6 molecules were present in the structure. While the IP6 molecules have high relative B-factors, they mediate crystal-packing contacts and are coordinated by a large number of basic and polar residues. For the final structure, atomic coordinates and TLS parameters were refined. Secondary structure restraints as implemented in PHENIX were used throughout refinement in order to maintain proper backbone geometry. Ramachandran statistics are 97.6% preferred, 2.4% allowed and 0% outliers as reported by Molprobity35.
Native data were collected on the Nup159/Δ90 Dbp5L327V/IP6/Gle1H337R/ADP crystals at a wavelength of 1.116 Å and processed in HKL2000. The structure was solved using PHENIX AutoMR by searching for a single copy each of Gle1, the Dbp5-NTD, the Dbp5-CTD and Nup159 (PDB: 1XIP)8, 33. For the final model, atomic coordinates, and TLS parameters were refined. Ramachandran statistics are 97.3% preferred, 2.7% allowed and 0% outliers as reported by Molprobity35.
Structural analysis
All structural analysis and rendering for figures was performed with PyMOL (Schrödinger, LLC). Solvent accessible electrostatics in Figure 1 was calculated using ABPS37. Energy minimized linear interpolations shown in Supplementary Movies 1–3 were made using a CNS script written by the Yale Morph Server38, 39
Complex formation and gel filtration
To assay for Dbp5-Gle1 complex formation, 100 μL of a binding reaction was applied to a Superdex 200 gel filtration column (GE Healthcare) in the presence of 30 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM DTT, 0.25 mM IP6, and 5% glycerol at 0.4 mL/min. Binding reactions contained Dbp5 (20 μM) and Gle1 (40 μM) mixed with column buffer.
ATPase assays
ATPase assays using Dbp5 were performed as previously described10. When present, Nup159 was added to a final concentration of 1 μM.
RNA binding and release assay
The RNA substrate used for the binding and release assays was a 29 nt ssRNA of the sequence 5′-GGGUAAAAAAAAAAAAAAAAAAAAAAAAA-3′for Dbp5 or a 25 nt ssRNA of the sequence 5′-GGGUACAACGGAAGACAGCAGAGAA for eIF4A carrying a fluorescein conjugated UTP made by in vitro transcription (MEGAshortscript Kit; Ambion). RNA binding assays were performed as previously described10, 40. To perform RNA release assays, Dbp5E240Q (1.0 μM) or eIF4AE172Q (4.0 μM), FITC-RNA (40 nM), and ATP (2 mM) +/− Gle1 (2.0 μM) or eIF4G (8.0 μM) were complexed for 5 minutes in a buffer containing 50 mM HEPES (pH 7.5), 140 mM KCl, 5 mM MgCl2, 1 mM DTT, 20% glycerol, 0.1mg/mL BSA, and 10 μM IP6 for Dbp5 or 20 mM MES (pH 6.0), 20 mM KOAc, 5 mM MgCl2, 1 mM DTT, 20% glycerol, and 0.1mg/mL BSA for eIF4A. After rapid 1:1 mixing of DEAD-box-RNA binding reactions with polyA-RNA (1.0 mg/mL) using a stopped flow apparatus (RX2000; Applied Photophysics) linked to a Fluoromax-3 fluorimeter (Horiba Jobin Yvon), fluorescence intensities were measured every 0.05 to 0.2 seconds depending on the reaction. To measure fluorescent polarization, samples were excited with vertically polarized light (492 nM), and emission (521 nM) of vertically (IVV) and horizontally (IVH) polarized light was measured independently in two consecutive runs. Anisotropy (r) was then calculated using the equation: r = (IVV − (G × IVH))/(IVV + (2 × G × IVH)), where G = IHV/IHH. Reported t1/2 values are an average of three independent experiments +/− standard deviation. Release curves were fit to a first order exponential decay curve using KaleidaGraph (Synergy Software).
MANT-ADP binding and release assays
MANT-ADP labeled at the 2′- or 3′ position of the base (Invitrogen) was used for the binding and release assays with Dbp5E240Q. MANT-ADP binding assays were performed in a manner similar to the RNA binding assay10. Briefly, Dbp5E240Q (0–2 μM) and MANT-ADP (100 nM) were mixed with buffer (50 mM HEPES (pH 7.5), 140 mM KCl, 5 mM MgCl2, 1 mM DTT, 20% glycerol, 0.1mg/mL BSA, and 10 μM IP6) and RNA (1mg/mL) or Gle1 (2 μM) as indicated. Following mixing, anisotropy was measured using a Fluoromax-3 fluorimeter (Horiba Jobin Yvon). To perform the release assay, Dbp5E240Q (1 μM) was complexed with MANT-ADP (100nM) for 5 minutes +/− Gle1 (2 μM) and/or RNA (1mg/mL) in a buffer containing 50 mM HEPES (7.5), 140 mM KCl, 5 mM MgCl2, 1 mM DTT, 20% glycerol, 0.1mg/mL BSA, and 10 μM IP6. After rapid 1:1 mixing of Dbp5-MANT-ADP reactions with excess ADP (2 mM) using a stopped flow apparatus (RX2000; Applied Photophysics) linked to a Fluoromax-3 fluorimeter (Horiba Jobin Yvon), fluorescence intensities were measured every 0.01 seconds using 370 nM (excitation) and 445 nM (emission) light. Fluorescent polarization and t1/2 values were calculated as described above in the RNA release assays.
Supplementary Material
Acknowledgments
We thank Jack Kirsch, Jeremy Thorner, Matt Welch, David Wemmer, Bryan Krantz, Julie Zorn, and all Berger and Weis lab members for helpful discussions and advice. We also thank John Kuriyan, Matt Welch, and Susan Marqusee for access to equipment and workspace; Jane Tanamachi, George Meigs, and James Holton at ALS beamline 8.3.1; and Nat Echols for assistance with programs from the Yale Morph Server. Research was supported by the G Harold and Leila Y Mathers foundation (JMB) and the NIH (KW R01GM58065 and RC1GM91533).
Footnotes
Author contributions
BM, NDT, JMB, and KW designed the experiments. Protein purifications for crystallization and biochemical assays were performed by BM. Crystal screening, optimization, and data collection were performed by BM and NDT. Data processing, structure solution and model building was performed by NDT. In vivo tests and in vitro RNA binding, ATPase assays, and other biochemical assays performed by BM with assistance from MS and KH. BM, NDT, JMB, and KW both analyzed and interpreted the data, and wrote the manuscript.
Author information
Coordinates and structure factors for Dbp5-CTDL327V/IP6/Gle1H337R, Dbp5-CTDL327V/IP6/Gle1WT, Δ90Dbp5L327V/RNA/ADP BeF3, Δ90Dbp5/RNA/ADP BeF3, Δ90Dbp5L327V/IP6/Gle1H337R/ADP, and Δ90Dbp5L327V/IP6/Gle1H337R/ADP/Nup159 have been deposited in the protein data bank under accession numbers 3PEU, 3PEV, 3PEW, 3PEY, 3PEX and 3PEZ; respectively.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Main Text References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 9.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 10.Weirich CS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 11.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 12.Dossani ZY, Weirich CS, Erzberger JP, Berger JM, Weis K. Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proc Natl Acad Sci U S A. 2009;106:16251–16256. doi: 10.1073/pnas.0902251106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korneeva NL, First EA, Benoit CA, Rhoads RE. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J Biol Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 16.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 17.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 19.Kadlec J, Guilligay D, Ravelli RB, Cusack S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA. 2006;12:1817–1824. doi: 10.1261/rna.177606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcazar-Roman AR, Bolger TA, Wente SR. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J Biol Chem. 2010 doi: 10.1074/jbc.M109.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schutz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marintchev A, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henn A, et al. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci U S A. 2010;107:4046–4050. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins R, et al. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Napetschnig J, et al. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci U S A. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 28.Fan JS, et al. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol. 2009;388:1–10. doi: 10.1016/j.jmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr Purif. 2000;19:312–318. doi: 10.1006/prep.2000.1251. [DOI] [PubMed] [Google Scholar]
- 30.MacDowell AA, et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. In: Methods in Enzymology. Charles W, Carter J, editors. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci U S A. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 37.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flores S, et al. The Database of Macromolecular Motions: new features added at the decade mark. Nucleic Acids Res. 2006;34:D296–301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Lorsch JR, Herschlag D. The DEAD box protein eIF4A.1 A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.