Abstract
The ability of cells to respire requires that mitochondria undergo fusion and fission of their outer and inner membranes. The means by which levels of fusion ‘machinery’ components are regulated and the molecular details of how fusion occurs are largely unknown. In Saccharomyces cerevisiae, a central component of the mitochondrial outer membrane (MOM) fusion machinery is the mitofusin Fzo1, a dynamin-like GTPase. We demonstrate that an early step in fusion, mitochondrial tethering, is dependent on the Fzo1 GTPase domain. Furthermore, the ubiquitin ligase SCFMdm30 (a SKP1–cullin-1–F-box complex that contains Mdm30 as the F-box protein), which targets Fzo1 for ubiquitylation and proteasomal degradation, is recruited to Fzo1 as a consequence of a GTPase-domain-dependent alteration in the mitofusin. Moreover, evidence is provided that neither Mdm30 nor proteasome activity are necessary for tethering of mitochondria. However, both Mdm30 and proteasomes are critical for MOM fusion. To better understand the requirement for the ubiquitin–proteasome system in mitochondrial fusion, we used the N-end rule system of degrons and determined that ongoing degradation of Fzo1 is important for mitochondrial morphology and respiration. These findings suggest a sequence of events in early mitochondrial fusion where Fzo1 GTPase-domain-dependent tethering leads to recruitment of SCFMdm30 and ubiquitin-mediated degradation of Fzo1, which facilitates mitochondrial fusion.
Key words: Oxidative phosphorylation, Fzo1p, Mdm30p, Membrane fusion, SCF, F-box
Introduction
Mitochondria are dynamic in nature and in Saccharomyces cerevisiae they constitute a single tubular network whose morphology is determined by the equilibrium between ongoing fusion and fission of mitochondrial membranes (Hoppins et al., 2007; Okamoto and Shaw, 2005; Westermann, 2010). This equilibrium is essential for faithful inheritance of mitochondrial DNA and oxidative phosphorylation (Hermann et al., 1998; Okamoto and Shaw, 2005; Westermann, 2010). In higher eukaryotes, a similar equilibrium between mitochondrial fusion and fission is observed, and shifts towards increased fission are associated with apoptosis (Liesa et al., 2009; Wasilewski and Scorrano, 2009).
Fusion and fission each require a distinct set of proteins that constitute the fusion and fission ‘machineries’ (Hoppins et al., 2007; Okamoto and Shaw, 2005; Westermann, 2010). Unlike other organelles that require small GTPases and SNAREs to initiate membrane fusion, mitochondria use dynamin-like large GTPases, the mitofusins (Mfn1 and Mfn2 in mammals; Fzo1 in yeast). The clinical significance of mitofusins is best demonstrated by the association between Mfn2 mutations, including GTPase domain mutations, and Charcot-Marie-Tooth Type 2A (CMT2A) neuropathy (Liesa et al., 2009; Wasilewski and Scorrano, 2009).
Mitofusins are characterized by an N-terminal GTPase domain and two C-terminal coiled-coil heptad repeats (HR1 and HR2) surrounding two transmembrane domains (Fig. 1A) (Heymann and Hinshaw, 2009). In yeast, an additional coiled-coil domain (HRN) is located N-terminal to the GTPase domain. Collectively the coiled-coil and GTPase domains are required to promote mitochondrial tethering and subsequent mitochondrial outer membrane (MOM) fusion (Griffin and Chan, 2006; Hermann et al., 1998; Ishihara et al., 2004; Koshiba et al., 2004). However, the sequence of events leading to fusion is poorly understood.
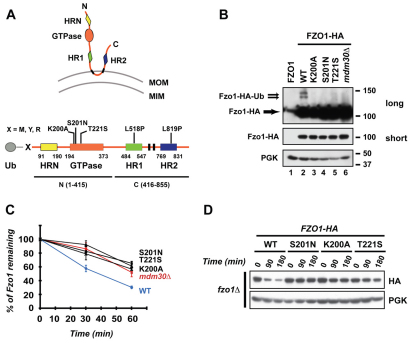
Fig. 1.
Integrity of Fzo1 GTPase domain is essential for Mdm30-mediated ubiquitylation and degradation of Fzo1. (A) Top, schematic representation of Fzo1 in the MOM. Domains of Fzo1 are indicated. MOM, mitochondrial outer membrane; MIM, mitochondrial inner membrane. Bottom, linear schematic of forms of Fzo1 used in this study. Fzo1 domains are indicated by boxes. Transmembrane domains are indicated in black. Grey circle, ubiquitin (Ub). X indicates either Met (M), Tyr (Y) or Arg (R) residues. Point mutations (above) and truncations (below) of Fzo1 used in the study are indicated. (B) Immunoblot of whole-cell extracts prepared from fzo1Δ (lanes 1–5) or fzo1Δ mdm30Δ (lane 6) cells expressing untagged Fzo1 (lane 1), WT (lanes 2 and 6) or GTPase mutants (lanes 3–5) of Fzo1–HA. Top, anti-HA long exposure; middle, anti-HA short exposure; bottom, anti-PGK (loading control). Molecular size markers (right) are in kDa. (C) Analysis of Fzo1–HA degradation by pulse-chase metabolic labeling using the same strains as in B. Error bars represent s.d. from three independent experiments. (D) Cycloheximide (CHX) chase performed in fzo1Δ strains expressing either WT, K200A, S201N or T221S mutants of Fzo1–HA. Immunoblotting was for HA or PGK.
In S. cerevisiae efficient mitochondrial fusion requires the F-box protein Mdm30, which binds Fzo1 (Durr et al., 2006; Escobar-Henriques et al., 2006; Fritz et al., 2003) and is a substrate recognition element of the family of multisubunit SCF (SKP1, cullin-1, F-box containing) ubiquitin ligases (Cohen et al., 2008; Muratani et al., 2005). SCFMdm30 (the SCF complex that contains Mdm30 as the F-box protein) promotes Fzo1 degradation by the ubiquitin–proteasome system (UPS) (Cohen et al., 2008). Deletion of FZO1 leads to the accumulation of fragmented mitochondria and loss of respiration (Hermann et al., 1998). Increased levels of Fzo1, as a result of either deletion of Mdm30 or overexpression of Fzo1, cause a characteristic phenotype of aggregated fragmented mitochondria. This is also associated with loss of respiration (Cohen et al., 2008; Durr et al., 2006; Escobar-Henriques et al., 2006; Fritz et al., 2003). These findings have led to the general belief that a primary role of Mdm30-mediated degradation is to maintain optimal steady state levels of Fzo1 for regulated mitochondrial fusion and respiration. However, the means by which Mdm30 is recruited to Fzo1 and whether the role of Mdm30 is limited to regulating steady state levels of Fzo1 has not been explored.
We now provide evidence that an early event in MOM fusion, tethering of mitochondria, requires the Fzo1 GTPase domain. GTPase-domain-dependent alterations in Fzo1, which probably occur concomitant with or downstream of mitochondrial tethering, allow for recruitment of Mdm30, which is required for MOM fusion. Furthermore, we demonstrate that, independently of steady state levels, ongoing proteasome-dependent degradation of Fzo1 is crucial for maintenance of mitochondrial morphology and for respiration. These findings suggest that targeting of Fzo1 for degradation by SCFMdm30 provides a critical link between mitochondrial tethering and later steps in mitochondrial fusion.
Results
An intact Fzo1 GTPase domain is essential for Mdm30-mediated ubiquitylation and degradation of Fzo1
To examine the relationship between GTPase activity and ubiquitylation, we evaluated three ‘classic’ GTPase inactivating mutations of Fzo1 (K200A, S201N and T221S) (Amiott et al., 2009; Hermann et al., 1998). Each mutation greatly diminished Mdm30-dependent ubiquitylation (Fig. 1B and supplementary material Fig. S1) resulting in stabilization of Fzo1 as assessed by pulse-chase metabolic labeling (Fig. 1C) and cycloheximide chase experiments (Fig. 1D). Together with our previous assessment of Fzo1 mutated in a position analogous to a CMT2A Mfn2 mutant (Fzo1 V327T) (Amiott et al., 2009), these findings establish that the integrity of the Fzo1 GTPase domain is essential for its Mdm30-dependent ubiquitylation and degradation.
The GTPase domain of Fzo1 is essential for binding of Mdm30 to the N-terminal half of the mitofusin
To further assess the relationship between the GTPase domain of Fzo1 and Mdm30-mediated ubiquitylation, we chose to evaluate the Fzo1 S201N GTPase mutant, which differs from other Fzo1 GTPase mutants as it is not complemented by co-expression of Fzo1 molecules with intact GTPase domains (Amiott et al., 2009; Griffin and Chan, 2006). As shown in Fig. 2A, Mdm30–Myc co-immunoprecipitated with wild-type (WT) Fzo1 tagged at its C-terminus with three tandem HA epitope tags (Fzo1–HA). By contrast, there was neither discernable association of Fzo1-S201N–HA with Mdm30 nor substantial ubiquitylation of this GTPase mutant (Fig. 2A, compare right two lanes). Mdm30 binding was also abolished using other Fzo1 GTPase mutants (supplementary material Fig. S2). These results indicate that an intact GTPase domain is essential for binding of Mdm30 to Fzo1. This loss of binding provides a molecular basis for the lack of ubiquitylation and the decreased rate of degradation observed with mutations in the Fzo1 GTPase domain.
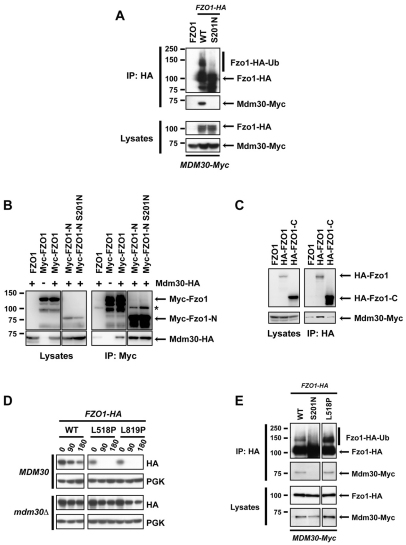
Fig. 2.
Accessibility of Mdm30 to its N-terminal Fzo1 binding site is controlled by the Fzo1 C-terminal region in a GTPase domain-dependent manner. Experiments were performed in DCY538 background cells. Myc–Fzo1-N and HA–Fzo1-C are both N-terminally tagged in B and C. N-terminally instead of C-terminally tagged versions of full-length Fzo1 were therefore used for consistency. (A) fzo1Δ mdm30Δ cells expressing untagged Fzo1 (FZO1), Fzo1–HA (WT) or Fzo1-S201N–HA (S201N) together with Mdm30 epitope-tagged at its C-terminus (Mdm30–Myc) were subjected to co-immunoprecipitation with anti-HA followed by immunoblotting as indicated and developed using ECL. Top panels, immunoprecipitates; bottom panels, lysates (10% input of IP). (B) Co-immunoprecipitations were performed as in A with strains that either express (+) or do not express (−) C-terminal HA-tagged Mdm30 and transformed with a plasmid expressing either WT or GTPase mutant (S201N) forms of either full-length or truncated (Myc–Fzo1-N, aa 1–415) Fzo1 bearing nine Myc epitope tags at their N-termini. Right panels, immunoprecipitates; left panels, lysates (10% input of IP). Incomplete reduction of a small amount of IgG heavy chain dimers that is variably observed is indicated (*). (C) Co-immunoprecipitations with anti-HA were performed as in A with strains co-expressing indicated N-terminal 6-HA-tagged forms of full-length or truncated (HA–Fzo1-C; aa 416–855) Fzo1 and C-terminal Myc-tagged Mdm30. Right panels, immunoprecipitates; left panels, lysates (10% input of IP). (D) Cycloheximide (CHX) chase performed in fzo1Δ (Top) and fzo1Δ mdm30Δ (bottom) strains expressing indicated versions of C-terminal HA-tagged forms of either WT Fzo1 or Fzo1 mutated in either its HR1 (L518P) or HR2 (L819P) domain. (E) fzo1Δ mdm30Δ cells expressing WT, S201N or L518P forms of Fzo1-HA together with Mdm30-Myc were subjected to co-IP with anti-HA followed by immunoblotting with anti-HA or anti-Myc and detection using ECL. Top two panels, immunoprecipitates; bottom two panels, lysates (10% input of IP).
To extend these observations, we assessed binding of Mdm30 to the N-terminal (aa 1–415) and C-terminal (aa 416–855) halves of Fzo1 that were tagged at their N-termini with nine tandem Myc and six HA epitope tags, respectively (see schematic, Fig. 1A). When expressed alone, neither the cytosolic Myc–Fzo1-N nor the transmembrane HA–Fzo1-C fragments complement the absence of full-length Fzo1 in fzo1Δ cells, as assessed by growth on glycerol (Griffin and Chan, 2006). However, when expressed together, these two polypeptides form a functional complex at the mitochondrial outer membrane that rescues mitochondrial fusion and respiration (Griffin and Chan, 2006).
As predicted, co-immunoprecipitation of Mdm30 was observed with the GTPase-containing N-terminal fragment (Myc–FZO1-N) but not with the C-terminal half of Fzo1 (HA–FZO1-C) (Fig. 2B,C). Thus, the cytosolic N-terminal 415 amino acids, which include the GTPase domain, are sufficient for Mdm30 binding. Surprisingly, however, introduction of the S201N mutation into this same N-terminal fragment (Myc–FZO1-N S201N) did not affect its capacity to bind Mdm30 (Fig. 2B). This was unexpected, as in the context of the full-length Fzo1, Mdm30 binding is disrupted with this mutation (Fig. 2A and supplementary material Fig. S2).
These findings, together with the known interaction between the N- and C-terminal halves of Fzo1 (Griffin and Chan, 2006), suggest a model in which an intact GTPase domain is required to relieve a steric block to Mdm30 binding imposed by the C-terminal HR1 and HR2-containing half of Fzo1.This model predicts that disruption of the structure of the C-terminal half of Fzo1, such that interactions with the N-terminal half of the molecule are interrupted, should result in increased Mdm30-mediated degradation. Consistent with this idea, mutations in either HR1 (L518P) or HR2 (L819P), which are each predicted to disrupt the structure of coiled-coils and abolish interactions with the N-terminal half of Fzo1 (Griffin and Chan, 2006), resulted in rapid degradation of Fzo1 (Fig. 2D, top HA panel). This degradation was largely Mdm30 dependent (Fig, 2D compare top and bottom HA panels) and correlated with binding of Mdm30 to Fzo1 and with Fzo1 ubiquitylation (Fig. 2E and data not shown). These observations suggest that an intact Fzo1 GTPase domain is required to induce an alteration in Fzo1, either within a single molecule or in the context of Fzo1 oligomers, such that an Mdm30 binding site in the N-terminal half of the molecule becomes accessible. Notably, Fzo1 bearing both the L518P and S201N mutations was also more rapidly degraded than WT Fzo1 (supplementary material Fig. S3). Importantly, however, this degradation is independent of Mdm30 and is consistent with a recently described mitochondrial quality control pathway (Heo et al., 2010).
Differential requirements for the Fzo1 GTPase domain, Mdm30 and proteasome activity for mitochondrial tethering and outer membrane fusion
To better understand the relationship between the Fzo1 GTPase domain, Mdm30 binding and mitochondrial fusion, we evaluated mitochondrial tethering. Tethering of mitochondria is an early step in fusion that, in mammals, is dependent on oligomerization of mitofusins on interacting mitochondria (i.e. in trans) (Ishihara et al., 2004; Koshiba et al., 2004). Pertinent to this is the finding that mutations in the GTPase domain of Fzo1 decrease its oligomerization (Griffin and Chan, 2006). Whether this decrease in Fzo1 oligomerization is reflective of a diminished association of molecules in trans, or of those on the same mitochondrion (in cis) is not established. To investigate the requirements for the GTPase domain of Fzo1 and for Mdm30 in tethering, we adapted an in vitro MOM fusion assay (Meeusen et al., 2004), which uses mitochondria-enriched fractions prepared from cells expressing either mito–GFP or mito–DsRed in the mitochondrial matrix (Fig. 3A). Using this assay, 11.5% of mitochondria from fzo1Δ cells were found to be apposed (Fig. 3B). Based on mammalian studies (Ishihara et al., 2004; Koshiba et al., 2004), this probably represents the assay background, although we cannot rule out that it corresponds to Fzo1-independent tethering mediated by unidentified factors. Expression of WT Fzo1 increased the percentage of apposed mitochondria to 27.5%. Significantly, mitochondria from cells expressing Fzo1-S201N had a level of apposed mitochondria that was similar to levels obtained with mitochondria lacking Fzo1. This is consistent with a requirement for an intact GTPase domain in trans-oligomerization of Fzo1 (Griffin and Chan, 2006) during early steps of mitochondrial fusion. In contrast to the GTPase mutation, loss of Mdm30 expression (mdm30Δ) had minimal, if any, effect on mitochondrial tethering, indicating that Mdm30 is dispensable for tethering. This is consistent with the finding that the capacity for Fzo1 to self-associate is independent of Mdm30 expression (supplementary material Fig. S4).
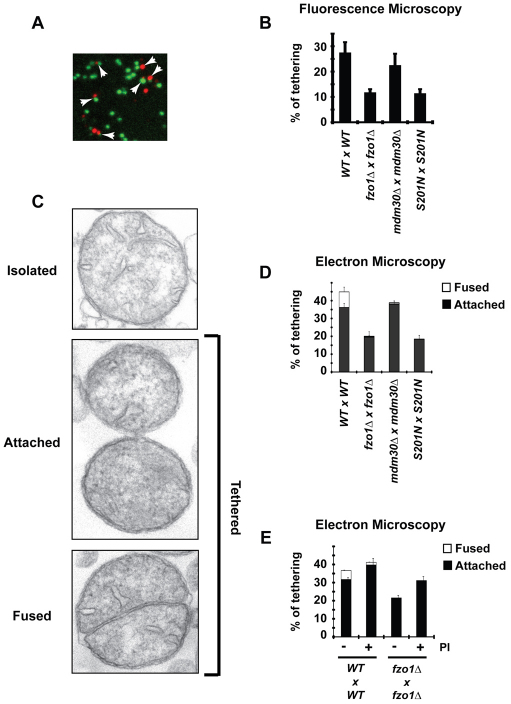
Fig. 3.
The GTPase domain of Fzo1 is required for mitochondrial tethering, whereas Mdm30 and proteasome activity are crucial for MOM fusion. (A) In vitro mitochondrial tethering assay, using WT Fzo1 strains, assessed by fluorescence microscopy. Red and green signals attached to each other were scored as tethered. Examples of ‘tethered’ mitochondria are indicated by arrows. (B) Percentage of tethered mitochondria observed by fluorescence microscopy in each reaction. Error bars represent the s.d. of two independent experiments. (C) In vitro mitochondrial tethering assay assessed by electron microscopy. Examples of the three categories of mitochondria observed: isolated (corresponding to content-dense organelles lacking any contact with neighboring mitochondria); attached (corresponding to mitochondria in physical contact through their outer membrane); fused (corresponding to two cristae surrounded by a single outer membrane). Among these three categories, attached and fused organelles were scored as tethered mitochondria. (D) Percentage of tethered mitochondria observed by electron microscopy in each reaction. Black: attached mitochondria; white: fused mitochondria. The amount of fused mitochondria seen in other conditions than WT reactions is less than 0.75%. Error bars represent the s.d. of fused (top) and attached (bottom) mitochondria from the mean of two independent experiments. (E) Samples were treated with (+) or without (−) proteasome inhibitors (PI) before initiation of the tethering assay. Error bars represent the s.d. of fused (top) and attached (bottom) mitochondria from two independent experiments.
To extend these observations, mitochondria were assessed by conventional transmission electron microscopy (EM) using the same assay conditions. Three distinct categories of mitochondria were observed (Fig. 3C): isolated mitochondria, attached mitochondria and mitochondria with fused outer membranes. The EM analysis yielded a striking correlation in the number of mitochondria scored as tethered compared with the fluorescence results (Fig. 3D). Consistent with the omission of green–green and red–red complexes in the fluorescence microscopy analysis (Fig. 3B), levels of tethered mitochondria were greater in the EM analysis. Importantly, however, mitochondria with fused outer membranes were exclusively observed in WT preparations. These EM results confirm that the GTPase domain of Fzo1 is required for efficient mitochondrial tethering and indicate that Mdm30 is required for fusion of outer membranes.
These findings imply that ubiquitylation and potentially proteasomal degradation of Fzo1 act at the stage of MOM fusion. To assess a potential role for proteasome activity, fzo1Δ and WT preparations were treated with inhibitors of the proteasome (Lactacystin+MG132) or with carrier (DMSO). Unexpectedly, the percentage of mitochondria scored as attached increased with proteasome inhibitors (PI) in both fzo1Δ and WT preparations. Strikingly, however, mitochondria with fused outer membranes decreased by approximately 70% in WT samples treated with proteasome inhibitor compared with WT samples treated with DMSO (Fig. 3E). This indicates that proteasome activity is required for MOM fusion and is consistent with a requirement for Mdm30 at this same step of mitochondrial fusion.
Ongoing degradation of Fzo1 is required for mitochondrial function
Our findings establish that the Fzo1 GTPase domain is required for mitochondrial tethering, Mdm30 recruitment and Fzo1 degradation mediated by SCFMdm30. Additionally, the EM data suggest that MOM fusion is dependent on Mdm30 and proteasome activity. For these reasons, we considered the possibility that, in addition to its role in the regulation of Fzo1 levels (Cohen et al., 2008; Durr et al., 2006; Escobar-Henriques et al., 2006; Fritz et al., 2003), ongoing degradation of Fzo1 might be linked to mitochondrial fusion. One way to address this possibility would be to modulate the degradation of Fzo1 independently of its steady state level. Towards this end, we turned to the N-end rule pathway system of degrons. In this pathway, which is well established to be mediated primarily by the N-end rule ubiquitin ligase Ubr1, the rate of UPS-dependent destruction of a test substrate is determined by the nature of its N-terminal amino acid (Varshavsky, 1996). Methionine (M), tyrosine (Y) and arginine (R) confer relative half-lives of M>>>Y>R to test substrates. These N-termini are engineered by fusing an N-terminal ubiquitin moiety to substrates, which is then rapidly cleaved by endogenous deubiquitylating enzymes.
C-terminally HA-tagged N-end rule forms of Fzo1 [Ub-M-Fzo1 (M-Fzo1), Ub-Y-Fzo1 (Y-Fzo1), and Ub-R-Fzo1 (R-Fzo1); Fig. 1A bottom] were expressed in an fzo1Δ mdm30Δ strain and their stabilities assessed by pulse-chase metabolic labeling (Fig. 4A). Consistent with N-end rule-mediated degradation, R-Fzo1 was highly unstable, turnover of Y-Fzo1 resembled that of WT Fzo1 in an Mdm30-expressing background, and M-Fzo1 was at least as stable as WT Fzo1 in an mdm30Δ background. Although the relative steady state levels of M-Fzo1 and R-Fzo1 were largely consistent with their observed stabilities (Fig. 4B), those of Y-Fzo1 and M-Fzo1 were not appreciably different from each other, despite their differences in stability. Similar steady state levels of Y-Fzo1 and M-Fzo1were also observed with untagged forms of the protein (Fig. 4C). The disparity between rate of degradation and steady state levels suggests a difference in rates of synthesis (Y-Fzo1>M-Fzo1), perhaps as a consequence of feedback regulation of the FZO1 promoter. The difference in synthesis was confirmed by a short (5 minute) pulse labeling, where the initial rate of accumulation of 35S-labeled Y-Fzo1 was greater than that of M-Fzo1 (supplementary material Fig. S5).
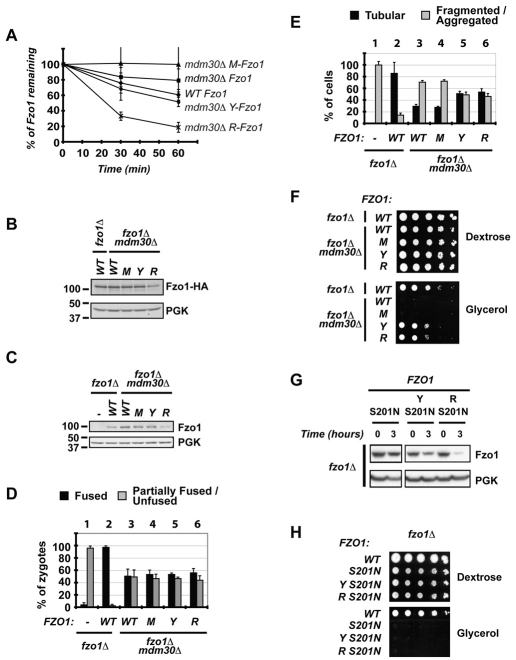
Fig. 4.
Degradation of Fzo1 by the N-end-rule pathway partially bypasses the requirement for Mdm30 in mitochondrial function. All experiments were performed in fzo1Δ or fzo1Δ mdm30Δ cells (DCY538 background) expressing untagged versions of Fzo1 unless indicated: (−), empty vector; (WT), Fzo1; (M), Ub–M-Fzo1; (Y), Ub–Y-Fzo1; (R), Ub–R-Fzo1. Error bars are the s.d. from three independent experiments. (A) Analysis of degradation of indicated forms of Fzo1–HA by [35S]Met metabolic labeling. (B) Immunoblot of steady state Fzo1–HA from whole cell extracts using anti-HA. PGK was used as a control for equal loading. (C) Immunoblot of steady state Fzo1 from whole cell extracts using anti-Fzo1. PGK was used as a control for equal loading. (D) In vivo mitochondrial fusion assay. Percentage of zygotes (derived from mating of DCY538 and JSY2294 strains expressing indicated versions of Fzo1) with fused or partially fused/unfused mitochondria. (E) Percentage of cells with tubular or fragmented/aggregated mitochondria. (F) Serial dilutions of indicated strains on dextrose or glycerol medium. (G) Cycloheximide (CHX) chase performed in fzo1Δ strains expressing S201N mutants of Fzo1 (untagged) fused, or not, to indicated N-end rule degrons (Y, R). Immunoblotting was for Fzo1 or PGK. (H) Serial dilutions of strains expressing WT untagged Fzo1 or the S201N mutants (as in C) on dextrose or glycerol medium.
M-Fzo1 and Y-Fzo1 show steady state levels that are similar to Fzo1 in mdm30Δ cells, whereas R-Fzo1 exhibits levels of Fzo1 that are significantly lower than levels in the WT (Fig. 2B,C). For this reason, if the steady state level of Fzo1 was the determining factor in mitochondrial function, no restoration of function would be expected with either M-Fzo1 or Y-Fzo1. To begin to assess the influence of levels versus turnover on function, we examined mitochondrial fusion in vivo. MATa and MATα strains expressing mito–GFP or mito–DsRed were mated and fusion was assessed in vivo 3–4 hours after mating. In Mdm30-expressing cells lacking Fzo1 (fzo1Δ), re-expression of WT Fzo1 resulted in almost complete fusion (Fig. 4D compare column pairs 1 and 2). In the absence of Mdm30 (mdm30Δ fzo1Δ), only 50% restoration of fusion was observed, regardless of whether WT Fzo1 or any of the N-end rule forms were expressed (Fig. 4D, compare 3–6 with 2). Interestingly, these negative findings obtained during the limited time frame of in vivo mitochondrial fusion assays (3–4 hours) stand in contrast to results obtained by assessing in vivo mitochondrial morphology at steady state. Compared with WT Fzo1 in cells expressing Mdm30 (fzo1Δ), where ~80% normal ‘tubular’ morphology was seen (Fig. 4E column pair 2), ~30% tubular morphology was observed in cells lacking Mdm30 (fzo1Δ mdm30Δ) in which either WT or the slowly degraded M-Fzo1 was expressed (Fig. 4E, compare 3 and 4 with 2). However, when either of the more rapidly degraded forms, Y-Fzo1 or R-Fzo1, was expressed, greater than 50% tubular morphology was observed (Fig. 4E, Figs 5 and 6). These results suggest an association between degradation of Fzo1 by the N-end rule and increased restoration of morphology, rather than simply a correlation between morphology and steady state levels of Fzo1. To determine whether this morphological data actually correlates with mitochondrial function, respiration was assessed. Both Y-Fzo1 and R-Fzo1 resulted in a substantial restoration of growth on glycerol in the absence of Mdm30 expression (fzo1Δ mdm30Δ; Fig. 4F, ‘Y’ and ‘R’), whereas no growth was observed with either WT Fzo1 or M-Fzo1 in the absence of Mdm30 expression (Fig. 4F, ‘WT’ and ‘M’). These findings indicate that, at least within the range of Fzo1 levels examined, maintenance of mitochondrial morphology and function do not merely correlate with Fzo1 steady state levels. Instead, these results strongly suggest that ongoing proteasomal degradation of Fzo1 is required for maintenance of mitochondrial morphology and for respiration.
A question of mechanistic and potentially clinical importance for CMT2A neuropathy is whether bypassing the requirement for the GTPase domain in Fzo1 degradation can restore mitochondrial function. As shown in Fig. 4G, the S201N mutation did not prevent degradation of the N-end rule substrates (Y-Fzo1-S201N and R-Fzo1-S201N). However, neither of these rapidly degraded proteins restored growth on glycerol (Fig. 4H). Therefore, independently of its role in Fzo1 degradation, the GTPase domain is essential for mitochondrial function, which is a finding that is consistent with its role in mitochondrial tethering.
Discussion
Mitochondrial fusion is crucial for maintenance of organelle integrity in both yeast and higher eukaryotes (Hoppins et al., 2007; Okamoto and Shaw, 2005; Westermann, 2010). Despite this, the mechanisms by which fusion occurs and how it is regulated are just being unraveled. This study offers new insights into the relationship between the GTPase domain of the yeast mitofusin Fzo1, the UPS and MOM fusion. In particular, evidence is provided that: (1) Fzo1-dependent mitochondrial tethering occurs in a GTPase-domain dependent manner; (2) Mdm30 recruitment to Fzo1 and SCFMdm30-dependent Fzo1 degradation occur in a GTPase-domain-dependent manner as a consequence of an alteration in the state of Fzo1; (3) Mdm30 and proteasomal activity are required for MOM fusion, but not for tethering of mitochondria or for the self-association of Fzo1 molecules; and (4) ubiquitylation and degradation of Fzo1 are integral to the process of mitochondrial fusion and do not simply regulate steady state levels of this protein.
There are, of course, several potential interpretations for these findings. A straightforward interpretation is that a functional GTPase domain of Fzo1, whether through binding or hydrolysis of nucleotide, facilitates a change from a ‘closed’ to an ‘open’ state that allows for oligomerization of Fzo1 molecules on adjacent mitochondria. Concomitant or subsequent to this, the Mdm30 binding site, which is located on the N-terminal half of Fzo1, becomes accessible to SCFMdm30. This leads to UPS-mediated degradation of Fzo1 and outer mitochondrial membrane fusion, through mechanisms that are yet to be fully determined.
Although this study does not provide biophysical evidence for this series of events, there are several important findings in the literature that lend credence to this interpretation. First, it has been established that oligomerization of Fzo1 and Mfn1 molecules is dependent on an intact GTPase domain (Griffin and Chan, 2006; Ishihara et al., 2004). Second, trans interactions between Fzo1 N- and C-terminal halves are abolished through mutations in the GTPase domain (Griffin and Chan, 2006), indicating a fundamental role for the GTPase domain in dynamic rearrangements within the Fzo1 polypeptide. Further support for the concept of a change from a closed to an open state comes from studies on bacterial dynamin-like protein (BDLP). This protein is a close relative of Fzo1, sharing 20% identity and 41% similarity with Fzo1. Binding of GTP analogs by BDLP results in a switch from a compact structure, seen with GDP binding, to an ‘open’ conformation that stimulates BDLP oligomerization and favors its ability to shape the morphology of liposomes (Low and Lowe, 2006; Low et al., 2009).
An intriguing aspect of the current study is the finding that, by targeting Fzo1 for degradation using the N-end rule, mitochondrial morphology and function was substantially restored in the absence of Mdm30. While these findings demonstrate the importance of Fzo1 degradation as part of the fusion process, they are also provocative, in that N-end-rule-mediated degradation does not fully restore mitochondrial function. There are several non-mutually exclusive explanations for this. One possibility is that ubiquitylation of other mitochondrial SCFMdm30 substrates, possibly Mdm34 (Durr et al., 2006; Ota et al., 2008), is required for reconstitution of WT function. Other possibilities relate to probable differences in sites of ubiquitylation and processivity of ubiquitylation when mediated by SCFMdm30 versus the N-end rule E3, Ubr1. Additionally, it is important to consider that although recruitment and ubiquitylation by SCFMdm30 is dependent on the state of Fzo1 (e.g. open conformation, oligomerization), Ubr1 is not subject to the same constraints. This is underscored by the finding that N-end-rule-mediated degradation does not require an intact GTPase domain. Thus, we suggest that the regulated recruitment of Mdm30 allows for degradation of the subset of Fzo1 molecules that are actively engaged in MOM fusion. This degradation occurs in an appropriately timed manner that would maximize the efficiency of mitochondrial fusion.
As noted above, how ubiquitylation and degradation of Fzo1 favor outer mitochondrial membrane fusion remains to be discerned. At one extreme, the ubiquitylated intermediate might perform a discrete role in, for example, stabilizing an open state conformation, allowing increased generation of anti-parallel coiled-coil intermediates between Fzo1 molecules (Ishihara et al., 2004; Koshiba et al., 2004) before proteasomal degradation. At the other end of the spectrum, ubiquitylation might simply provide a means to eliminate Fzo1 from the MOM to allow completion of fusion. Intriguingly, obligate degradation of the fusion machinery occurs in at least one other membrane fusion system. The UPS, acting through yet unknown ligases, promotes degradation of the Rab GTPase Ypt7, which is required for the initiation of SNARE-mediated vacuolar fusion (Kleijnen et al., 2007). The significant differences between membrane fusion mediated by SNAREs and mitofusins preclude mechanistic generalizations. Regardless, it is intriguing that in both cases, the UPS disposes of a GTPase activity that is required to initiate the fusion process.
A critical mechanistic question, mentioned above, that arises from our work as well as that of others (Ishihara et al., 2004; Koshiba et al., 2004), relates to the exact role of the GTPase domain. Although we know that GTPase mutations that decrease or eliminate Mdm30 binding and Fzo1 function exhibit loss of GTPase activity, the nature of Fzo1 and other dynamin-like molecules have not yet allowed an analysis of whether, in vivo, it is the binding of the guanine nucleotide or the hydrolysis of the γ-phosphate bond that is crucial to their function in mitochondrial fusion. Clearly, there are many interesting questions that require further detailed investigation.
Materials and Methods
Plasmids, yeast strains and media
The plasmids and S. cerevisiae strains used in this study are listed in supplementary material Table S1. Standard methods were used for growth, transformation and genetic manipulation of S. cerevisiae. Minimal media supplemented with either 2% dextrose or 2% glycerol were prepared as described (Sherman et al., 1986). In the indicated strains (see supplementary material Table S1), MDM30 was chromosomally deleted, as previously described (Longtine et al., 1998).
All strains were generated using a plasmid-shuffle strategy. Briefly, fzo1Δ and fzo1Δ mdm30Δ strains carrying a URA-based FZO1 plasmid (pRS416-FZO1) were transformed with various FZO1 plasmids under TRP1 or LYS selection. Ten colonies were systematically isolated on dextrose –TRP –URA or dextrose –LYS –URA medium and replica-plated on corresponding glycerol or 5-fluoroorotic acid medium. Strains cured from the pRS416-FZO1 plasmid were then patched on dextrose or glycerol medium lacking tryptophan or lysine. The glycerol growth phenotypes of strains with or without the pRS416-FZO1 plasmid were reproducibly observed in 80–100% of the colonies tested after 4 days of growth at 30°C. Representative colonies tested for expression of FZO1 encoded by TRP or LYS-based plasmids were used for subsequent experiments.
Immunoblots and antibodies
Proteins resolved by SDS-PAGE were immunoblotted and developed using ECL or fluorescent analysis. Proteins were revealed using polyclonal antibodies raised against Fzo1 residues 1–343 (Janet Shaw, University of Utah, Salt Lake City, UT) or monoclonal antibodies against HA and Myc epitopes (12CA5 and 9E10; Santa Cruz Biotechnology), phosphoglucokinase (PGK, Monoclonal 22C5; Molecular Probes), flavoprotein subunit of succinate dehydrogenase (SDH1; Agnes Delahodde, IGM, Orsay, France) or anti-ubiquitin (P4D1; Santa Cruz Biotechnology). Appropriate HRP-conjugated secondary antibodies were used for detection with ECL Plus (GE Healthcare) followed by exposure to film or analysis using Genesnap software for G-box (Syngene). Alternatively, fluorescent secondary antibodies (IRDye 800 anti-mouse or IRDye 680 anti-rabbit; LiCor) were used and proteins detected and quantified using an Odyssey scanner and Odyssey 3.0 analysis software (LiCor). Figures were assembled in Adobe Photoshop with only linear adjustments of contrast and brightness.
Yeast extracts and cycloheximide chase
Cells grown in dextrose medium were collected during the exponential growth phase. Total protein extracts were prepared by the NaOH–trichloroacetic acid (TCA) lysis method (Avaro et al., 2002). To monitor constitutive turnover of Fzo1, cycloheximide (CHX) was added to yeast cultures growing at 37°C to a final concentration of 100 μg/ml. Total protein extracts were prepared at the indicated time points after addition of CHX.
Metabolic labeling
For pulse-chase experiments, cells grown in dextrose medium were collected during the exponential growth phase, incubated for 50 minutes at 23°C in 1 ml labeling medium lacking methionine (Dextrose –Met), and pulsed with 25 μCi of [35S]methionine (Perkin-Elmer Cetus) per OD of cells for 15 minutes at 30°C followed by 15 minutes at 37°C. After addition of 1 ml chase medium (dextrose–Met supplemented with 6 mg/ml methionine and 2 mg/ml BSA), cells were incubated at 37°C. Two OD600 units of cells were collected immediately (time=0) and at indicated time points and treated as described previously (Moreau et al., 1997). Results were quantified by Storm Phosphorimager and ImageQuant software (GE Healthcare). Error bars were determined by calculating the s.d. of three independent experiments. To measure the initial rates of synthesis by metabolic labeling, cells were grown and collected as above, incubated for 5 minutes at 28°C in 1 ml labeling medium lacking methionine (Dextrose –Met), and pulsed with 25 μCi of [35S]methionine (Perkin-Elmer Cetus) per OD600 units of cells for 5 minutes at 28°C. Two OD600 units of cells were then collected and processed as above. Error bars were determined by calculating the s.d. of two independent experiments.
Immunoprecipitation
For co-immunoprecipitation assays, spheroplasts were generated from 100 OD600 units of cells grown to mid-log phase and treated with 75U zymolyase (Zymo Research) for 45 minutes at 30°C as described previously (Ingerman et al., 2005). Spheroplasts were resuspended in 1 ml IP buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.6% Triton X-100, 10% glycerol, and protease inhibitors (Complete Mini Protease Inhibitor; Roche)] and incubated at 4°C for 30 minutes. Lysates were cleared by centrifugation at 13,000 g for 30 minutes and supernatants were incubated with anti-HA Affinity Matrix (Roche) or anti-Myc (9E10; Santa Cruz Biotechnology) pre-bound to protein G beads (GE Healthcare) for 1 hour at 4°C. Beads were washed with IP buffer and immunoprecipitated proteins were eluted by heating in SDS sample buffer before resolution by SDS-PAGE and immunoblotting with anti-HA or anti-Myc with development using ECL. Forms of Mdm30 mutated in its F-box domain (Cohen et al., 2008) were used in Fig. 3B,C to preclude binding to core SCF components.
For detection of Fzo1–HA ubiquitylation, yeast extracts were prepared using the NaOH–TCA lysis method (Avaro et al., 2002). Extracts were then boiled for 10 minutes at 70°C in SDS sample buffer and insoluble material removed by centrifugation for 5 minutes at 13,000 g. Resulting supernatants were diluted tenfold in IP buffer, and immunoprecipitations were performed as described above.
Glycerol growth analysis
For glycerol growth assays, cultures grown overnight in Dextrose medium were pelleted, resuspended at OD600=1, and diluted 1:10 five times in water. 3 μl of the dilutions were spotted on plates and grown for 4 days at 30°C.
Microscopy analysis of mitochondrial morphology
Mitochondrial morphology was scored in WT and mutant cells expressing matrix-targeted green fluorescent protein (mito-GFP; DCY538, supplementary material Table S1). Strains were grown in selective dextrose medium and scored in mid-log phase (OD600=0.7–1.0). Morphology phenotypes were assessed in at least 100 cells in three independent experiments. Data reported are the mean and s.d. of all experiments. Cells were visualized on a Zeiss Axioplan 2 imaging microscope (Carl Zeiss) with a 100× oil immersion objective.
In vivo mitochondrial fusion assay
Mitochondrial fusion in zygotes was examined essentially as described (Mozdy et al., 2000). Mitochondria in MATa or MATα haploid fzo1Δ or fzo1Δ mdm30Δ parents (DCY538 and JSY2294 backgrounds expressing WT or mutant Fzo1 protein) were labeled with mito–GFP or mito–RFP. Large-budded zygotes were scored for mitochondrial fusion 3–4 hours after mating. Mitochondrial fusion was quantified in 50–100 zygotes per strain in three separate experiments. Data reported are the mean and s.d. of all experiments.
In vitro tethering assay
For tethering reactions, mitochondria-enriched fractions were prepared from 500 OD600 units of DCY538 background cells expressing matrix-targeted GFP from their genome and JSY2293 background cells transformed with a plasmid containing matrix-targeted dsRed T4 [mdsRed T4, created by replacing GFP in pYX232-mtGFP (Westermann and Neupert, 2000) with dsRed T4 (Bevis and Glick, 2002)]. Briefly, 0.15 mg of total protein (as determined by Bradford assay) from mitochondria-enriched fractions obtained from WT, fzo1 S201N, fzo1Δ or mdm30Δ cells expressing either mito–GFP or mito–DsRed were mixed together (0.3 mg of protein total), concentrated by centrifugation, incubated for 30 minutes on ice and resuspended in NMIB buffer [0.6 M sorbitol, 5 mM MgCl2, 50 mM KCl, 100 mM potassium acetate, 20 mM HEPES, pH 7.4, 50 mM iodoacetamide (Sigma) and protease inhibitors (Complete Mini Protease Inhibitor; Roche)]. Importantly, canonical in vitro fusion reactions (Meeusen et al., 2004) could not be performed because fzo1 S201N, fzo1Δ and mdm30Δ cells do not grow in glycerol medium. For analysis of tethering by confocal fluorescence microscopy, aliquots from each reaction were fixed by resuspension in 2× volume of 8% paraformaldehyde in PBS, pH 7.4, for a minimum of 20 minutes and immobilized on microscope slides by mixing with an equal volume of 2% low melting point agarose in NMIB. The level of mitochondrial tethering in each reaction was then determined by calculating the ratio of red and green signals attached to each other over the total number of mitochondria (n>300). Error bars were determined by calculating the s.d. of two independent experiments. For analysis of tethering by electron microscopy, aliquots from each reaction were processed as described previously (Meeusen et al., 2004). Briefly, mitochondria were fixed in suspension by the addition of fixative (3% paraformaldehyde, 1.5% glutaraldehyde, 2.5% sucrose, contained in 100 mM cacodylate, pH 7.4, 4°C) for up to 24 hours. Mitochondria were washed in 100 mM cacodylate; spun into a tight pellet; and post-fixed in Palade's OsO4 for 1 hour, 4°C; and subsequently en bloc stained in Kellenberger's uranyl acetate overnight. The pellets were dehydrated through a graded series of ethanol; infiltrated with EPON; and allowed to polymerized 24–48 hours at 60°C. 80 nm sections were cut on a Leica UCT ultramicrotome; collected onto 400 mesh high-transmission grids; post-stained with lead citrate and uranyl acetate; and analyzed on a Tecnai 12 TWIN TEM operating at 100 kV. Images were collected with a Soft Imaging System Megaview III digital camera; and figures assembled in Adobe Photoshop with only linear adjustments of contrast and brightness. The level of mitochondrial tethering in each reaction was determined by calculating the ratio of attached and fused mitochondria over the total number of mitochondria (n>100). In experiments involving proteasome inhibition samples were treated with 50 μM each of lactacystin (Boston Biochem) and MG132 (Calbiochem) or with carrier for 30 minutes at 4°C before concentration by centrifugation and incubation on ice, as above. MG132 and lactacystin at 50 μM were found to each inhibit proteasome activity by at least 75% in mitochondrial-enriched fractions as assessed using the proteasome substrate Z-GGL-AMC (Enzo Life Sciences; data not shown).
Supplementary Material
Acknowledgments
We thank Greg J. Hermann (University of Utah) for generating anti-Fzo1 antisera and David C. Chan (California Institute of Technology), Benjamin S. Glick (University of Chicago), Agnes Delahodde (Institut de Génétique et Microbiologie) and Eric E. Griffin (California Institute of Technology) for generously providing reagents critical to this study and Y. C. Tsai for assistance with assessment of proteasome inhibition. We thank T. J. Turbyville and members of the Weissman and Shaw laboratories for discussions and N. Belgareh-Touze, Z. Kostova, M. B. Metzger and M. Nejati for review of this manuscript. E.A.A. was supported by grants from the United Mitochondrial Disease Foundation (UMDF# 08-064 Mitochondrial Fusion Defects in Neurological Disease) and NIH (T32 Institutional Training Grant # 5 T32 HD07576-22). Research in the Shaw laboratory and Weissman laboratories is supported by NIH GM53466 and the National Institutes of Health, National Cancer Institute, Center for Cancer Research, respectively. This work was also supported by a grant to A.M.W. from the Michael J. Fox Foundation for Parkinson's Research; a grant to A.M.W. and M.H.G. from the USA-Israel Bi-National Science Foundation (BSF); and a NIH grant to J.M.M. (NCRR grant 1 S10 RR023454-01). Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/9/1403/DC1
References
- Amiott E. A., Cohen M. M., Saint-Georges Y., Weissman A. M., Shaw J. M. (2009). A mutation associated with CMT2A neuropathy causes defects in Fzo1 GTP hydrolysis, ubiquitylation, and protein turnover. Mol Biol. Cell 20, 5026-5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avaro S., Belgareh-Touze N., Sibella-Arguelles C., Volland C., Haguenauer-Tsapis R. (2002). Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast 19, 351-371 [DOI] [PubMed] [Google Scholar]
- Bevis B. J., Glick B. S. (2002). Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20, 83-87 [DOI] [PubMed] [Google Scholar]
- Cohen M. M., Leboucher G. P., Livnat-Levanon N., Glickman M. H., Weissman A. M. (2008). Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 19, 2457-2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr M., Escobar-Henriques M., Merz S., Geimer S., Langer T., Westermann B. (2006). Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol. Biol. Cell 17, 3745-3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. (2006). Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 173, 645-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S., Weinbach N., Westermann B. (2003). Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell 14, 2303-2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. E., Chan D. C. (2006). Domain interactions within Fzo1 oligomers are essential for mitochondrial fusion. J. Biol. Chem. 281, 16599-16606 [DOI] [PubMed] [Google Scholar]
- Heo J. M., Livnat-Levanon N., Taylor E. B., Jones K. T., Dephoure N., Ring J., Xie J., Brodsky J. L., Madeo F., Gygi S. P., et al. (2010). A stress-responsive system for mitochondrial protein degradation. Mol. Cell 40, 465-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann J. A., Hinshaw J. E. (2009). Dynamins at a glance. J. Cell Sci. 122, 3427-3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. (2007). The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751-780 [DOI] [PubMed] [Google Scholar]
- Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J. (2005). Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Eura Y., Mihara K. (2004). Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 117, 6535-6546 [DOI] [PubMed] [Google Scholar]
- Kleijnen M. F., Kirkpatrick D. S., Gygi S. P. (2007). The ubiquitin-proteasome system regulates membrane fusion of yeast vacuoles. EMBO J. 26, 275-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S. A., Kaiser J. T., Chen H., McCaffery J. M., Chan D. C. (2004). Structural basis of mitochondrial tethering by mitofusin complexes. Science 305, 858-862 [DOI] [PubMed] [Google Scholar]
- Liesa M., Palacin M., Zorzano A. (2009). Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89, 799-845 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961 [DOI] [PubMed] [Google Scholar]
- Low H. H., Lowe J. (2006). A bacterial dynamin-like protein. Nature 444, 766-769 [DOI] [PubMed] [Google Scholar]
- Low H. H., Sachse C., Amos L. A., Lowe J. (2009). Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139, 1342-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J. M., Nunnari J. (2004). Mitochondrial fusion intermediates revealed in vitro. Science 305, 1747-1752 [DOI] [PubMed] [Google Scholar]
- Moreau V., Galan J. M., Devilliers G., Haguenauer-Tsapis R., Winsor B. (1997). The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol. Biol. Cell 8, 1361-1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M., Shaw J. M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Kung C., Shokat K. M., Tansey W. P. (2005). The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell 120, 887-899 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. (2005). Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39, 503-536 [DOI] [PubMed] [Google Scholar]
- Ota K., Kito K., Okada S., Ito T. (2008). A proteomic screen reveals the mitochondrial outer membrane protein Mdm34p as an essential target of the F-box protein Mdm30p. Genes Cells 13, 1075-1085 [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. (eds) (1986). Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; [Google Scholar]
- Varshavsky A. (1996). The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93, 12142-12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewski M., Scorrano L. (2009). The changing shape of mitochondrial apoptosis. Trends Endocrinol. Metab. 20, 287-294 [DOI] [PubMed] [Google Scholar]
- Westermann B. (2010). Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872-884 [DOI] [PubMed] [Google Scholar]
- Westermann B., Neupert W. (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421-1427 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.