Abstract
Whereas glycoproteomic studies provide unique opportunities for cancer research, it has been necessary to develop specific methods for analysis of oncologically interesting glycoproteins. We describe a general, multimethodological approach for quantitative glycoproteomic analysis of fucosylated glycoproteins in human blood serum. A total of 136 putative fucosylated glycoproteins were identified with very high confidence in three clinically relevant sample pools (N=5 for each), with a mean coefficient of variation of 3.1% observed for replicate analyses. Two samples were collected from subjects diagnosed with esophagus disease states, high-grade dysplasia (HGD) plus esophageal adenocarcinoma (EAC), while the third sample was representative of a disease-free (DF) condition. Some glycoproteins, observed to be significantly upregulated in EAC, i.e. more than 2-fold higher than in the DF condition, are briefly discussed. Further investigation will be necessary to validate these findings; however, the method itself is demonstrated to be an effective tool for quantitative glycoproteomics of clinical samples.
Introduction
As a set of strategies gradually developed to study the phenotypic expression of genes, proteomics methodologies have been utilized to generate massive, information-rich datasets [1]. The extreme complexity of most biological materials has driven researchers to develop specialized techniques that facilitate focused investigation of interesting subproteomes. Several subproteomes have been defined by the type of post-translational modifications (PTMs) present, since PTMs have been shown to significantly impact protein function. They also present chemical moieties that can be targeted by various types of separation techniques for a more focused investigation. Example PTMs include phosphorylation, ubiquitination, and glycosylation [2–4]. The authors have had a particular interest in glycosylation, i.e. enzymatic addition of a complex carbohydrate to a protein, because glycans have been historically implicated as the initial points of contact in both intermolecular and intercellular interactions [5].
Changes in glycosylation have been implicated in the onset of several diseases, including different types of cancer [6–9]. In particular, changes in the structure and abundance of fucosylated glycans have been linked to the progression of breast, esophageal, and liver cancers [6, 10–12]. It has been shown that particular types of glycosylation can be targeted for enrichment through the use of lectins, proteins that exhibit affinity toward specific glycans [13–15]. With the intention of targeting fucosylated glycoproteins as the molecules of potentially high significance in activities that coincide with the onset of certain types of cancer, we have developed an approach for label-free, quantitative glycoproteomics in which we have employed two lectins, Aleuria aurantia lectin (AAL) and Lotus tetragonolobus agglutinin (LTA), to enrich fucosylated glycoproteins present in human blood serum and then performed further protein fractionation with liquid chromatography (LC) using superficially porous, reversed-phase packing. Finally, trypsin-digested proteins are analyzed through the generated peptides by LC-MS/MS with a high-resolution mass spectrometer. In this publication, the value of this general approach is being demonstrated through an initial study of esophageal adenocarcinoma (EAC) and a related condition, high-grade dysplasia (HGD), while the results are being compared to a disease-free (DF) condition. It is important to note that this is a small-scale study, and that a larger study involving significantly more samples will be the next step in our ongoing work in this area.
Esophageal adenocarcinoma is a cancer that originates in the epithelial membrane lining the esophagus. The five-year relative survival rate for cancer of the esophagus from 1999–2005 was only 16.8% [16]. However, only the early detection of cancer in situ, synonymous with HGD, has allowed for effective treatment by surgical resection with excellent prognoses [17]. Unfortunately, early detection remains difficult with the current methods limited to computer tomography [18] (CT scan) and endoscopic ultrasound (EUS) [19], with EUS being the better of the two techniques, providing between 70–80% accuracy for regional nodal staging. The discovery of glycoprotein markers in blood serum could potentially lead to the development of assays with similar or better sensitivity and accuracy for clinical screening.
Experimental Procedures
Pooled Human Blood Serum Samples
This study was approved by the Indiana University institutional review board. A set of serum samples from patients diagnosed with high grade dysplasia (HGD), and esophageal adenocarcinoma (EAC) were collected along with a group of disease free (DF) individuals that were used as a control. Venous blood samples were taken in the morning's fasting state, being collected with the minimal stasis in evacuated tubes. After at least 30 min, but within 2 h, the tubes were centrifuged at 20 °C for 12 min at 1200 g, and the sera were stored in plastic vials at −80 °C until the time of glycoproteomics analyses. 50-μL aliquots of each of the human blood serum samples were pooled according to disease state (N=5 for each sample pool): EAC, HGD, and DF.
Immunoaffinity Chromatography
The depletion of seven highly abundant proteins, namely albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin and fibrinogen from serum samples, was performed using the Multiple Affinity Removal System (MARS) Hu-7 column purchased from Agilent Technologies (Palo Alto, CA) and utilized according to the manufacturer's instructions. Briefly, pooled human serum samples were diluted 4-times with Buffer A, and applied to a cellulose acetate spin filter (0.22 μm) from Corning, Inc. (Corning, NY). A 200-μL sample was then injected onto the MARS column with an ÄKTA purifier liquid chromatograph from GE Healthcare (Piscataway, NJ), and the flow-through fraction was collected at a flow rate of 1 mL/min for 1.1 minutes. Whereas a total of 250 μL of blood serum (50 μL per subject) was immunodepleted for each disease state, the pooled samples were combined and desalted in a spin concentrator with three 2-mL washes of 50 mM ammonium bicarbonate by centrifugation at 4500 g before being concentrated to a final volume of 500 μL in 50 mM ammonium bicarbonate.
Serial Lectin Affinity Chromatography
Prior to their use, agarose-bound lectins purchased from Vector Laboratories (Burlingame, CA) were prepared by washing three times with 500 μL of the binding buffer (Tris, pH 7.5, 0.15 M NaCl, 0.1 M Ca2+, 0.08% NaN3) to remove lactose used for stabilizing the media during storage. Each depleted blood serum pool was subjected to a BCA protein assay obtained from Pierce (Rockford, IL) as a kit to determine protein concentration (data not shown). For each disease state, a sample volume containing 1 mg of protein was added to a 250-μL gel bed of agarose beads coupled to Aleuria aurantia lectin (AAL) (3mg/mL, lectin/gel volume) in a 1.5 mL tube. The sample volume was brought to 500 μL with the binding buffer. The sample-lectin mixture was subsequently incubated at 4 °C with gentle agitation for 18 hours. Next, the AAL-unbound fraction was removed and applied to a 270-μL agarose bed coupled to Lotus tetragonolobus lectin (LTL) (3mg/mL, lectin/gel volume), while the mixture was incubated again at 4 °C with gentle agitation for 18 hours. Following a rinse with 500 μL of deionized water, all lectin-bound proteins were eluted in the same way with a 500-μL aliquot of 0.1 M acetic acid at 4 °C with agitation for 1 hour. The gel beds were washed a second time with 500 μL of 0.1 M acetic acid and the two elution volumes were filtered with a particle filter (0.22 μm), then combined and frozen at −70 °C. Frozen elution fractions were lyophilized and resuspended in 100 μL of 50 mM ammonium bicarbonate. The AAL and LTL fractions were then combined to create a serial lectin-enriched mixture of glycoproteins for each disease state, DF, HGD, and EAC. Lectin-enriched glycoprotein samples were subjected to BCA Protein Assay (see supporting information Table S1) to determine protein concentration prior to protein fractionation as described below.
Reversed-phase Protein Fractionation with a Superficially Porous Stationary Phase
A volume of lectin-enriched sample that corresponded to 100 μg of glycoproteins was vacuum-centrifuged and resuspended in 100 μL of 8 M urea. The sample was then injected onto a superficially porous Halo C8 column (Advanced Materials Technology, Wilmington, DE) (2.7 μm, 1.7 μm fused-core and a 0.5 μm porous shell, 4.6 mm × 10 cm) using a Dionex 680 HPLC instrument (Sunnyvale, CA). The column was heated to 45 °C to increase the efficiency of separation. The solvent system consisted of two solvents, 0.1 % trifluoroacetic acid (TFA) in water (solvent A) and 0.1 % TFA in acetonitrile (solvent B). The sample was loaded onto the Halo column in 3 % solvent B, then separated over a stepwise gradient from 3–15 % B in 6 minutes, 15–55 % B in 40 minutes, 55–100 % B in 45 minutes, held at 100 % B for 4 minutes, and then readjusted to 3 % solvent B over 15 minutes. Separation was performed at a flow rate of 0.75 mL/min. Proteins were collected in fifteen 2-min fractions, beginning at 28 min of retention time. Fractions were vacuum-centrifuged and resuspended in 20 μL 50 mM ammonium bicarbonate.
Trypsin Digestion
Samples were denatured initially in 8 M urea prior to protein fractionation, as described above. The protein fractions resuspended in 20 μL of 50 mM ammonium bicarbonate (also as described above) were reduced in 5 mM dithiothreitol (DTT) at 60 °C for 30 minutes. Next, the samples were alkylated in 20 mM iodoacetamide (IAA) at room temperature in the dark for 30 min. Following alkylation, a 0.5-μL aliquot of 200 mM DTT was added to quench the alkylation reaction and the sample was incubated for an additional 30 minutes at room temperature. Finally, a 1-μL aliquot of trypsin (1 mg/mL) was added to each fraction to ensure a minimum enzyme-to-substrate ratio of 1:20, and the samples were incubated overnight (ca. 18 hours) at 37 °C.
Analysis of Tryptic Peptides by Nano-LC-ESI-MS/MS
Peptide digests were separated by reversed-phase liquid chromatography (RPLC) with an Ultimate 3000 LC system (Dionex, Sunnyvale, CA) fitted with a capillary column (5 μm, 0.075 mm × 15 cm) packed in-house with Jupiter Proteo C12 silica (Phenomenex, Torrance, California). The mobile-phase solvent system consisted of an aqueous solution containing 3 % acetonitrile, 0.1 % formic acid (FA) in water (solvent A) and an organic solution containing 0.1 % FA in acetonitrile (solvent B). Peptides were eluted from the column using a stepwise gradient from 3 to 55 % B over 45 min, 55 to 80 % B over 10 min, 80 % B for 10 min, then 80 to 3 % B over an additional 5 min to re-establish the initial conditions. A 45-minute, high organic washing step was performed after each experiment to ensure that the results were not influenced by possible carryover. The liquid chromatography system was coupled to an LTQ-Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA) via the nanospray ESI connection. Ionized peptides were subjected to high-resolution MS scans in the Orbitrap mass analyzer and the five most abundant precursor ions were selected for collision-induced dissociation (CID) experiments. A dynamic exclusion window of 30 seconds was utilized to prevent dominant precursor ions from being selected more than once for CID in the described time frame, allowing for the fragmentation of less abundant precursors.
Database Searching and Protein Quantification
RAW files generated by Xcaliber software v2.0.7 (Thermo Scientific) were used to create MASCOT generic files (.MGF) for database searching with MASCOT. The .MGF files were generated using TURBO RAW2MGF, a utility developed in-house as part of a software bundle [20]. The MASCOT search engine was used to search the UniProt protein database, and search results were filtered with ProteinParser according to the following parameters: 1 missed cleavage allowed from tryptic digestion, ± 0.03 m/z tolerance for precursors, +2 and +3 charges, oxidation of methionine (variable modification), ion score ≥ 30, expect ≤ 0.1, accept only bold red queries, and a minimum peptide mass of 600.00 Da. Additionally, .MGF files were searched against a randomized version of the UniProt database that was previously generated using the decoy database generator utility available from Matrix Science (Boston, MA) and filtered using the same parameters as described above for the regular searches. The identified peptides from each LC-MS run were combined into a master peptide file [20], which was used for quantification of proteins as described previously. Briefly, instrument raw files were converted from .RAW format to .mzXML using the ReAdW utility that is freely available at http://sourceforge.net/projects/sashimi/files. LC peak areas for identified peptides were then defined by their edges and integrated using ProteinQuant [20]. The following parameters were defined in ProteinQuant: ±0.03 m/z, an apex reassignment window of ± 3.5 minutes, peak width ≤ 1 minute with an intensity threshold of 3-times the baseline intensity, and peptide areas were normalized to the total area for all identified peptides. Protein areas were reported as the sum of all peptide areas identified for each protein.
Results and Discussion
General Considerations
Quantitative glycoproteomics measurements are inherently challenging to perform. Each preparation step in a proteomic experiment provides opportunities for compound measurement discrepancies introduced in the previous steps. The desire for a robust, controlled proteomic sample preparation methodology is easily strained by the need to target glycoproteins, and, in particular, the subset of glycoproteins that are pertinent to a given study. Several approaches have been developed to fill this need, including hydrazide coupling, agarose enrichment, and lectin affinity techniques [15, 21–24], As described by Aebersold and coworkers, hydrazide coupling provides a means to covalently link cis diols, which are first oxidized to aldehydes with periodate, to hydrazide groups that are attached to the support media [21]. This approach is attractive because it utilizes well-understood chemistry to immobilize analytes, and it also allows the researcher to subject the covalently linked glycoproteins to a harsh wash, significantly limiting the presence of nonspecifically bound molecules in the eluted fraction. A second method has been reported [22], in which agarose gel was used to enrich glycoproteins through the use of a normal-phase solvent system. Briefly, the protein mixture was applied to an agarose gel bed in butanol/ethanol/water (4:1:1 v/v), which provided a relatively nonpolar solution environment that encouraged hydrophilic interaction between glycoproteins and the agarose media. After washing steps to remove non-interactive proteins, presumably non-glycosylated ones, the enriched glycoproteins were eluted with a polar solvent containing ethanol/water (1:1 v/v). Analyte recovery for this method was reported to be 30–50%. While both of these techniques provide a general method for glycoprotein enrichment, only the third strategy, namely lectin affinity, provides the potential to target subsets of glycans that may be differentially expressed in a particular disease state, in a specific way. Lectin affinity chromatography, as it was performed here, has been previously shown to be a repeatable enrichment technique for a comparison of multiple samples that had been individually enriched and quantified in separate label-free proteomic experiments [25]. To illustrate this unique benefit, we have utilized two lectins (AAL and LTA), in a serial fashion, to enrich fucosylated glycoproteins, which have repeatedly been implicated in the progression of several different types of cancers [6, 10–11].
Controlling Multistep Label-free Experiments for Quantification
A clear challenge for label-free proteomics has been the need to normalize measurements made across several LC-MS/MS experiments for comparison [26]. A plethora of techniques for normalization have been reported, including the use of a global normalization factor [27], spiked peptides derived from a standard protein or a set of proteins [28], and a separate reference LC-MS/MS data file used to perform linear regression with a set of “housekeeping” proteins present in each sample [29]. An approach similar to the first possibility, i.e. normalization to all measured features has been utilized in our experiments. Through first compiling a master file of all identified peptides, including their m/z and retention time information, it is possible to measure the relative abundance of each peptide contained therein as described previously [20]. Such an approach requires reproducible chromatography, and it becomes more effective with the use of MS at high resolution. Not surprisingly, the recent commercial availability of several high-resolution instruments has prompted a renewed interest in the label-free proteomics techniques, which benefit greatly from these technological advancements.
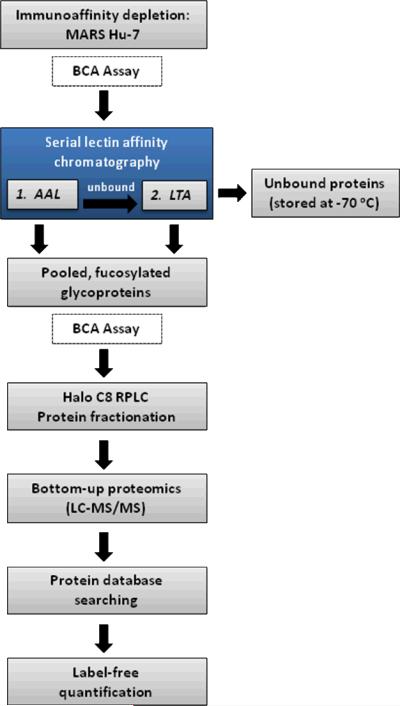
The multi-step methodologies used to target glycoproteins require corresponding measures be taken to ensure that each step in the process is controlled to avoid experimental bias. The BCA protein assay has been used here to mass-normalize the sample for each of the three pooled sera twice—once prior to serial lectin enrichment, and a second time before reversed-phase protein fractionation (as indicated in Figure 1)—to ensure that the downstream LC-MS/MS analyses will generate comparable ion currents. Since it is expected that a different amount of fucosylated glycoproteins will be bound to AAL and LTA lectins for each of the sample pools (as unusual fucosylation has been known to change as a function of disease progression), BCA analysis following this step ensured that the same amount of protein was subjected to the reversed-phase fractionation.
Figure 1.
Quantitative glycoproteomic workflow as applied to enrichment of fucosylated glycoproteins.
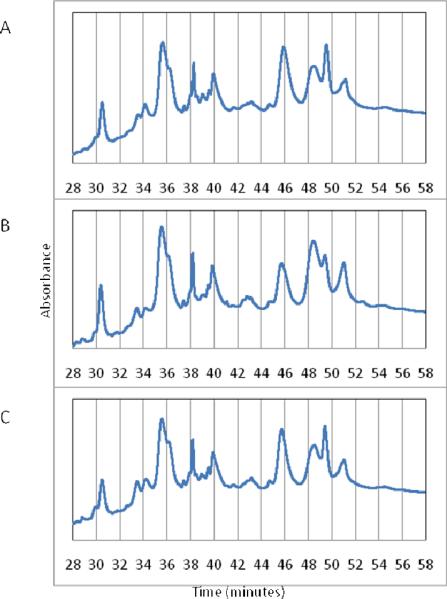
Next, it was necessary to consider that small shifts in the retention time during fractionation could potentially change the fraction in which a particular glycoprotein becomes collected. These small shifts, if they occurred near the edge (start or end) of a fraction, will change the fraction in which a particular glycoprotein was collected. It was possible to account for these shifts quantitatively by summing the LC-MS protein signal across all collected fractions, but it must be acknowledged that any shifts which resulted in elution prior to collection of the first fraction or after collection of the final one would have introduced a significant bias in the procedure. Furthermore, significant shifts in the chromatographic alignment would bias downstream global normalization of LC-MS peptide data for individual fractions. Therefore, for such an approach to work for a glycoprotein separation, it was necessary for the chromatograms to be comparable over the span of the collection window. Moreover, proteins with elution profiles that overlapped the beginning of the first fraction, or the ending of the last one, would have to be considered incomplete and not suitable for quantitation. To reduce chromatographic shifting, a blank sample was run prior to each of the three fractionation experiments, ensuring that initial fractionation conditions were highly similar. Possible changes in protein fractionation due to the reasons mentioned above notwithstanding, the chromatograms for each of the three pooled samples separated here were quite comparable (Figure 2).
Figure 2.
Reversed-phase protein fractionation chromatograms for a) the disease-free pool (DF), b) high-grade dysplasia (HGD), and c) esophageal adenocarcinoma (EAC). Vertical gridlines indicate the edges of 2-min fractions.
Label-free Protein Quantitation
Proteins were quantified by summing the peak areas for extracted ion chromatograms (± 0.03 m/z) of the identified peptides. Understandably, this method will not provide a totally accurate comparison for two separate LC-MS/MS experiments, in which different peptides are identified, because each peptide has a unique ionization efficiency determined by the gas-phase basicity, and, perhaps of greater significance, an LC-MS/MS experiment that results in a higher number of peptide identifications will result in a protein measurement that is based on a higher number of extracted ion profiles. However, the use of a master peptide file will address both of these concerns by ensuring that each LC-MS/MS data set is mined for all peptides that are endogenous to the sample [20]. In this way, each protein measurement is computed from the summation of the same identified peptides, thereby facilitating cross-experiment comparison of measurements. This method is not without limitation, as it cannot be applied to a comparison of two samples that have significantly different protein compositions, nor can it be used in cases where separation conditions vary greatly, although this second limitation can be partly compensated with increased resolution and accuracy of peptide m/z measurements, as discussed by Smith and coworkers [30].
Profiling of AAL/LTA Enriched Glycoproteins
The MASCOT parameters used for peptide identification (described in detail above) were very stringent, including an ion score ≥ 30 and a peptide mass tolerance of ± 0.03 Da. A decoy UniProt database in which all peptide sequences were randomly shuffled was used to estimate the false discovery rate, and the search resulted in zero peptides accepted as positive identifications, so we could conservatively estimate the false discovery rate to be below 0.1%. A total of 136 glycoproteins and glycoprotein families were quantified from 1224 unique peptide identifications. Each sample pool was measured in triplicate by LC-MS/MS, and the mean CV for all 408 protein measurements was 3.14 %. The range of protein areas measured spanned over five orders of magnitude, 3.61 × 10−5 to 1.26 (normalized values), and while this is not directly reflective of the range of endogenous protein abundances, it suggests that the methodology is very suitable for the analysis of glycoproteins with considerably different natural abundances in serum. Additionally, the quality of measurements was observed to be consistent over this range, as can be seen in the two graphs presented in Figure 3. Glycoproteomic experiments are attractive only because they offer the ability to measure large numbers of analytes simultaneously and with extremely high confidence of identification. The low mean CV for the entire group of measurements reported herein, coupled with the wide range of measured protein areas, exemplifies the potential for this experimental approach to provide useful data for future targeted research. To the best of our knowledge, these measurements represent some of the highest quality relative quantitative measurements for LC-MS of glycoproteins.
Figure 3.
Relative abundance comparison of fucosylated glycoproteins for a) the ten highest area proteins and b) ten lowest area proteins. Error bars indicate standard deviation in the measurement (N=3).
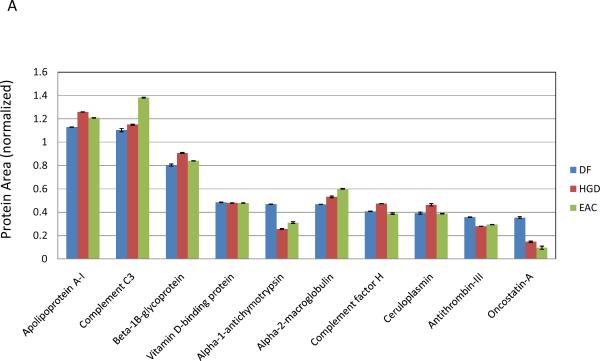
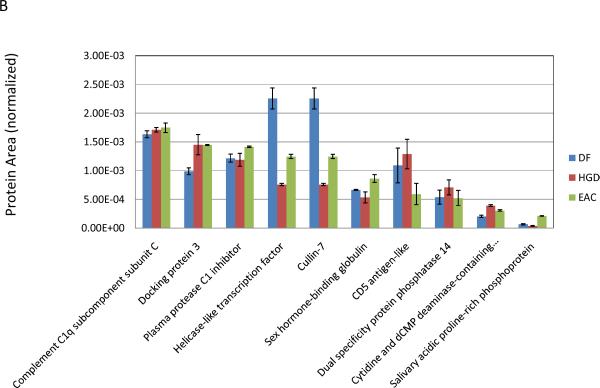
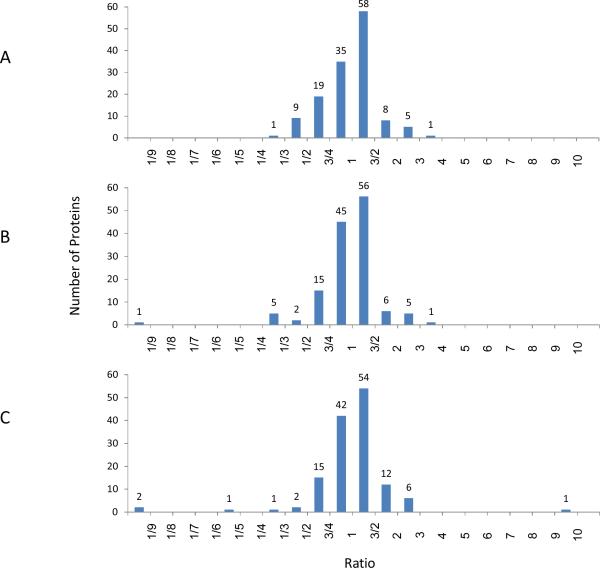
Three ratios, HGD:DF, EAC:DF, and EAC:HGD, have been calculated from the protein area values presented in the supplemental table (S2). For each of the three comparisons, the percentage of glycoproteins that were observed to be within 1.5- and 2-fold difference in their relative intensity was calculated; they were 68 and 89% for HGD:DF, 74 and 90% for EAC:DF, and 70 and 90% for EAC:HGD for 1.5- and 2-fold differences, respectively. The distribution of expression ratios was visualized with three histograms (Figure 4). With a threshold of 2-fold difference set for identification of upregulated proteins, it can be seen that the majority of protein concentration levels remain relatively stable. Still a number of differentially expressed analytes were observed, including seven proteins that were upregulated greater than 2-fold in EAC compared to the DF group, and of these seven proteins, six were also over-expressed in HGD. HGD is a condition described as a “carcinoma in situ”, and in 30–50% of subjects diagnosed with HGD, the condition portends EAC (i.e. metastasis of dysplastic cells) [31]. It is therefore reasonable that most proteins present at raised levels in EAC were also observed in somewhat lesser, but still increased levels in HGD. Proteins that were upregulated greater than 2-fold in EAC are summarized in Table 1, with the corresponding observation in HGD. Although this data set is not sufficiently large to make any statistically significant observations regarding changes in protein expression, it offered a valuable survey of a subset of glycoproteins that could be used to design a well-informed, targeted study of potentially interesting glycoproteins.
Figure 4.
Histograms that illustrate the distribution of protein ratios for the three comparisons: a) HGD:DF, b) EAC:DF, and c) EAC:HGD. A trend of increased abundance of fucosylated glycoproteins was observed in both HGD and EAC, as compared to DF.
Table 1.
Protein signals, coefficients of variation (CV) (N=3) and relative abundance ratios for the six glycoproteins upregulated more than 2-fold in EAC with respect to DF.
| Protein | DF | HGD | EAC | HGD:DF | EAC:DF | EAC:HGD | |||
|---|---|---|---|---|---|---|---|---|---|
| Signal | CV (%) | Signal | CV (%) | Signal | CV (%) | ||||
| Collagen alpha-1(I) chain | 2.40E-03 | 2.21 | 5.13E-03 | 2.19 | 9.00E-03 | 0.92 | 2.14 | 3.75 | 1.76 |
| EMILIN-2 | 7.81E-03 | 0.28 | 1.59E-02 | 1.60 | 2.00E-02 | 2.15 | 2.04 | 2.56 | 1.25 |
| Fetuin-B | 1.41E-03 | 24.86 | 1.93E-03 | 12.89 | 3.39E-03 | 9.27 | 1.37 | 2.41 | 1.76 |
| Beta-2-glycoprotein 1 | 1.13E-01 | 0.99 | 2.23E-01 | 0.54 | 2.63E-01 | 0.64 | 1.97 | 2.32 | 1.18 |
| Kynurenine--oxoglutarate transaminase 3 | 3.40E-03 | 1.86 | 7.41E-03 | 2.68 | 7.45E-03 | 5.13 | 2.18 | 2.19 | 1.01 |
| Ig kappa chain V–III region B6 | 1.31E-02 | 1.49 | 1.45E-02 | 2.99 | 2.79E-02 | 3.24 | 1.11 | 2.13 | 1.93 |
Fucosylated Glycoproteins Dysregulated in HGD and EAC
While the results observed here have yet to be validated through additional samples, several of the proteins listed in Table S2 have been previously reported to be upregulated as a result of inflammatory response, which is often observed as coincident with cancer and other chronic disease conditions. These include some fucosylated acute-phase glycoproteins such as haptoglobin and α-1-acid glycoprotein, which were observed upregulated and with altered glycosylation by Rudd and coworkers previously [32]. While these proteins could be of high interest for future studies of EAC, they are generally accepted to be dysregulated in cancer conditions as well as in general inflammatory response, so they will not be addressed further in this discussion. In addition to the acute-phase glycoproteins, a number of new candidate markers were also identified. A brief discussion of some of the most notable observations is provided below.
Fetuin-B
Fetuin B is a glycoprotein and is a member of the cystatin protein family, specifically one of the type 3 cystatins. Its specific function is still unclear, though several cystatins have been implicated as regulators of angiogenesis and thereby associated with cancer metastasis [33]. In this study, it was observed to be somewhat over-expressed in HGD (1.37, HGD:DF), but significantly more so in EAC (2.41, EAC:DF). A difference of this magnitude is readily identifiable by most established protein quantitation techniques, and while fetuin-B may not be specific for EAC, the magnitude of over-expression could be an indicator of the disease progression.
EMILIN-2
Elastin microfibril interface located protein 2 (EMILIN-2) is an extracellular matrix glycoprotein, which has been recently reported to regulate the extrinsic apoptotic pathway [34] and to be a negative regulator of tumor cell growth. Our observation that it is upregulated in HGD and EAC could indicate that it is a component of the host immune response to dysplastic cell proliferation. The extent to which EMILIN-2 is observed upregulated as a result of the lectin enrichment performed here could be tested by comparison to a proteomic analysis of the entire serum proteome, provided that peptides from highly abundant proteins did not mask EMILIN-2 peptides during the electrospray process. More directly, an enzyme-linked immunosorbent assay (ELISA) could be used to target the protein with little bias toward its glycosylation, assuming the antibody binding was not inhibited by altered glycosylation.
Collagen alpha-1(I) chain
Another extracellular matrix protein, collagen type 1 is a structural protein to which tissue cells adhere for stability. It has a single known site of N-linked glycosylation at residue 1365. Over-expression of type 1 collagen has been linked to angiogenesis and the development of invasive melanoma [35]. It has also been implicated in a number of other cancers, including breast, skin, colon, and prostate cancers [36–40].
Conclusions
Shotgun proteomics, while a valuable tool for measuring a large number of proteins simultaneously, has repeatedly been stifled by its many limitations, primarily as they relate to the challenge of studying lower-abundance proteins in a biological mixture. This limitation has driven researchers to explore possibilities that somewhat lessen the complexity of these mixtures and, at the same time, allow us to target the proteins of highest interest. Glycoproteins, recognized as one of the most diverse groups of post-translationally modified proteins, remain an attractive subset of the proteome for continued investigation. Through lectin enrichment chromatography, it is possible to further focus a proteomic investigation to look at a specific subset of glycoproteins according to a specific type of glycosylation. Here, we have targeted fucosylation, employing AAL and LTA lectins for the unique affinity each has to particular glycosidic linkages of fucose as a substituent in N-linked glycans. Through our enrichment procedures, we have been able to quantify 136 glycoproteins and glycoprotein families with an average CV of 3.14%. The subset of glycoproteins quantified here has spanned a range of measured values greater than 5 orders of magnitude, indicating that removal of a significant number of high-abundance proteins from human serum in tandem with a further protein fractionation can expose a wide dynamic range of potentially interesting analytes. That being said, a method such as this involves significant sample preparation, and it would not be attractive for large-scale analyses of many dozens or hundreds of samples. It has merit as a survey tool that will allow researchers to identify potentially interesting glycoproteins that can subsequently be measured through the established assay techniques such as ELISA.
The glycoproteomic method has been applied to an initial study of HGD and EAC, and its utility has been demonstrated. Several potentially interesting proteins have been identified in this study. We believe they represent good candidates for future studies of serum markers for EAC progression.
Supplementary Material
Acknowledgements
This work was primarily supported by grant No. CA128535 from the National Cancer Institute, U.S. Department of Health and Human Services. Further support was provided by NIH/NCRR - National Center for Glycomics and Glycoproteomics (NCGG), Grant No. RR018942. The authors appreciate the opportunity to use the instrumental facilities available at Indiana University through the METACyt Biochemical Analysis Center.
References
- [1].Kumar C, Mann M. FEBS Lett. 2009;583:1703–1712. doi: 10.1016/j.febslet.2009.03.035. [DOI] [PubMed] [Google Scholar]
- [2].Wilkinson KD. Anticancer Drug. Des. 1987;2:211–229. [PubMed] [Google Scholar]
- [3].Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Burnett G, Kennedy EP. J. Biol. Chem. 1954;211:969–980. [PubMed] [Google Scholar]
- [5].Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- [6].Kyselova Z, Mechref Y, Kang P, Goetz JA, et al. Clin. Chem. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- [7].Saldova R, Wormald MR, Dwek RA, Rudd PM. Dis. Markers. 2008;25:219–232. doi: 10.1155/2008/601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goldman R, Ressom HW, Varghese SA, Bascug G, et al. Cancer Biomarkers. 2008;4:143–144. [Google Scholar]
- [9].Tabares G, Radcliffe CM, Barrabes S, Ramirez M, et al. Glycobiology. 2006;16:132–145. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- [10].Mechref Y, Hussein A, Bekesova S, Pungpapong V, et al. J. Proteome Res. 2009;8:2656–2666. doi: 10.1021/pr8008385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dai Z, Zhou J, Qiu SJ, Liu YK, Fan J. Electrophoresis. 2009;30:2957–2966. doi: 10.1002/elps.200900064. [DOI] [PubMed] [Google Scholar]
- [12].Hamid UMA, Royle L, Saldova R, Radcliffe CM, et al. Glycobiology. 2008;18:1105–1118. doi: 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- [13].Cummings RD, Kornfeld S. J. Biol. Chem. 1982;257:1235–1240. [PubMed] [Google Scholar]
- [14].Plavina T, Wakshull E, Hancock WS, Hincapie M. J. Proteome Res. 2007;6:662–671. doi: 10.1021/pr060413k. [DOI] [PubMed] [Google Scholar]
- [15].Madera M, Mechref Y, Novotny MV. Anal. Chem. 2005;77:4081–4090. doi: 10.1021/ac050222l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Horner MJ, Ries LAG, Krapcho M, Neyman N, et al. SEER Cancer Statistics Review 1975–2006 (webpage) National Cancer Institute; 2009. http://seer.cancer.gov. [Google Scholar]
- [17].Esophageal Cancer Treatment: General Information (webpage) National Cancer Institute; 2009. http://www.cancer.gov. [Google Scholar]
- [18].Ziegler K, Sanft C, Zeitz M, Friedrich M, et al. Gut. 1991;32:16–20. doi: 10.1136/gut.32.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vazquez-Sequeiros E, Norton ID, Clain JE, Wang KK, et al. Gastrointest. Endosc. 2001;53:751–757. doi: 10.1067/mge.2001.112741. [DOI] [PubMed] [Google Scholar]
- [20].Mann B, Madera M, Sheng Q, Tang H, Mechref Y, Novotny MV. Rapid Commun. Mass Spectrom. 2008;22:3823–3834. doi: 10.1002/rcm.3781. [DOI] [PubMed] [Google Scholar]
- [21].Zhang H, Li X.-j., Martin DB, Aebersold R. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- [22].Wada Y, Tajiri M, Yoshida S. Anal. Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- [23].Cummings RD, Kornfeld S. J. Biol. Chem. 1982;257:11235–11240. [PubMed] [Google Scholar]
- [24].Yang Z, Hancock WS. J. Chromatogr., A. 2004;1053:79–88. [PubMed] [Google Scholar]
- [25].Madera M, Mann B, Mechref Y, Novotny MV. J. Sep. Sci. 2008;31:2722–2732. doi: 10.1002/jssc.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Callister SJ, Barry RC, Adkins JN, Johnson ET, et al. 2006:277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang WX, Zhou HH, Lin H, Roy S, et al. Anal. Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- [28].Riter LSH, Barry D, Gooding, Karen M, Julian, Randall K. J. Proteome Res. 2005;4:153–160. doi: 10.1021/pr049819s. [DOI] [PubMed] [Google Scholar]
- [29].Fang RH, Elias DA, Monroe ME, Shen YF, et al. Mol. Cell. Proteomics. 2006;5:714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- [30].Pasa-Tolic L, Masselon C, Barry RC, Shen YF, Smith RD. Biotechniques. 2004;37:621–+. doi: 10.2144/04374RV01. [DOI] [PubMed] [Google Scholar]
- [31].Miller CT, Moy JR, Lin L, Schipper M, et al. Clin. Cancer Res. 2003;9:4819–4825. [PubMed] [Google Scholar]
- [32].Saldova R, Royle L, Radcliffe CM, Hamid UMA, et al. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- [33].Lee C, Bongcam-Rudloff E, Sollner C, Jahnen-Dechent W, Claesson-Welsh L. Front. Biosci. 2009;14:2911–2922. doi: 10.2741/3422. [DOI] [PubMed] [Google Scholar]
- [34].Mongiat M, Ligresti G, Marastoni S, Lorenzon E, et al. Mol. Cell. Biol. 2007;27:7176–7187. doi: 10.1128/MCB.00696-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].van Kempen LCLT, Rijntjes J, Mamor-Cornelissen I, Vincent-Naulleau S, et al. Int. J. Cancer. 2008;122:1019–1029. doi: 10.1002/ijc.23147. [DOI] [PubMed] [Google Scholar]
- [36].RonnovJessen L, Petersen OW, Bissell MJ. Physiol. Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- [37].Ohtani H. Pathol. Int. 1998;48:1–9. doi: 10.1111/j.1440-1827.1998.tb03820.x. [DOI] [PubMed] [Google Scholar]
- [38].Tuxhorn JA, Ayala GE, Rowley DR. J. Urol. 2001;166:2472–2483. [PubMed] [Google Scholar]
- [39].Jacobs TW, Schnitt SJ, Tan XL, Brown LF. Hum. Pathol. 2002;33:29–38. doi: 10.1053/hupa.2002.30190. [DOI] [PubMed] [Google Scholar]
- [40].van Kempen LCLT, Rijntjes J, Claes A, Blokx WAM, et al. J. Pathol. 2004;204:333–339. doi: 10.1002/path.1659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.