Abstract
Regulation of chromatin structure involves histone post-translational modifications which can modulate intrinsic properties of the chromatin fiber to change the chromatin state. We used chemically defined nucleosome arrays to demonstrate that H2B ubiquitylation (uH2B), a modification associated with transcription, interferes with chromatin compaction and leads to an open and biochemically accessible fiber conformation. Importantly, these effects were specific for ubiquitin, as compaction of chromatin modified with a similar ubiquitin-sized protein, Hub1, was only weakly affected. Applying a fluorescence based method we found that uH2B acts through a mechanism distinct from H4 tail acetylation (acH4), a modification known to disrupt chromatin folding. Finally, incorporation of both uH2B and acH4 in nucleosomes resulted in synergistic inhibition of higher order chromatin structure formation, possibly a result of their distinct mode of action.
Tight control of chromatin structure is central to gene regulation, as chromatin occludes DNA access to protein effectors. Nucleosomes, representing the lowest level of chromatin organization, form arrays connected by stretches of linker DNA. At physiological ion concentration, nucleosome arrays compact into fibers with a helical architecture and a diameter of around 30 nm. In the following, we apply the term “chromatin fiber compaction” to refer to the formation of 30 nm fibers. Chromatin fibers are highly dynamic in nature 1-4 and can interact further to form larger superstructures, thereby allowing tight packing of the DNA 5. The fiber compaction process is driven by internucleosomal interactions. These include contacts between the N-terminal tail of H4 with an acidic patch on H2A 6-8 and direct interactions between the C-terminal helices of H2B 9.
Chromatin compaction can be regulated by histone post-translational modifications 10. One such modification is H4 K16 acetylation which interferes with fiber compaction 11,12 and which is associated with active chromatin 13. Likewise, ubiquitylation of H2B at its C-terminal helix (uH2B) 14 is associated with transcription, occurs at promoters and within gene coding regions 15-17 and global gene expression is altered upon disruption of the modification 18,19. Several non-exclusive models have been proposed for the function of uH2B, including a role in nucleosome stabilization 20, H2A/H2B dimer removal 21 or nucleosome reassembly 22 during transcription elongation. Further, it has been hypothesized that uH2B might locally “pry open” chromatin structure, thereby increasing nucleosome and DNA access to downstream factors 23,24. Indeed, inspection of a model of the chromatin fiber 9 suggests that uH2B is incompatible with nucleosome stacking (Fig. 1a). Moreover, deubiquitylation of uH2B was found to be required for heterochromatin spreading and DNA methylation 25. Nonetheless, a detailed analysis of the specific effect of H2B ubiquitylation on chromatin structure has not been reported and so any inhibitory role of the modification on chromatin compaction remains speculative. In this study, we exploited our ability to chemically synthesize uH2B in order to investigate the conformation and biochemical accessibility of unmodified and ubiquitylated chromatin fibers in solution. Using these reagents in conjunction with a novel fluorescence anisotropy based assay, we found that uH2B interferes with chromatin fiber compaction and inter-fiber interactions. Moreover, additional structure-function studies revealed that this is a specific property of ubiquitin rather than simply the addition of steric bulk.
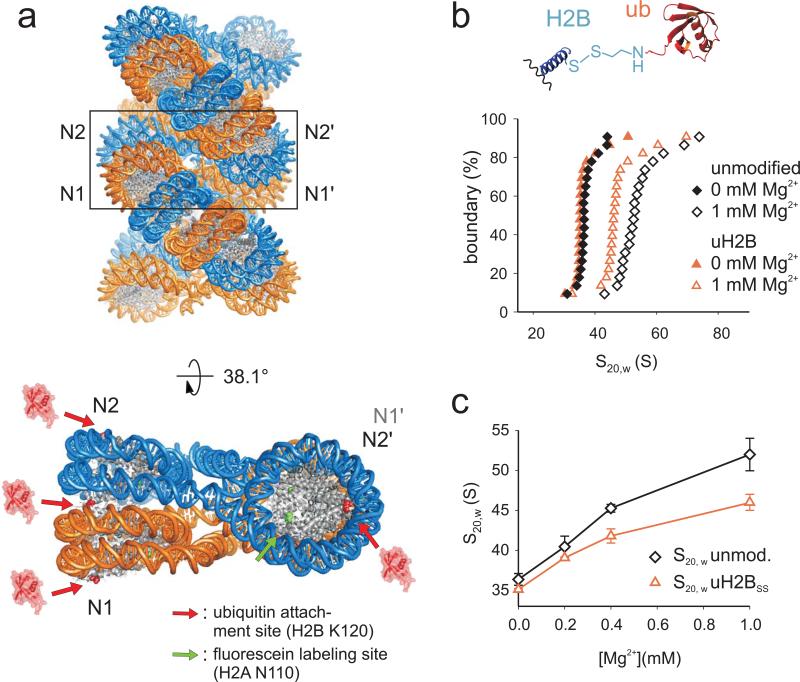
Figure 1. H2B ubiquitylation impairs fiber folding.
a, Ubiquitylated H2B appears incompatible with nucleosome stacking. Upper panel: Model of a 30 nm chromatin fiber (1ZBB, ref. 9). Lower panel: The structure of a tetranucleosome unit extracted from the fiber and rotated by 38.1° 9. The sites of ubiquitin attachment are shown in red, whereas the fluorescein labeling site is shown in green. b, Sedimentation coefficient distributions for unmodified and uH2BSS containing chromatin arrays (black and red symbols) are determined by sedimentation velocity experiments and van Holde-Weischet analysis at 0 and 1 mM Mg2+ (solid and open symbols). Inset: ubiquitin is attached via disulfide based coupling chemistry 26. c, S20,w values of unmodified (black) and uH2BSS containing arrays (red) are shown as a function of Mg2+ concentration. Error bars, standard deviation (n=3).
Results
H2B ubiquitylation impairs chromatin fiber compaction
We set out to characterize the effect of uH2B on chromatin fiber conformation in a chemically defined model system. We recently developed a disulfide-directed methodology for the site-specific modification of H2B by ubiquitin (uH2BSS, Fig. 1b, inset and Supplementary Fig. 1) 26. This structural analogue of uH2B is readily accessible and was designed such that the ubiquitin moiety can be selectively removed under mild reducing conditions. We generated nucleosome arrays with and without uH2B using a DNA template containing 12 copies of the 177 base pair “601” nucleosome positioning sequence 7,27 (Supplementary Fig. 2). Restriction enzyme digests of the arrays demonstrated full nucleosome occupancy, and partial digestion with micrococcal nuclease confirmed 12 positioned nucleosomes (Supplementary Fig. 2).
To determine differences in compaction, we performed sedimentation velocity experiments with unmodified and uH2BSS containing arrays over a range of Mg2+ concentrations from 0 to 1 mM (Fig. 1b, c). The latter condition induces formation of maximally compacted fibers similar to condensed chromatin in vivo, even in the absence of linker histone H1 5. In solutions devoid of Mg2+, unmodified and uH2BSS arrays sedimented at 36 ± 0.8 S and 35 ± 0.3 S, respectively. These values are compatible with an extended beads-on-a-string conformation 7,11. At 1 mM Mg2+ the unmodified arrays were fully compacted at 52 ± 2 S, whereas the uH2BSS containing arrays only reached a sedimentation coefficient of 46 ± 1 S. The latter value is indicative of a significant reduction in array compaction (p=0.03, Student's two tailed t-test, n=3), comparable to the effect of H4 tail acetylation 11 or H4 tail removal 7.
To ascertain whether the ubiquitylated fibers also exhibit increased biochemical accessibility, we investigated the activity of the methyltransferase Disruptor of telomeric silencing-1 (hDot1L) toward unmodified and ubiquitylated arrays. In humans, the hDot1L enzyme is responsible for methylation of H3 K79 on the nucleosomal surface, and is directly stimulated by uH2B 28. In unmodified arrays, hDot1L activity was reduced by about 40% in comparison with mono-nucleosomal substrates due to array compaction (Fig. 2a-c, lanes 1, 2). Conversely, the methyltransferase activity toward ubiquitylated arrays and mono-nucleosomes was found to be similar (Fig. 2a-c, lanes 3, 4). Thus, uH2B mediated array decompaction allows unhindered access of hDot1L to the nucleosomal surface and H3 K79.
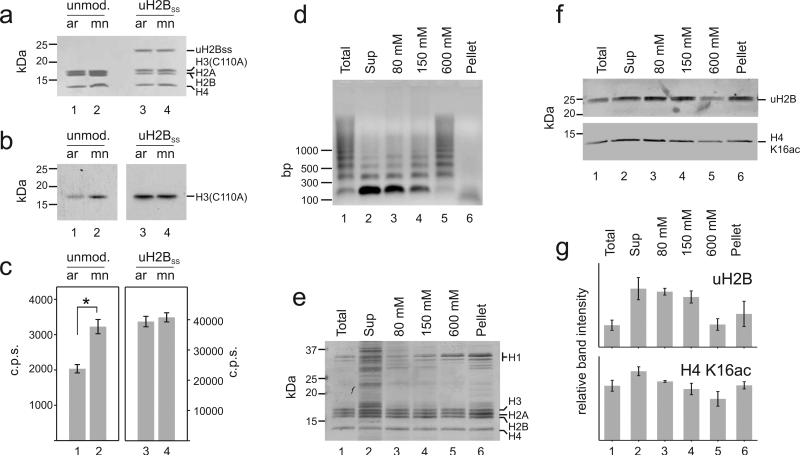
Figure 2. H2B ubiquitylation increases chromatin accessibility in vitro and in vivo.
a-c, Unmodified arrays (ar) (lane 1) and mono-nucleosomes (mn) (lane 2), or H2B ubiquitylated arrays (lane 3) and mono-nucleosomes (lane 4) were used as substrates for hDot1L methyltransferase assays with 3H-SAM at 1 mM Mg2+. Histones were separated by SDS-PAGE, (a) stained with Coomassie Brilliant Blue and (b) probed for 3H-methyl incorporation by fluorography. c, Quantification of methylation was performed by p81 filter binding followed by liquid scintillation counting. Student's two tailed t-test: *: p = 0.0001; Error bars, SEM (n=9). The fluorography represents two different exposures of the film: 24 h, left panel and 3 h, right panel. For verification of the modification site and full gels, see Supplementary Fig. 3. d-g Chromatin from micrococcal nuclease digested nuclei of NIH/3T3 fibroblasts was successively extracted with increasing concentrations of NaCl. d, DNA purified from MNase digested nuclei (lane 1), from the supernatant after centrifugation of the nuclei (lane 2), from 80, 150 and 600 mM salt extracted chromatin (lanes 3, 4, 5) and from the insoluble pellet (lane 6) was separated on an agarose gel and stained with Ethidium bromide. Histones were acid extracted and equal amounts were separated by SDS-PAGE and either (e) stained with Coomassie Brilliant Blue or (f) analyzed by Western blotting with antibodies against uH2B and H4 K16ac. For full gels see Supplementary Fig. 3. g, uH2B and H4 K16ac levels from two independent experiments were quantified by densitometry. Error bars, SEM (n=2).
Increased accessibility of chromatin due to H2B ubiquitylation can also be probed in vivo by measuring sensitivity to nucleases. Therefore, we partially digested chromatin of NIH/3T3 fibroblasts with micrococcal nuclease and successively extracted chromatin fragments using increasing salt concentration up to 600 mM NaCl (Fig. 2d, e). Highly digested chromatin, which can be extracted with 80 - 150 mM NaCl, has been shown to represent predominantly active chromatin by sequencing methods 29. uH2B was found to be enriched in linker histone depleted micrococcal nuclease sensitive regions (Fig. 2f, g, lanes 2-4 vs. lane 5), indicating its positioning in open and active chromatin. A similar trend was found for H4 K16 acetylation. This correlation, and others like it 30,31, further corroborates our in vitro findings about a function of uH2B in modulating chromatin structure.
Chromatin compaction monitored by fluorescence anisotropy
Having established that H2B ubiquitylation results in chromatin decompaction to a similar degree as H4 tail acetylation, we wondered if these two very different post-translational modifications may function through distinct mechanisms. Fluorescence homo resonance energy transfer (homo-RET) has been widely applied to study protein structure, oligomerization and interactions in vitro and in vivo 32-35. Inspired by this work, we developed a homo-RET based method to gain higher resolution information regarding the conformation of chromatin fibers. Fluorescein chromophores attached to the nucleosomal surface undergo homo-RET with partners upon compaction of the fiber (Fig. 3a). When excited with polarized light this results in a decrease in emission steady state anisotropy (SSA) as a function of internucleosomal distance 36. Importantly, homo-RET yields complementary information to sedimentation experiments, as it reports directly on internucleosomal distances, whereas the latter method measures the overall shape of the fiber. Therefore, the combined information from both methods can be used to gain insight about the ensemble of fiber conformations at different Mg2+ concentrations.
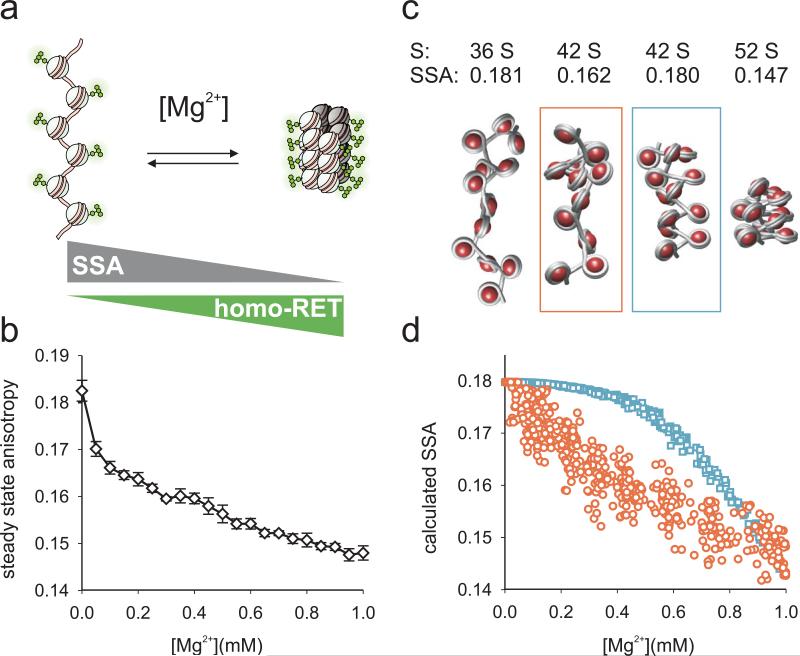
Figure 3. A fluorescence method to monitor chromatin fiber folding reveals conformational heterogeneity at intermediate chromatin fiber compaction.
a, Compaction upon addition of divalent cations results in a loss of fluorescence steady state anisotropy (SSA) due to internucleosomal homo-RET between fluorescein moieties. b, Experimental SSA data obtained from fluorescently labeled 12-mer nucleosomal arrays as a function of Mg2+ concentration. Error bars, standard deviation (n=3). c, Representative structures of arrays in different compaction states. S20,w and SSA values are calculated from the structures as described in the supplementary information. The two middle structures share the same sedimentation constant, whereas the conformationally heterogenous chain configuration (red) leads to more RET than the conformationally homogenous chain (blue). d, Calculated SSA values from 500 randomly generated array conformations from either heterogenous (red) or homogenous chains (blue).
Based on the structure of a tetranucleosome 9 and preliminary experimental evaluation of several candidate sites, we chose position 110 in histone H2A as the fluorophore attachment point (Fig. 1a). Recombinant H2A bearing the required N110C mutation was labeled with fluorescein maleimide and incorporated into nucleosome arrays (Supplementary Figs. 1, 2). First, we validated the method using otherwise unmodified arrays. Indeed, as the Mg2+ concentration was increased from 0 to 1 mM we observed a decrease in SSA (Fig. 3b). The magnitude of the change in SSA upon array compaction correlated with the extent of labeling (Supplementary Fig. 4). Moreover, we observed no change in SSA for mono-nucleosomes in a similar experiment (Supplementary Fig. 4). Importantly, due to the inability of the fluorescein probes to self-quench through physical contact, absolute emission intensity remained nearly constant throughout the experiment and we determined no change in the fluorescence lifetime of the system as a function of Mg2+ concentration (Supplementary Fig. 5). We however observed self-quenching at other labeling sites in our initial evaluation. Collectively, these observations are consistent with homo-RET being the dominant contributor to the decrease of SSA upon array compaction.
Chromatin compacts through heterogenous intermediates
The shape of the observed SSA trace warrants further discussion. To gain information about the conformational distribution of unmodified chromatin chains giving rise to the observed signal and to reconcile fluorescence and sedimentation data, we applied computational modeling. We implemented a two-angle chromatin model 37 to calculate sedimentation coefficients. To describe energy migration between fluorophores and to calculate SSA values for given chromatin configurations, we applied a cluster theory developed by Knox 38, and Runnels and Scarlata 32. We calculated transitions by generating ensembles of fiber structures and tested two compaction models: i) a homogenous model where chain parameters were gradually changed from decompacted to compacted values and ii) a heterogenous model where local inter-nucleosomal contacts were randomly introduced and extended along the chains (Fig. 3c, Supplementary Fig. 6 and supplementary discussion). Comparing experimental and calculated traces, we could reproduce the measured SSA data by the heterogenous model (Fig. 3d), where a significant number of fluorophores are within range for RET even at low overall compaction (Supplementary Fig. 6). In contrast, homogenous fiber compaction led to a low number of fluorophores within RET distance and thus unrealistically low levels of RET and high SSA. We conclude that at intermediate compaction, chromatin fibers are heterogenous, containing both clusters of inter-nucleosomal contacts and extended regions. The initial decay of the experimental SSA trace therefore monitors the formation of local interactions, although the arrays likely retain substantial conformational freedom under these conditions. At higher Mg2+ concentrations a further decrease in SSA can then be attributed to the establishment of increasingly compact, regular fibers.
uH2B and acH4 act through distinct mechanisms
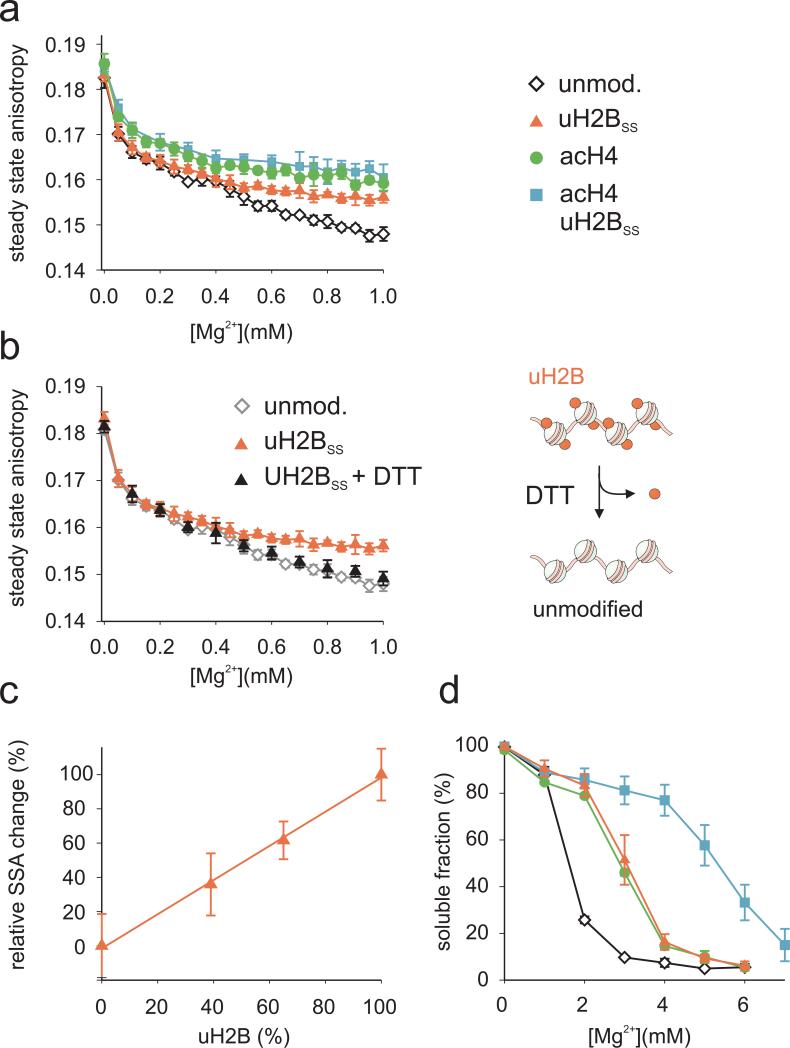
We next utilized our SSA based approach to study the effect of ubiquitylation and acetylation on the different stages of chromatin fiber compaction. We used a traceless expressed protein ligation strategy 28 to produce hyperacetylated H4 (acH4) bearing the acetyl mark on the N-terminus as well as on lysines 5, 8, 12, 16 and 20 (Supplementary Fig. 1). We reconstituted arrays containing either uH2BSS, acH4 or both modifications and assayed array quality by restriction enzyme and micrococcal nuclease digests (Supplementary Fig. 2). As expected, acetylation of the H4 tail impaired array compaction, manifested by higher SSA values compared to unmodified chromatin (Fig. 4a). Ubiquitylation also led to a pronounced increase in SSA compared to unmodified arrays. However, in this case the SSA measurements only diverged at Mg2+ concentrations above ~0.4 mM (Fig. 4a). Nonetheless, at 1 mM Mg2+ the ubiquitylated arrays exhibited SSA values similar to the acH4 containing arrays. The different shape of the SSA traces suggests that the two modifications inhibit array compaction through distinct mechanisms. To establish that ubiquitylation alone is responsible for the observed modulation of array compaction, we took advantage of the reversible disulfide linkage between ubiquitin and H2B. Reduction with dithiothreitol (DTT) allowed complete chemical deubiquitylation of reconstituted arrays (Supplementary Fig. 7) resulting again in full compaction (Fig. 4b). Further, it is of interest to determine the extent of H2B ubiquitylation required for measurable chromatin decompaction. We therefore assembled arrays where ubiquitylated H2B was titrated in at different levels, and performed compaction assays (Supplementary Fig. 8). The SSA transition endpoints at 1 mM Mg2+ increase monotonously with increasing uH2BSS content (Fig. 4c). Thus, local concentration of uH2B, for example on an active gene, can result in a substantial change in chromatin conformation. Finally, the simultaneously ubiquitylated and acetylated array exhibited a compaction behavior that was indistinguishable from the acH4 array (Fig 4a) and removal of ubiquitin with DTT had no further effect (Supplementary Fig. 7). This suggests that the effect of H4 acetylation on chromatin fiber compaction dominates over that of H2B ubiquitylation and that the effects are not additive.
Figure 4. H2B ubiquitylation and acH4 have distinct effects on fiber folding and higher order formation.
a, Chromatin folding as a function of Mg2+ concentration was assessed by measuring SSA for unmodified (black), H2B ubiquitylated (red), H4 acetylated (green) and uH2BSS/acH4 containing (blue) nucleosomal arrays. b, Refolded arrays containing uH2BSS were chemically deubiquitylated by reduction of the disulfide bond with dithiothreitol (DTT). Compaction was then determined at various Mg2+ concentrations by measuring SSA (black). For comparison, the SSA traces of unmodified and ubiquitylated arrays are shown in grey and red, respectively. Error bars, standard deviation (n=2-4). c, Chromatin folding upon Mg2+ addition was determined by measuring SSA for arrays containing increasing amounts of uH2BSS. The relative SSA change at 1 mM Mg2+ is shown as a function of uH2BSS content. The solid line is a linear fit to the data. Error bars, standard deviation (n=4-6). d, Arrays containing unmodified (black), H2B ubiquitylated (red), H4 acetylated (green) and doubly acetylated/ubiquitylated (blue) nucleosomes were incubated in the presence of indicated concentrations of Mg2+ and oligomers were removed by centrifugation. The amount of arrays remaining in solution was determined by UV absorption. Errors bars, SEM (n=2-4).
uH2B and acH4 synergistically impair inter-fiber interactions
In interphase chromosomes, extensive inter-strand interactions in combination with structural proteins stabilize higher order chromatin structure. Interdigitation of nucleosomes from different fibers has been proposed as a possible mechanism for these inter-fiber interactions 39. The intrinsic self-oligomerization of chromatin fibers in vitro mimics long-range intraand inter-fiber interactions 5. We therefore investigated the effects of ubiquitylation and acetylation on fiber oligomerization at Mg2+ concentrations beyond ~1.5 mM. Consistent with previous studies 11, we found that the midpoint for array association is shifted to around ~3 mM Mg2+ upon acetylation of H4 (Fig. 4d). Introduction of the ubiquitin moiety had a similar repressive effect on array oligomerization. Strikingly, the occurrence of both chromatin marks on the same array profoundly reduced inter-fiber interactions, reflected in a midpoint for the transition at 5.5 mM Mg2+ (Fig. 4d). Thus, H2B ubiquitylation acts synergistically with H4 tail acetylation within the same array to impede fiber oligomerization, further underlining different modes of action for these two histone modifications. The observed cooperativity may have important implications for the maintenance of open chromatin around actively transcribed genes, since both modifications are localized in such regions (Fig. 2f,g and ref. 15,16,40,41).
Fiber disruption is a specific property of ubiquitin
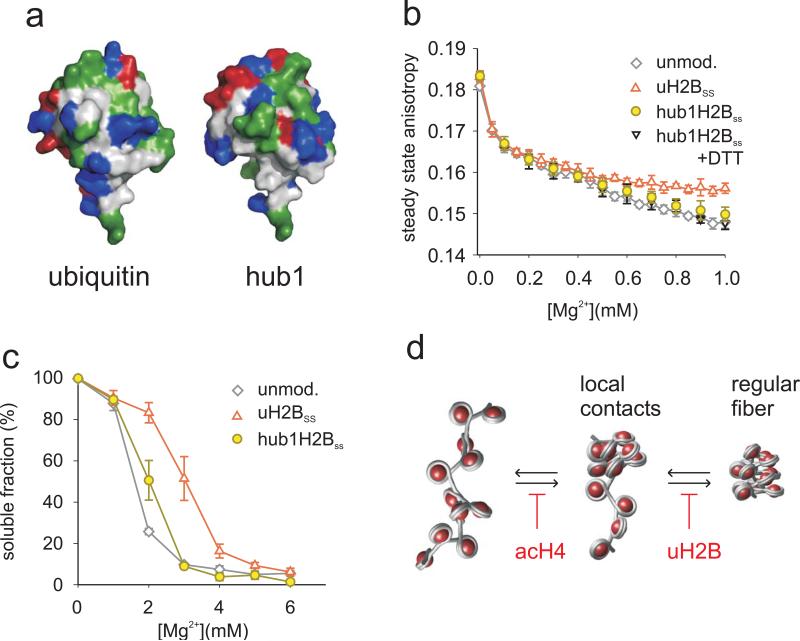
Finally, we investigated whether the effect of uH2B on chromatin structure is dependent on specific features of the ubiquitin protein or is simply a consequence of added steric bulk. Hub1 is a ubiquitin-like protein in yeast 42 which shares 23% sequence identity with ubiquitin. Hub1 and ubiquitin share the same fold but have markedly different surface residues (Fig. 5a). We thus linked Hub1, which had been extended by the ubiquitin specific C-terminal RGG sequence, to H2B at position 120 (hub1-H2BSS) applying the disulfide coupling strategy (Supplementary Figs. 1, 2). Importantly, chromatin fiber compaction was only weakly affected by hub1-H2BSS incorporation (Fig. 5b and Supplementary Fig. 9). Moreover, inter-fiber interactions were largely unperturbed by the Hub1 modification in otherwise unmodified arrays (Fig. 5c). Interestingly, in H4 acetylated arrays, we observed some cooperativity between Hub1 and acH4 in impairing inter-fiber interactions, albeit to a lesser degree than with uH2B (Supplementary Fig. 9). In accordance with these results, hDot1L methyltransferase activity was reduced upon array compaction at 1 mM Mg2+, demonstrating reduced substrate accessibility (Supplementary Fig. 10). These observations point toward a specific structural effect of uH2B in chromatin that depends on features unique to the ubiquitin moiety.
Figure 5. Fiber decompaction is a specific property of the ubiquitin protein.
a, Surface rendering of the X-ray structure of ubiquitin (1UBQ, left) 49 and the NMR structure of Hub1 (1M94, right, including the C-terminal RGG residues) 50. The color code shows hydrophobic (grey), hydrophilic (green), positively charged (blue) and negatively charged residues (red). b, Compaction behavior of arrays containing unmodified (grey), H2B ubiquitylated (red) and hub1-H2BSS containing arrays in the absence (yellow) or presence (black) of DTT was determined by SSA. Error bars, standard deviation (n=2-4). c, Oligomerization of arrays containing unmodified (grey), H2B ubiquitylated (red) and hub1-H2BSS containing arrays (yellow) was assessed by determining Mg2+ dependent solubility. Errors bars, SEM (n=3-4). d, A model of the distinct effects of uH2B and acH4 on fiber folding (for details see text).
Discussion
Higher order chromatin structure is involved in gene regulation by restricting access to the DNA and interfering with processes requiring mobilization of nucleosomes, such as transcription. Compacted chromatin is highly dynamic on biological timescales and its structure can be modulated by histone post-translational modifications – either by altering its intrinsic properties or by generating anchoring points for further structural proteins 43. To date, intrinsic structural effects have been demonstrated for acetylation of K16 in the H4 tail, leading to fiber decompaction 11, and for tri-methylation of K20 in H4 resulting in increased folding 44. Ubiquitylation of H2B has long been proposed to result in fiber decompaction, due to its size and positioning on the nucleosomal surface 23,24. However, a detailed study of the different effects of uH2B on chromatin structure, dynamics and function has been difficult, as its availability from natural sources is low. Using a defined model system and chemically modified histones we were able to demonstrate that uH2B inhibits both nucleosome array folding, as well as inter-fiber oligomerization, the latter in a cooperative fashion with H4 acetylation on the same array. Consistent with this, nucleosome arrays containing uH2B possess a more biochemically accessible fiber conformation, as demonstrated by our studies using hDot1L. We note, however, that these experiments do not rule out the possibility that binding of hDot1L to ubiquitylated arrays could have an additional effect on local fiber structure.
Several methods have been applied to investigate chromatin higher-order structure formation, including electron microscopy, sedimentation velocity measurements and, most recently, single molecule force spectroscopy 1,3. These methods are powerful, but either require sample fixation, large sample amounts or are technically challenging. Herein, we present a complementary method based on homo-RET between nucleosomes, which directly reports on inter-nucleosomal distance changes in equilibrium. Applying this method, we were able to show that divalent cation induced chromatin fiber compaction involves conformationally heterogenous intermediates, which are differentially affected by uH2B and acH4.
Based on our data we propose a model for fiber decompaction by H2B ubiquitylation, distinct from the effect of H4 tail acetylation. acH4 affects compaction throughout the folding transition, presumably by weakening H4 tail binding to the H2A acidic patch 12. This results in a reduction of closely interacting nucleosomes at a given Mg2+ concentration (Fig. 5d), and prevents full fiber folding due to counteracting electrostatic repulsion and thermal fluctuations. In contrast, our measurements show that uH2B only interferes with the later stages of compaction. At low ionic strength, transient interactions between nucleosomes in ubiquitylated arrays are not impaired, as reflected by sedimentation coefficients and SSA values comparable to unmodified fibers. This can be attributed to sufficient conformational freedom in these local contacts to accommodate the ubiquitin moiety. However, upon further compaction regular fiber packing is impaired and defects in nucleosome stacking may lead to fiber instability and local unfolding (Fig. 5d). The two different modes of action of uH2B and acH4 are further reflected in the cooperativity of these modifications in preventing inter-strand interactions. Finally, we found that the similar sized protein Hub1 could not substitute for ubiquitin in impairing chromatin folding. We therefore hypothesize that specific interactions between ubiquitin and the nucleosomal surface around its anchoring point may be required to prevent escape of ubiquitin from the interface between nucleosomes during compaction. Consistent with this idea, ubiquitylation of H2A, a modification associated with heterochromatin and situated at the opposite side of the nucleosomal surface, does not appear to hinder fiber compaction 45. It remains to be seen exactly how ubiquitin, when properly localized on the nucleosome, impedes chromatin fiber compaction. We can envision several models for this effect (see Supplementary Fig. S11) including ubiquitin imposing steric hindrance. Further experiments are required to discriminate between these possible mechanisms. In summary, our results establish a novel function for uH2B in disrupting local chromatin structure and add to the understanding of how combinations of histone post-translational modifications allow fine-tuning of local chromatin fiber compaction as well as higher order structure. By extension, we propose that the increased local accessibility of ubiquitylated nucleosomes might also facilitate chaperone mediated nucleosome dis- and reassembly during transcription 21,22, in agreement with in vivo observations on the function of this modification.
Methods
Cell assays, preparation of modified proteins and DNA and computational modeling are described in the Supplementary Methods.
Array preparation
Fluorescein labeled H2A was obtained by reacting recombinant H2A(N110C) with fluorescein maleimide. uH2BSS and hub1-H2BSS were produced according to literature protocols 26. Acetylated H4 was generated by expressed protein ligation of a synthetic acetylated peptide N-fragment and a recombinant C-fragment. The purified ligation product was desulfurized to yield the native acH4 protein. Histone octamers were assembled as previously described 26 and reconstituted into nucleosome arrays by step-wise dialysis using 12-177-601 DNA.
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed as described 7, with minor modifications. Samples were prepared in measurement buffer (10 mM Tris, pH 7.8, 10 mM KCl) containing 0.1 mM EDTA for 0 mM Mg2+ or measurement buffer containing various concentrations of Mg2+. The initial absorbance at 260 nm was 0.8 at a sample volume of 400 μL. Samples were equilibrated under vacuum for 1 h at 20° C in the centrifuge (Beckman XL-1 analytical ultracentrifuge with absorbance optics (Beckman Coulter Inc., Brea, CA)) prior to sedimentation at 12,000 rpm in 12 mm double sector cells and an An-60 Ti rotor (8-place). Absorbance was measured at 260 nm in continuous scan mode. The data was analyzed using UltraScan 9.9 rev 863 46. Boundary fractions were calculated by enhanced van Holde & Weischet analysis 47. S20,w values were calculated by adjusting for a partial specific volume of 0.622 mL/g for chromatin and buffer viscosity. Mean S20,w values were obtained by averaging over the dominant homogenous fraction. To be able to directly compare S20,w values of ubiquitylated and unmodified arrays, the averaged sedimentation coefficients were corrected for the different molecular weights of the arrays as described previously 7.
hDot1L methyltransferase assays
Full-length hDot1L was purified from a baculovirus expression system as previously described 28,48. In a typical methyl-transferase assay, 12 pmol of nucleosomes in arrays or mono-nucleosomes were equilibrated in 35 μL of assay buffer (20 mM Tris, pH 7.9, 1 mM MgCl2, 50mM KCl). After reaction initiation by addition of 0.3 pmol hDot1 and [3H]-S-adenosyl methionine (5.6 μCi, 560 pmol) the reaction was incubated at 30° C for 1 h. Subsequently, 30 μL of each sample was mixed with SDS loading buffer. The histones were separated by SDS-PAGE followed by visualization by Coomassie Brilliant Blue staining. Furthermore, the gel was incubated in Amplify solution for 15 min followed by drying and visualization by fluorography. The residual 5 μL of the reaction mixture were mixed with 5 μL of 20 mM EDTA solution, spotted on Whatman p81 filter paper and washed three times with NaHCO3 solution (pH 9) followed by air-drying. Ready Safe Liquid Scintillation Cocktail (Beckman Coulter, Brea, CA) was added and the samples were counted with a LKB Wallac 1209 RackBeta Primo Liquid scintillation counter.
Fluorescence Spectroscopy
Fluorescence measurements were performed in measurement buffer (10 mM Tris, pH 7.8, 10 mM KCl) containing appropriate amounts of MgCl2. For measurements in the absence of Mg2+, the measurement buffer contained 0.1 mM EDTA. All buffers were degassed prior to measurement by purging with Ar. Nucleosomal array or mono-nucleosome concentrations were 50 nM (per nucleosome) for all measurements. Samples containing different Mg2+ concentrations were prepared fresh by mixing an equivalent volume of array or mono-nucleosome stock (100 nM per nucleosome) with measurement buffer containing twice the final amount of MgCl2, followed by a 15 min equilibration step at 22.5°C prior to measurement. Measurements were performed on a Fluorolog-3 instrument (HORIBA Jobin Yvon, Edison NJ) equipped with automated dual polarizers and using a Sub-Micro Fluorometer cell (Starna Cells, Atascadero, CA) with 10 mm path length. The excitation wavelength was 480 nm with a band width of 5 nm and emission was recorded at 520 nM with 5 nm band width. 6 – 8 measurements were taken per sample with an integration time of 5 s for V/V, V/H, H/H and H/V polarizer settings (excitation/emission, V: vertical polarization, H: horizontal polarization). The final anisotropy r was calculated by the formula r = (IVV-G*IVH)/(IVV+2*G*IVH), with G = IHV/IHH.
Disulfide Reduction
To reduce the uH2BSS disulfide, an uH2BSS array stock was incubated in the presence of 10 mM DTT for 30 min on ice prior to sample preparation and fluorescence measurements. The reduction of the ubiquitin – H2B disulfide bond was confirmed by SDS-PAGE followed by Coomassie Brilliant Blue staining (Supplementary Fig. 7).
Oligomerization assays
Chromatin oligomerization assays were performed as described in ref. 7 with some modifications. Reconstituted arrays were dialyzed into measurement buffer (10 mM Tris, pH 7.8, 10 mM KCl). The samples were subsequently mixed with MgCl2 containing stock solution at twice the final concentration, incubated for 10 min at 22.5°C and then centrifuged at 16,000 × g for 10 min. The amounts of arrays remaining in solution were determined by UV absorption at 260 nm.
Statistics
The experimental values were compared using the two tailed Student's t-test. In the figures, statistical significance was indicated with an asterisk if p < 0.05.
Supplementary Material
Acknowledgements
. We thank H. Deng and H. Yu (The Rockefeller University Proteomics Resource Center) for mass spectrometric analysis of histones, J. Kim and R. Subramanian for assistance with preparing hDot1L, K. Chiang for help with DNA preparation, H. Yang and D. Montiel for help with fluorescence lifetime measurements and A. Ruthenburg, R. Sadeh, P. Moyle and M. Vila-Perello for assistance with cell experiments and discussions. This work was funded by the U.S. National Institute of Health (Grant Number RC2CA148354) and the Starr Cancer Consortium. B.F. was funded by the Swiss National Science Foundation (Nr. PBBSA-118839 and PA00P3_129130/1) and by the Novartis Foundation.
Footnotes
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci U S A. 2000;97:127–32. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol. 2008;379:772–86. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruithof M, et al. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol. 2009;16:534–40. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 4.Poirier MG, Oh E, Tims HS, Widom J. Dynamics and function of compact nucleosome arrays. Nat Struct Mol Biol. 2009;16:938–44. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–92. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 6.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 8.Chodaparambil JV, et al. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat Struct Mol Biol. 2007;14:1105–7. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 10.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 11.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 12.Robinson PJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–25. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. Embo J. 1997;16:2054–60. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West MH, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980;8:4671–80. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao T, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–51. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minsky N, et al. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–8. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu B, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–11. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Shema E, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–76. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–91. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavri R, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–17. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–4. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 24.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 25.Sridhar VV, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–8. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide directed histone ubiquitylation reveals plasticity in hDot1L stimulation. Nat Chem Biol. 2010;6:267–9. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 27.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 28.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–6. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–9. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958–63. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- 31.Delcuve GP, Davie JR. Chromatin structure of erythroid-specific genes of immature and mature chicken erythrocytes. Biochem J. 1989;263:179–86. doi: 10.1042/bj2630179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Runnels LW, Scarlata SF. Theory and application of fluorescence homotransfer to melittin oligomerization. Biophys J. 1995;69:1569–83. doi: 10.1016/S0006-3495(95)80030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergstrom F, Hagglof P, Karolin J, Ny T, Johansson LB. The use of site-directed fluorophore labeling and donor-donor energy migration to investigate solution structure and dynamics in proteins. Proc Natl Acad Sci U S A. 1999;96:12477–81. doi: 10.1073/pnas.96.22.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautier I, et al. Homo-FRET microscopy in living cells to measure monomer-dimer transition of GFP-tagged proteins. Biophys J. 2001;80:3000–8. doi: 10.1016/S0006-3495(01)76265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thaler C, Koushik SV, Puhl HL, 3rd, Blank PS, Vogel SS. Structural rearrangement of CaMKIIalpha catalytic domains encodes activation. Proc Natl Acad Sci U S A. 2009;106:6369–74. doi: 10.1073/pnas.0901913106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawski A. Excitation energy transfer and its manifestation in isotropic media. Photochem. Photobiol. 1983;38:487–504. [Google Scholar]
- 37.Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993;90:9021–5. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knox R. Theory of polarization quenching by excitation transfer. Physica. 1968;39:361–386. [Google Scholar]
- 39.Woodcock CL, Horowitz RA. Chromatin organization re-viewed. Trends Cell Biol. 1995;5:272–7. doi: 10.1016/s0962-8924(00)89038-8. [DOI] [PubMed] [Google Scholar]
- 40.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Koch CM, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dittmar GA, Wilkinson CR, Jedrzejewski PT, Finley D. Role of a ubiquitin-like modification in polarized morphogenesis. Science. 2002;295:2442–6. doi: 10.1126/science.1069989. [DOI] [PubMed] [Google Scholar]
- 43.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu X, et al. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol. 2008;15:1122–4. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jason LJM, Moore SC, Ausio J, Lindsey G. Magnesium-dependent association and folding of oligonucleosomes reconstituted with ubiquitinated H2A. J. Biol. Chem. 2001;276:14597–14601. doi: 10.1074/jbc.M011153200. [DOI] [PubMed] [Google Scholar]
- 46.Demeler B. UltraScan version 9.9 rev 863. A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments. The University of Texas Health Science Center at San Antonio, Department of Biochemistry; 2009. http:/www.ultrascan.uthscsa.edu. [Google Scholar]
- 47.Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem. 2004;335:279–88. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 48.McGinty RK, et al. Structure activity analysis of semisynthetic nucleosomes: Mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol. 2009 doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 50.Ramelot TA, et al. Solution structure of the yeast ubiquitin-like modifier protein Hub1. J Struct Funct Genomics. 2003;4:25–30. doi: 10.1023/a:1024674220425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.