Abstract
Background
Today many large mammals live in small, fragmented populations, but it is often unclear whether this subdivision is the result of long-term or recent events. Demographic modeling using genetic data can estimate changes in long-term population sizes while temporal sampling provides a way to compare genetic variation present today with that sampled in the past. In order to better understand the dynamics associated with the divergences of great ape populations, these analytical approaches were applied to western gorillas (Gorilla gorilla) and in particular to the isolated and Critically Endangered Cross River gorilla subspecies (G. g. diehli).
Results
We used microsatellite genotypes from museum specimens and contemporary samples of Cross River gorillas to infer both the long-term and recent population history. We find that Cross River gorillas diverged from the ancestral western gorilla population ~17,800 years ago (95% HDI: 760, 63,245 years). However, gene flow ceased only ~420 years ago (95% HDI: 200, 16,256 years), followed by a bottleneck beginning ~320 years ago (95% HDI: 200, 2,825 years) that caused a 60-fold decrease in the effective population size of Cross River gorillas. Direct comparison of heterozygosity estimates from museum and contemporary samples suggests a loss of genetic variation over the last 100 years.
Conclusions
The composite history of western gorillas could plausibly be explained by climatic oscillations inducing environmental changes in western equatorial Africa that would have allowed gorilla populations to expand over time but ultimately isolate the Cross River gorillas, which thereafter exhibited a dramatic population size reduction. The recent decrease in the Cross River population is accordingly most likely attributable to increasing anthropogenic pressure over the last several hundred years. Isolation of diverging populations with prolonged concomitant gene flow, but not secondary admixture, appears to be a typical characteristic of the population histories of African great apes, including gorillas, chimpanzees and bonobos.
Background
How species form and change over time is one of the central preoccupations of evolutionary biology (e.g [1,2]). Many species exist today in distinct populations whose size and interconnectivity have changed over time, in response to both recent and ancient climate changes and human activities. New advances in the analysis of patterns of genetic variation found in contemporary populations allow the disentanglement of the combined effects of these forces to reveal the recent and long-term histories of those populations. Population histories, including the timing of population divergences, the direction and extent of migration and estimates of effective population sizes, can be effectively inferred using explicit modeling approaches based upon coalescent theory (e.g. [3]).
These approaches combine information from neutrally-evolving loci sampled across the genome and employ models that can allow for a variety of population divergence scenarios. The classic allopatric model of speciation suggests that new species form when populations become isolated from one another [4]. However, subdivision of species or populations may not happen abruptly or completely, and divergence can occur even while some degree of gene flow may continue. Models of isolation that incorporate migration must, therefore, also be tested [5]. In addition, gene flow, even among evolutionarily divergent lineages, is routine at the contact zones of many taxa including lizards, ungulates and primates (e.g., baboons [6,7], macaques [8]), with convincing evidence even suggesting that some species arose from hybridization of two different species [9,10]. Hence, other speciation models also allow for the possibility of resumption of gene flow following population divergence.
In order to gain an appreciation for the tempo, mode, timings and ultimate causes of species and population divergences, it is advisable to first focus on representatives of a group of closely related species in order to uncover general trends. As our closest living relatives, the African great apes are particularly fascinating and have been well-studied in the past. For example, the closely-related chimpanzee (Pan troglodytes) and bonobo (Pan paniscus) species are suggested to have diverged some 1.5 million years ago (mya), and, not surprisingly given their long-term separation by the formidable barrier of the Congo River, there are no signs of recent gene flow between the species, although ancient gene flow was likely [11,12]. The three regional populations or subspecies of chimpanzees diverged on the order of half a million years ago, but the eastern and central populations exchange migrants at appreciable rates, while recent unidirectional migration from western to central chimpanzees is also inferred [12,13]. The central chimpanzee (Pan troglodytes troglodytes) population is suggested to be the ancestral population of all chimpanzees based on the findings of its larger effective population size and signals of population bottlenecks associated with the divergence of the other subspecies [12]. Overall, the Pan species and chimpanzee subspecies appear to have evolved following processes of isolation, limited migration, and population expansions.
Like chimpanzees, gorillas occur in central equatorial Africa but are somewhat less widespread. The contemporary distribution of gorillas features a pronounced gap of several hundred kilometers between western gorillas (Gorilla gorilla) and eastern gorillas (G. beringei) (Figure 1), yet these two species retain behavioral and morphological similarities and were until recently considered subspecies of the same species [14,15]. In previous work, we inferred from genetic data that the initial population divergence of western and eastern gorillas might have occurred approximately 0.9-1.6 mya [16], but that gene flow in both directions had persisted until as recently as 78,000 years ago [17]. Whereas these studies contributed to our understanding of the evolutionary history of gorillas on an interspecific level, little is known about population size changes in the past and the intraspecific evolutionary dynamics of gorilla populations. Intriguingly, a recent survey of skull and dental morphology of museum specimens suggested that some observed craniofacial anomalies may represent the outcome of secondary admixture between separate populations in both western and eastern gorillas, but this scenario has not yet been explicitly tested with genetic data [18]. While analyses of genetic data from great apes tend to favor population histories featuring isolation with migration rather than secondary introgression, such admixture is suspected to play a role in the evolution of various old world monkey taxa [19] as well as in the recent history of modern humans and Neanderthals [20].
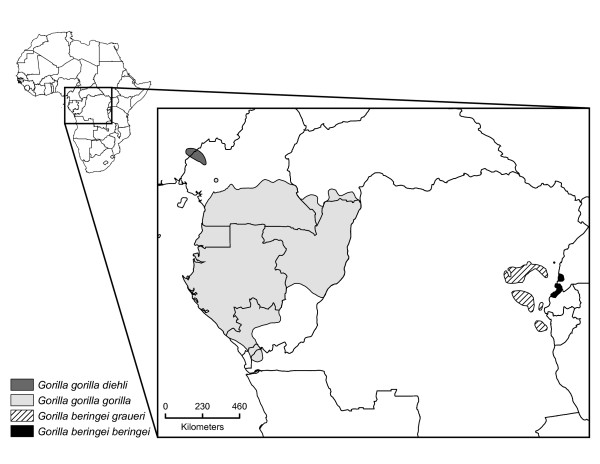
Figure 1.
Gorilla distribution map. Approximate current distribution of gorillas in Equatorial Africa. Gorilla gorilla gorilla refers to western lowland gorillas and Gorilla gorilla diehli to Cross River gorillas. East African gorillas are divided into Gorilla beringei beringei, also known as mountain gorillas and Gorilla beringei graueri, known as eastern lowland gorillas.
The focus of this study is a comprehensive assessment of the evolutionary histories of the broadly distributed population of western lowland gorillas (G. g. gorilla) and the range restricted, Critically Endangered Cross River gorillas (G. g. diehli). The current census population size of western lowland gorillas is on the order of tens of thousands (IUCN Red List of Threatened Species™). In contrast, Cross River gorillas consist of approximately 200-300 individuals in fragmented populations in a highland region on the Nigeria-Cameroon border [21]. Previous genetic analyses of Cross River gorillas using multiple autosomal microsatellite loci applied to DNA derived from non-invasively collected fecal samples suggested a recent and/or severe population decrease [22]. Here we build upon this work by using DNA obtained from museum specimens to directly compare the genetic diversity observed today with that existing approximately 100 years ago. We next employ a highly efficient coalescent-based approach that takes advantage of both our temporal sampling scheme and use of multiple independently-evolving loci. This allows us to reconstruct the evolutionary relationship of Cross River and western lowland gorillas by estimating the time of their divergence, the occurrence and duration of gene flow between them, and their current and ancestral effective population sizes. To evaluate different evolutionary scenarios we specifically test two alternative models, one of which allows for divergence in the presence of gene flow [23] while the other encompasses a secondary admixture scenario by permitting divergence in isolation and subsequent resumption of genetic exchange or admixture between the two populations [18].
Results and Discussion
Recent changes in genetic diversity
In order to assess the patterns of genetic diversity within and between two temporally spaced Cross River gorilla population samples, we generated microsatellite genotypes for 14 approximately 100-year-old Cross River museum specimens (Additional file 1: Table S1) and compared these data with published results from 71 non-invasively collected contemporary Cross River gorillas [24].
The low concentration and fragmented nature of DNA extracted from sources such as fecal samples or historical remains can lead to inaccurate microsatellite genotyping through the stochastic nonamplification of an allele at a heterozygous locus ('allelic dropout') or consistent nonamplification of an allele due to mutations at a priming site ('null alleles') [25,26]. Such errors can potentially bias conclusions concerning a population's genetic diversity or demographic history. We found no evidence of problems with any of the loci amplified in the historical samples, while two loci in the published modern Cross River data set potentially contained null alleles. This suggestion of null alleles disappeared when sampling localities were considered and the pattern was attributed to the existence of genetic structure in contemporary Cross River gorillas [24]. The analyses presented here provide further evidence for this conclusion. For instance, patterns of Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium differed between the two data sets. Specifically, three loci from the modern data set (D5S1470, D7S817, D8S1106) deviated significantly from HWE in a locus-wise probability test, a test that was also highly significant when performed across all loci (corrected p < 0.001). Furthermore, the modern Cross River samples approached a significant 'deficit of heterozygotes' when using a global test over all loci (corrected p = 0.071). Significant linkage disequilibrium was observed for 14 of the 28 locus by locus comparisons in the modern data set (corrected p < 0.0042), but for none of the loci in the historical data, when corrected for multiple testing. In addition, although not statistically significant, the mean observed heterozygosity (Ho) was higher in the historical dataset than the modern one (0.732 and 0.631, respectively) and FIS values increased from -0.05 in the historical to a significant 0.072 in the contemporary sample (SI: Tables S2 and S3). In summary, these results are most readily explained by a Wahlund effect due to pooling of the samples from the structured, contemporary population [27].
The average expected heterozygosity (He) was 0.701 and 0.679 for the historical and contemporary samples, respectively (SI: Table S2). Because the sample sizes differed, we resampled 14 genotypes from the contemporary samples 10,000 times and estimated mean He. We obtained a higher or equal mean He than observed in the historical sample in only 10.6% of the cases. Consistent with this result we observed a trend towards higher allelic richness in the historical samples compared to the modern samples in five of eight loci (SI: Table S3). A test for population differentiation assuming the same distribution of alleles within the two populations approached significance (p = 0.063), and is in agreement with the low FST value (FST = 0.0071, p = 0.199). In sum, the lack of statistical significance of the comparisons of the standard summary statistics may reflect low power due to the limited number of samples and loci, while the tendency towards higher variation in the past may reflect genetic drift or a population size decrease in the Cross River gorillas over the last 100 years, a finding that has been observed in other large mammals such as whales and orangutans and is usually attributed to increasing anthropogenic pressure [28,29].
Temporal estimates of effective population sizes
Estimates of a population's effective size (Ne) - the size of an idealized Wright-Fisher population [30,31] resulting in the same genetic variability as observed in the population examined [32] - highlight the strength of genetic drift acting on the genetic variation and is considered in assessments of a population's viability [33]. We used multiple analytical approaches to estimate the effective population sizes of the Cross River gorillas. First we used short-term estimators based on the changes in allele frequencies over the last hundred years. Two such moment estimators of Ne yielded values of 221 and 276, while a pseudo-likelihood estimator produced a similar value of 244 (Table 1).
Table 1.
Estimates of the short-term effective population size (Ne) of the Cross River gorilla population
| Method | Ne point estimator | 95% boundaries | |
|---|---|---|---|
| Moment estimators | 2211 | n.d. | |
| 2762 | 57 - infinity | ||
| Pseudo-likelihood | 2443 | 86 - infinity | |
| Coalescence | 1,6264 | 165 - infinity | |
| 1,1425 | 193 - 2,792 |
As complex models use more information, it has been suggested that likelihood- and coalescence-based methods may outperform moment estimators of effective population sizes in precision and accuracy [34,35]. Our Ne estimates derived from two probabilistic approaches yielded substantially higher values of 1,626 and 1,142 (Table 1). However, complex coalescence-based models are highly dependent on underlying assumptions. The apparent discrepancy between the various Ne estimators might be aggravated by the complexity of gorillas' life history and demography and potentially reflect deviations from a standard Wright-Fischer model. One of the most unrealistic assumptions is that of panmixia in the focal population, a situation rarely applicable to social animals. Second, the assumption of discrete generations is often violated due to the existence of age-structured, social groups containing several overlapping generations. This is especially the case in great apes, where closely related individuals (parent-offspring pairs) are found together in one group and dispersal distances are likely limited [36,37]. In sum, our results are consistent with the varied sensitivities of Ne estimators and confirm the utility of comparing results from multiple estimators [38] or using modeling approaches to infer demographic histories.
Demographic simulations and gorilla population histories
In order to investigate the deeper demographic history of Cross River gorillas we compared two potential demographic scenarios: a divergence with secondary admixture model and a model of isolation with migration (Figure 2). Both models allowed for a severe, recent population decline in the Cross River population and incorporated contemporary and museum samples appropriately.
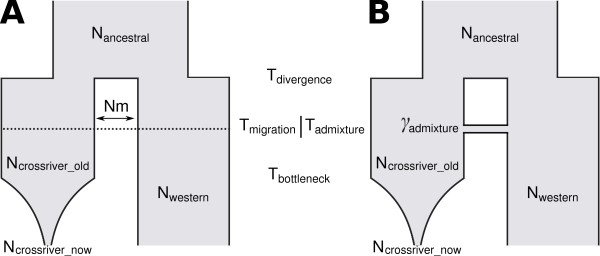
Figure 2.
Schemes of the demographic models. Schemes of the models describing the evolutionary history of western gorillas. A) Divergence accompanied with gene flow model; B) Divergence and secondary admixture model. For a detailed description see text and Supplementary Information.
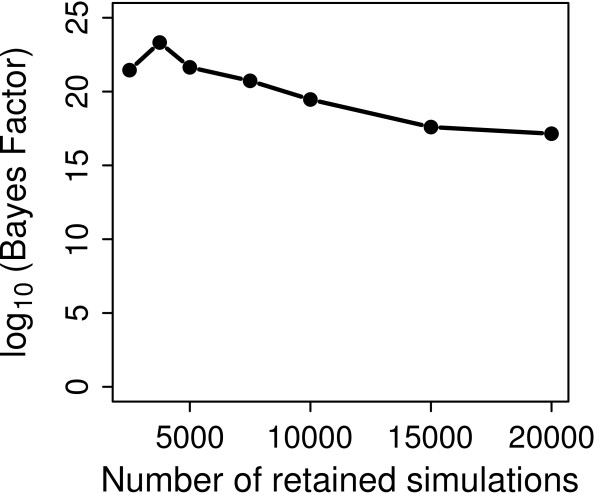
It has recently been suggested that the existence of genetic structure in a population might generate "spurious" bottleneck signals in demographic simulations [39-41]. This erroneous inference can be understood by considering the coalescent process in a subdivided population. As shown by [42], the coalescence process may be divided into two distinct phases: During the "scattering phase", lineages either coalesce or migrate quickly to demes other than those being sampled, until there is a single lineage left per deme. In the "collecting phase", the remaining lineages follow a standard coalescence process, like in a single population, but on a different time scale. If many samples are drawn from the same deme, the rate of coalescent events in the scattering phase is large, resulting in a genealogy that will look much like that expected after a recent bottleneck [5,39]. Since we found limited population structure in the contemporary sample, only one haplotype per sampled individual was considered in all our demographic simulations. This effectively reduces the number of alleles sampled from the same deme and thus reduces the potential bias in our inference. Despite the resulting reduced sample size we could safely reject the secondary admixture model in favor of the model of isolation with migration (log10Bayes factor ~ 22) (Figure 3).
Figure 3.
Bayes factor robustness. Given are estimates of the Bayes factor on the log10 scale when comparing the migration model to the secondary admixture model (see text for further details), according to the number of retained simulations.
We next estimated parameters of the preferred isolation with migration model, including the timing of divergence between the Cross River and western lowland gorillas, timing of cessation of gene flow between these populations, onset of population size changes and the contemporary and ancestral effective population sizes (Figure 2A).
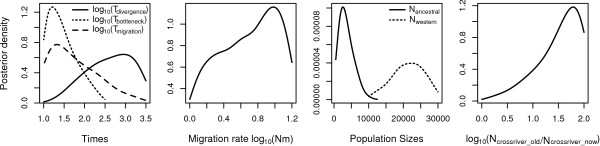
The effective size of the ancestral population of Cross River and western lowland gorillas was estimated at 2,547 (95% HDI: 500, 7,684; Figure 4 and Table 2). This ancestral population may have diverged into Cross River and western lowland gorillas at least 890 generations or 17,800 years ago (95% HDI: 760, 63,245 years). The effective population size of the western lowland gorillas subsequently increased approximately 9-fold to an estimated current day 22,376 (95% HDI: 12,879, 29,532). The effective size of the ancestral Cross River gorilla population was also large initially, probably caused by an expansion in space and numbers but beginning at about 320 years ago (95% HDI: 200, 2,825 years) it decreased by approximately 60-fold to 271 (95% HDI: 122, 300). We observed a signature of substantial gene flow between western and Cross River gorillas that continued after the initial divergence (2Nm ≈ 9.5 or roughly 4.5 individuals per generation) but ceased at around 21 generations or 420 years ago (95% HDI: 200, 16,256 years), at approximately the same time as the inferred onset of the population bottleneck.
Figure 4.
Posterior distribution of demographic parameters. Posterior distribution of demographic parameters as estimated in the divergence with gene flow model including migration between Cross River and western lowland gorillas (log10[Nm]), ratio of ancestral to current effective size of Cross River gorillas (log10[NCR OLD/NCR NOW], effective population sizes and times of various demographic events, respectively. Times are given in log10-scale of generations and population sizes refer to the effective size of the respective population.
Table 2.
Estimates of parameters of the demographic model applied to Cross River and western lowland gorillas
| Prior distribution | min | max | Posterior mode | HDI 50b | HDI 90b | HDI 95b | |
|---|---|---|---|---|---|---|---|
| 2Nm | Loguniform | 1 | 15.85 | 9.55 | [4.57, 13.8] | [1.58, 15.84] | [1.32, 15.85] |
| Ncrossriver_old/Ncrossriver_now) | Loguniform | 1 | 100 | 61.7 | [33.1, 93.3] | [10, 100] | [4.2, 100] |
| Ncrossriver_now | N(200, 100) a | 68 | 300 | 271 | [223, 292] | [146, 300] | [122, 300] |
| Nancestral | Uniform | 500 | 25,000 | 2,547 | [1,383, 4,032] | [500, 6,681] | [500, 7,684] |
| Nwestern | N(24,000, 5000) a | 10,000 | 30,000 | 22,376 | [18,765, 25,319] | [14,217, 28,930] | [12,879, 29,532] |
| Tdivergence | Loguniform | 10 | 3,162 | 891 | [269, 1,738] | [60.3, 3,090] | [38, 3.162] |
| Tbottleneck | Loguniform | 10 | 316 | 16 | [33.1, 93.3] | [10, 97.7] | [10, 141] |
| Tmigration | Loguniform | 10 | 3,162 | 21 | [11.7, 60.3] | [10, 446.7] | [10, 812] |
Ncrossriver_now, Nancestral, Nwestern represent the effective population sizes of Cross River, ancestral and western lowland gorillas respectively. Timings in generations were estimated on the log10 scale and indicate the divergence (Tdivergence), the onset of the bottleneck (Tbottlneck) and the cessation of migration (Tmigration). The number of diploid individuals exchanged between the populations was also estimated on the log10 scale as 2 Nm. For parameters estimated on the log10 scale we chose uniform priors on the same scale.
a Corresponds to a normal distribution of the form N(μ, σ) truncated at [min, max]
b The high posterior density interval HDI is chosen as the smallest continuous interval spanning 50% of the posterior surface. The other HDI are chosen accordingly.
The large credibility intervals on our posterior parameters suggest that further investigations using more loci or individuals may be useful to refine the demographic model. However, we rigorously validated our inferences and found only slight biases for some of the estimates (see SI: Supplementary Figure.3): an overestimation of the current Cross River population size, an underestimation of the divergence time and a more global overestimation of the ancestral gorilla population size over the entire parameter space. These minor biases do not substantially influence our inferences, which are associated with large posterior density intervals but nonetheless reveal a strong recent decrease in the effective size of the Cross River population.
The current analysis contributes to a composite view of gorilla population history based on genetic data (Table 3). An initial split of western and eastern gorillas about one million years ago was accompanied by bidirectional gene flow until as recently as 78,000 years ago [17]. Some 17,800 (95% HDI 760, 63,245) years ago, western lowland and Cross River gorillas diverged but substantial gene flow between the two western gorilla subspecies ceased only about 420 (95% HDI 200, 16,256) years ago. A marked decline of Cross River gorilla population size began only about a hundred years later, resulting in a current estimated effective population size of 271 (95% HDI 122-300). Such a pronounced, recent reduction in size accounts for the high concordance of effective population and census size as it drastically reduces variation and prolongs gene coalescences. These demographic events, along with the signals of likely population structure in the ancestral western gorilla population [17], are consistent with a scenario of changing climate conditions over the last tens of thousands of years that led to repeated expansion and contraction of forests, and hence of forest-dwelling ape populations, as well as more recent increased anthropogenic impact.
Table 3.
Estimates of effective population sizes for the indicated gorilla populations
| Population | Method | Ne estimates |
|---|---|---|
| Cross River1 | temporal | ~250 |
| Cross River1 | ABC | 271 |
| Western lowland1 | ABC | 22,376 |
| Western gorillas2* | IM | 17,700 |
| Western gorillas2* | θW | 24,100 |
| Western gorillas3* | MIMAR | 13,000 |
| Western lowland and Cross River (ancestral) 1 | ABC | 2,547 |
Africa's climate during the late Pleistocene was characterized by oscillations of aridity and humidity leading to a continuous cycle of forest expansion and contraction and the creation of forest refugia during arid phases [43]. Patterns of genetic diversity in gorillas and other mammals have been suggested to reflect the effects of refugial fragmentation during the last glacial maximum [44,45], and one such forest refugia has been suggested to have existed in the Cross River area. Consequently, the isolation of this region from other forest refugia in western equatorial Africa might have promoted the divergence of Cross River gorillas from their ancestral population ~17,800 years ago. Although the mode estimate of the eventual cessation of gene flow between western lowland and Cross River gorillas is rather recent, its 95% HPD interval includes major climatic changes in the region around 3,000-2,500 years ago that led to increased aridity and the expansive replacement of forests by grasslands [46,47].
In addition to climate change, human activities have likely had an increasing impact upon the Cross River gorillas in recent times. Initial settlement of the northwestern region of Cameroon by Bantu agriculturalists has been estimated to have coincided with contraction of forests in the region and occurred as early as 2,500 years ago [47]. Aside from habitat destruction, hunting is another anthropogenic threat that has likely caused the apparent population size decrease of Cross River gorillas over the last centuries. Firearms were introduced to the region beginning in the 18th century and increased in both numbers and sophistication starting in the mid to late 19th century [48]. Additionally, beginning in the late 19th century, the region underwent a rapid growth in human population density, increasing tenfold between 1900 and 2000 [49]. These two factors together almost certainly created a significant intensification of hunting pressure on large mammals like gorillas [21,22]. Notably, the inferred population size decrease in the history of Cross River gorillas by a factor of ~60 translates into a loss of ~23% of the gorilla population per generation since the onset of the bottleneck some 16 generations ago. Although this is a substantial decrease in size, other equatorial African gorilla populations have been described to suffer even more dramatic size reductions. In 2003, Walsh and colleagues reported a 56% decline in the population size of gorillas in Gabon over the last 20 years (or approximately a single generation) [50]. The authors concluded that the cause of this severe population size crash is a synergism between human activities, including hunting, habitat loss due to deforestation and disease (see also [51]) and strongly emphasized the need for effective and immediate conservation actions in order to ensure the long-term survival of gorilla populations.
Hunting of gorillas continues today and represents one of the greatest threats to their persistence [21]. Particularly for small populations that may have gone through recent reductions in size, effective enforcement of anti-poaching laws and maintenance of existing habitat to allow the population to stabilize and expand is crucial to their future survival [21]. While such measures will facilitate the short-term survival of Cross River gorillas, the impact of reduced levels of genetic diversity on their long-term viability remains unclear. The weak signal for loss of genetic variation in the Cross River gorilla population is consistent with the recent population bottleneck we identified, as the number of generations elapsed may not yet have been sufficient to significantly reduce levels of heterozygosity or allelic richness. The relatively short duration but dramatic severity of the bottleneck also explains the concordance between estimated census population size and our estimates of Ne, which often differ by up to a factor of ten in wildlife populations [31]. Given this relationship between N and Ne, it is possible that if the population were allowed to expand, the loss of diversity could be arrested [e.g., 29].
Conclusion
This work builds upon previous studies of the evolutionary relationships between western and eastern gorilla species [17], between Pan species [11], and among chimpanzee subspecies [12] in showing the prevalence of long-term post-divergence gene flow in African ape population histories. It is further apparent that African ape populations were subject to changes in size in the past, with evidence for both size decreases and expansions, and these are likely linked to changes in habitat caused by climate oscillations during the Pleistocene. It is plausible that habitat changes might also facilitate secondary introgression of already divergent populations, and human mediated habitat fragmentation has been invoked to explain numerous cases of secondary admixture in old world monkeys [19]. However, we find no support in our data for the postulated recent occurrence of secondary admixture of western lowland and Cross River gorillas [18]. Instead, we find that gene flow accompanied the divergence of western lowland and Cross River gorillas until just 400 or so years ago, which rather supports a scenario in which intensifying human activities may have increased the isolation of ape populations. The lack of a signal of secondary admixture in African apes is in notable contrast to the recent finding of substantive levels of introgression between the extinct Neanderthals and modern humans who were ancestral to populations found outside of Africa [20].
Coalescence-based approaches to inferring population histories using genetic data can reveal complex patterns of isolation with persistent migration or secondary contact that are difficult to reconcile with a view of evolutionary relationships as bifurcating processes that can be represented in a tree-like form. These results, along with earlier work on the patterns of morphological and phylogenetic relationships among baboon taxa, emphasize the challenges inherent to applying subspecies and species designations to dynamic entities like populations of organisms [7,52].
Methods
Sampling, DNA extraction and genotyping
Thirty-five gorilla specimens from the Cross River region on the border of Cameroon and Nigeria were obtained from the Museum of Natural History in Berlin, Germany. Twenty-six of the samples came from Ossidinge, a former missionary station located on the Cross River in Cameroon (5°15' - 6°15' N and 8°50' - 9°50' E; [53]). Although collected on expeditions in 1904-1907, some of these skulls were apparently obtained from local people and thus might have been older but not substantially so. The collection was transferred to Berlin in the early 1900s and stored at the Museum of Natural History since then.
Approximately 150 mg segments of tooth roots were extracted in physically-isolated laboratories dedicated to ancient DNA work following the protocol suggested in [54]. We first assessed whether we had recovered amplifiable nuclear DNA by attempting to amplify three autosomal microsatellites as well as a segment of the amelogenin gene in separate standard PCR reactions (see SI). The 14 samples for which at least two loci successfully amplified were then processed in a two-step multiplex approach [55] using the three original and five additional microsatellites (see SI).
Genotypes from 71 contemporary gorillas were generated using non-invasively collected fecal samples. The data from the Cross River population came from a previously published dataset [24]. For the demographic simulations we used genotypes from 92 western gorillas (Gorilla gorilla gorilla) (11 Cameroonian described in [17] and 81 from Gabon [56]). We included individuals from several locations in order to recover the genetic variation present in extant western gorillas. The data used from published work [17,24,56] employed noninvasive fecal samples from contemporary gorillas, collected in accordance with all relevant governmental guidelines and so further permissions were not necessary for the study presented here.
Data analysis
We previously estimated the amount of repetition necessary to produce genotypes significantly likely to be free of errors due to allelic dropout or spurious alleles [55]. An individual genotype was considered to be confirmed when a homozygous allele was detected in at least six independent multiplex PCR replicates or when each of two apparently heterozygous alleles was observed in at least two multiplex PCR replicates [55]. Summary statistics were calculated as outlined in the Supplementary Information. We furthermore investigated whether or not individual loci in both the historical and the modern Cross River gorilla dataset show evidence for null alleles, small allele dominance (allelic dropout) or mis-scoring due to stuttering by using the program MICROCHECKER v2.2.3 [57].
We applied a randomization strategy in order to assess whether there was a significant difference in levels of genetic variation in the historical samples relative to the modern ones. We randomly drew a sample equal in size to the historical gorilla sample from the modern gorilla genotypes, and repeated this 10,000 times and computed the average expected heterozygosity (He) over all loci which provides an unbiased estimate. We evaluated the significance level by estimating the percentage of resampling steps that provided a higher or equal He than observed in the historical Cross River gorilla population.
An alternative approach of assessing levels of genetic variation while accounting for different sample sizes [58] is provided by allelic richness measurements as calculated with FSTAT v.2.9.3.2. We also used this program to calculate FIS values [59] in order to estimate the probability of allelic identity due to population substructure or potential inbreeding.
Effective population size
To generate short-term estimates of the effective population size (Ne), we assumed that Cross River gorillas constitute an isolated population during the time spanned by our sampling (100 years). In general all models follow the same basic assumptions: the focal population is expected to be in panmixia, discrete generations exist and mutation, selection and migration are negligible compared to genetic drift, thus representing a simplified Wright-Fisher population [30,31]. Short-term Ne estimators are based on the variance of allele frequency changes over time and hence provide a harmonic mean of Ne for the time elapsed between the two sampling periods (for comprehensive reviews see [60]).
We calculated two moment estimators, one using the program MNE 1.0 [61,62] and applying Nei and Tajima's sampling scheme 1 [62]. The authors suggested two exclusive sampling schemes: plan 1 assumes that gene sampling happened after reproduction from a population in which census size approximates the effective size; plan 2 instead is based on the assumption that the sampled individuals do not contribute to the next generation and the census size is much larger than Ne. The second moment estimator was calculated according to Waples' formula [63] also adopting Nei and Tajima's sampling plan 1. As simulations have shown that moment estimators may suffer from imprecision and overestimation of Ne (e.g. [35]) whenever genetic drift is strong and genetic markers with high diversities are used, we accompanied these two moment estimators with three probabilistic ones. We used a pseudo-likelihood approach [61] as implemented in MNE 1.0 that also applies Nei and Tajima's sampling plan 1 but reduces multiallelic loci into a biallelic state by choosing one and pooling all other alleles, respectively. A second approach that utilizes the full allele spectra for multiallelic markers is based on a Monte Carlo evaluation to compute the likelihood of Ne given the data and adopting sampling plan 2 according to Nei and Tajima (CoNe; [34,64]). Finally, we used a Bayesian approach coupled with Markov Chain Monte Carlo simulations that is based on the approximation of the gene genealogy by the coalescence principle and not the population gene frequency as in [64] (tm3.1; [34]). The authors implemented sampling plan 2 into their model.
When using these probabilistic approaches we commonly applied a prior Ne-max of 3,000 and minimum run time of one million steps.
Demographic inferences
We first assumed an evolutionary model in which the Cross River population diverged from the western gorilla population Tdivergence generations ago (Figure 2A). The two populations exchanged Nm migrants per generation until Tmigration generations ago with Tmigration ≤ Tdivergence. The effective population sizes of the ancestral gorilla population (Nancestral) and western lowland gorillas (Nwestern) after the divergence of the Cross River population are assumed to be constant but are allowed to be different. The effective size of the Cross River population Ncrossriver_old is also assumed to be constant until Tbottleneck generations when an exponential decrease began lasting until today and finally reducing the effective population size to Ncrossriver_now. All microsatellite loci were simulated assuming a stepwise mutation model with mutation rate μ with a prior distribution N (0.0004, 0.0001) truncated at 0.0002 and 0.0006 [12,65]. All prior distributions used for this model are given in Table 2 and the effective sizes are based on existing knowledge on the census size of the current Cross River population (< 300 individuals) or published estimates for western gorillas [13,17].
We also considered an alternative scenario, wherein the Cross River and the western gorillas did not exchange any migrants after their initial divergence. However, at a given time in the past (Tadmixture) a fraction of the Cross River population (γadmixture) was replaced by immigrants from the western gorillas (Figure 2B) and thus mirroring a hybridization scenario. For parameters common to both models we used the same prior distributions (Table 2). The prior distributions of Tadmixture, Tmigration and the prior distribution on (γadmixture) were chosen uniform within (0,1).
We used the software Serial SimCoal [66], a modified version of the program SimCoal [67], specifically designed to simulate temporally-spaced samples using a coalescent framework. The simulated data were then implemented in an Approximate Bayesian Computations framework (ABC) [68] in order to estimate parameters of the above model. We extended a methodology recently introduced and described in detail [69] to take the independence of the K studied loci explicitly into account (SI). This allowed us to reduce the necessary computations to a fraction of 1/K since we simulated a single locus rather than the complete set of observed loci, as was done previously. We successfully validated our approach (SI) using the methodology introduced in [39]. However, due to the low number of loci we report slight biases for some of our estimates: overestimation of Ncrossriver_now, underestimation of Tdivergence and a more global overestimation of Nancestral over the entire parameter space. Model comparison was performed using Bayes factors as described in [69], but adapted to the case of independent loci (SI).
Our inference relied upon the following summary statistics computed on the observed and simulated microsatellite loci: the variance in repeat length within each population and the difference in mean repeat length between all pairs of populations. We assumed a recently refined gorilla generation time of 20 years (IUCN Red List of Threatened Species™) and thus the historical samples were collected five generations before present.
Authors' contributions
OT, DW and LV designed research; OT, DW, MS, CL performed research; OT, DW, MA, KG, analyzed data; LV provided materials; OT, DW, MA, RAB and LV wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information. Here we provide additional information regarding laboratory methods and data analyses.
Contributor Information
Olaf Thalmann, Email: olatha@utu.fi.
Daniel Wegmann, Email: daniel.wegmann@ucla.edu.
Marie Spitzner, Email: spitzner_marie@yahoo.de.
Mimi Arandjelovic, Email: arandjel@eva.mpg.de.
Katerina Guschanski, Email: katerina.guschanski@unil.ch.
Christoph Leuenberger, Email: christoph.leuenberger@unifr.ch.
Richard A Bergl, Email: Richard.bergl@nczoo.org.
Linda Vigilant, Email: vigilant@eva.mpg.de.
Acknowledgements
We thank R. Asher and M. Willborn from the Natural History Museum in Berlin, Germany for allowing and assisting in sample collection. We thank M. Hofreiter for suggesting the multiplex approach and for fruitful discussions, C. Boesch for constructive comments on the project, and H. Siedel and A. Abraham for technical assistance in the lab and R. Mundry for statistical advice. OT is grateful to R. Wayne, S. Price, M. Gray, H. Thomassen and D. Schwochow for constructive comments on an earlier version of the manuscript and valuable discussions. This study was financed by the Max Planck Society and the Deutsche Forschungsgemeinschaft (VI 229/2-1).
References
- Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- Avise JC. Molecular markers, natural history and evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- Marjoram P, Tavare S. Modern computational approaches for analyzing molecular genetic variation data. Nat Rev Genet. 2006;7(10):759–770. doi: 10.1038/nrg1961. [DOI] [PubMed] [Google Scholar]
- Mayr E. Systematics and the Origin of Species. Cambridge: Harvard University Press; 1942. [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov Chain Monte Carlo approach. Genetics. 2001;158(2):885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Conroy JE, Jolly CJ. Changes in the structure of the baboon hybrid zone in the Awash National Park, Ethiopia. Vol. 71. Wiley Subscription Services, Inc., A Wiley Company; 1986. pp. 337–350. [Google Scholar]
- Jolly CJ. A proper study for mankind: Analogies from the Papionin monkeys and their implications for human evolution. Am J Phys Anthropol. 2001;116(S33):177–204. doi: 10.1002/ajpa.10021. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Supriatna J, Melnick DJ. Hybridization and population genetics of two Macaque species in Sulawesi, Indonesia. Evolution. 2001;55(8):1686–1702. doi: 10.1111/j.0014-3820.2001.tb00688.x. [DOI] [PubMed] [Google Scholar]
- Tosi AJ, Coke CS. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol Phylogenet Evol. 2007;42(2):498–504. doi: 10.1016/j.ympev.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Burrell AS, Jolly CJ, Tosi AJ, Disotell TR. Mitochondrial evidence for the hybrid origin of the kipunji, Rungwecebus kipunji (Primates: Papioni) Mol Phylogenet Evol. 2009;51(2):340–348. doi: 10.1016/j.ympev.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Won Y-J, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22(2):297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Excoffier L. Bayesian Inference of the Demographic History of Chimpanzees. Mol Biol Evol. 2010;27(6):1425–1435. doi: 10.1093/molbev/msq028. [DOI] [PubMed] [Google Scholar]
- Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17(10):1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt AH, Stewart KJ. Gorilla society: What we know and don't know. Evolutionary Anthropology: Issues, News, and Reviews. 2007;16(4):147–158. doi: 10.1002/evan.20142. [DOI] [Google Scholar]
- Groves CP. Ecology and taxonomy of the gorilla. Nature. 1967;213(5079):890–893. doi: 10.1038/213890a0. [DOI] [PubMed] [Google Scholar]
- Thalmann O, Serre D, Hofreiter M, Lukas D, Eriksson J, Vigilant L. Nuclear insertions help and hinder inference of the evolutionary history of gorilla mtDNA. Mol Ecol. 2005;14(1):179–188. doi: 10.1111/j.1365-294X.2004.02382.x. [DOI] [PubMed] [Google Scholar]
- Thalmann O, Fischer A, Lankester F, Paabo S, Vigilant L. The complex evolutionary history of gorillas: Insights from genomic data. Mol Biol Evol. 2007;24(1):146–158. doi: 10.1093/molbev/msl160. [DOI] [PubMed] [Google Scholar]
- Ackermann RR, Bishop JM. Morphological and molecular evidence reveals recent hybridization between Gorilla taxa. Evolution. 2010;64(1):271–290. doi: 10.1111/j.1558-5646.2009.00858.x. [DOI] [PubMed] [Google Scholar]
- Detwiler KM, Burrell AS, Jolly CJ. Conservation Implications of Hybridization in African Cercopithecine Monkeys. Int J Primatol. 2005;26(3):661–684. doi: 10.1007/s10764-005-4372-0. [DOI] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y. et al. A Draft Sequence of the Neandertal Genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates J, Sunderland-Groves J, Bergl R, Dunn A, Nicholas A, Takang E, Omeni F, Imong I, Fotso R, Nkembi L, Regional action plan for the conservation of the cross river gorilla (Gorilla gorilla diehli) Arlington, VA, USA: IUCN/SSC Primate Specialist Group and Conservation International; 2007. [Google Scholar]
- Bergl RA, Bradley BJ, Nsubuga AM, Vigilant L. Effects of habitat fragmentation, population size and demographic history on genetic diversity: The Cross River gorilla in a comparative context. Am J Primatol. 2008;70(9):848–859. doi: 10.1002/ajp.20559. [DOI] [PubMed] [Google Scholar]
- Nosil P. Speciation with gene flow could be common. Mol Ecol. 2008;17(9):2103–2106. doi: 10.1111/j.1365-294X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- Bergl RA, Vigilant L. Genetic analysis reveals population structure and recent migration within the highly fragmented range of the Cross River gorilla (Gorilla gorilla diehli) Mol Ecol. 2007;16(3):501–516. doi: 10.1111/j.1365-294X.2006.03159.x. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24(16):3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, Chambers KE, Boesch C, Vigilant L. Quantitative PCR analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzes (Pan troglodytes verus) Mol Ecol. 2001;10:1835–1844. doi: 10.1046/j.0962-1083.2001.01308.x. [DOI] [PubMed] [Google Scholar]
- Wahlund S. Zusammensetzung von Population und Korrelationserscheinung vom Standpunkt der Veerbungslehre aus betrachtet. Hereditas. 1928;11:65–106. doi: 10.1111/j.1601-5223.1928.tb02483.x. [DOI] [Google Scholar]
- Alter SE, Rynes E, Palumbi SR. DNA evidence for historic population size and past ecosystem impacts of gray whales. Proc Natl Acad Sci USA. 2007;104(38):15162–15167. doi: 10.1073/pnas.0706056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens B, Chikhi L, Ancrenaz M, Lackman-Ancrenaz I, Andau P, Bruford MW. Genetic signature of anthropogenic population collapse in orang-utans. PLoS Biology. 2006;4(2) doi: 10.1371/journal.pbio.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford: Clarendon; 1930. [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An Introduction to Population Genetic Theory. New York: Harper and Row; 1970. [Google Scholar]
- Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet Res. 1995;66:95–107. doi: 10.1017/S0016672300034455. [DOI] [PubMed] [Google Scholar]
- Berthier P, Beaumont MA, Cornuet J-M, Luikart G. Likelihood-based estimation of the effective population size using temporal changes in allele frequencies: A genealogical approach. Genetics. 2002;160(2):741–751. doi: 10.1093/genetics/160.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Estimation of effective population sizes from data on genetic markers. Philos Trans R Soc Lond B Biol Sci. 2005;360:1395–1409. doi: 10.1098/rstb.2005.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley BJ, Doran-Sheehy DM, Lukas D, Boesch C, Vigilant L. Dispersed male networks in western gorillas. Curr Biol. 2004;14(6):510–513. doi: 10.1016/j.cub.2004.02.062. [DOI] [PubMed] [Google Scholar]
- Langergraber K, Mitani J, Vigilant L. Kinship and social bonds in female chimpanzees (Pan troglodytes) Am J Primatol. 2009;71:840–851. doi: 10.1002/ajp.20711. [DOI] [PubMed] [Google Scholar]
- Miller CR, Waits LP. The history of effective population size and genetic diversity in the Yellowstone grizzly (Ursus arctos): Implications for conservation. Proc Natl Acad Sci USA. 2003;100(7):4334–4339. doi: 10.1073/pnas.0735531100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM, Wegmann D, Excoffier L. Distinguishing between population bottleneck and population subdivision by a Bayesian model choice procedure. Mol Ecol. 2010;19(21):4648–4660. doi: 10.1111/j.1365-294X.2010.04783.x. [DOI] [PubMed] [Google Scholar]
- Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA. The Confounding Effects of Population Structure, Genetic Diversity and the Sampling Scheme on the Detection and Quantification of Population Size Changes. Genetics. 2010;186(3):983–995. doi: 10.1534/genetics.110.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Beaumont MA. Statistical inferences in phylogeography. Vol. 18. Blackwell Publishing Ltd; 2009. pp. 1034–1047. [DOI] [PubMed] [Google Scholar]
- Wakeley J. Nonequilibrium Migration in Human History. Genetics. 1999;153(4):1863–1871. doi: 10.1093/genetics/153.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E, O'Hara RJ. The biogeographic evidence supporting the Pleistocene forest refuge hypothesis. Evolution. 1986;40:55–67. doi: 10.2307/2408603. [DOI] [PubMed] [Google Scholar]
- Anthony NM, Johnson-Bawe M, Jeffery K, Clifford SL, Abernethy KA, Tutin CE, Lahm SA, White LJT, Utley JF, Wickings JE. et al. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc Natl Acad Sci USA. 2007;104(51):20432–20436. doi: 10.1073/pnas.0704816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okello JBA, Wittemyer G, Rasmussen HB, Arctander P, Nyakaana S, Douglas-Hamilton I, Siegismund HR. Effective population size dynamics reveal impacts of historic climatic events and recent anthropogenic pressure in African elephants. Mol Ecol. 2008;17(17):3788–3799. doi: 10.1111/j.1365-294X.2008.03871.x. [DOI] [PubMed] [Google Scholar]
- Vincens A, Schwartz D, Elenga H, Reynaud-Farrera I, Alexandre A, Bertaux J, Mariotti A, Martin L, Meunier J-D, Nguetsop F. et al. Forest response to climate changes in Atlantic Equatorial Africa during the last 4000 years BP and inheritance on the modern landscapes. J Biogeogr. 1999;26(4):879–885. doi: 10.1046/j.1365-2699.1999.00333.x. [DOI] [Google Scholar]
- Ngomanda A, Neumann K, Schweizer A, Maley J. Seasonality change and the third millenium BP rainforest crisis in southern Cameroon (Central Africa) Quaternary Res. 2009;71(3):307–318. doi: 10.1016/j.yqres.2008.12.002. [DOI] [Google Scholar]
- Law R. Horses, Firearms and Political power in pre-colonial West Africa. Past and Present. 1976;72:112–132. doi: 10.1093/past/72.1.112. [DOI] [Google Scholar]
- Hayashi R. Union for African Population studies - 5th African Population Conference. Arusha, Tanzania; 2007. Long-term population dynamics in Africa: A perspective from the urban structure. [Google Scholar]
- Walsh PD, Abernethy KA, Bermejo M, Beyers R, De Wachter P, Akou ME, Huijbregts B, Mambounga DI, Toham AK, Kilbourn AM. et al. Catastrophic ape decline in western equatorial Africa. Nature. 2003;422(6932):611–614. doi: 10.1038/nature01566. [DOI] [PubMed] [Google Scholar]
- Walsh PD, Biek R, Real LA. Wave-Like Spread of Ebola Zaire. PLoS Biology. 2005;3(11) doi: 10.1371/journal.pbio.0030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. On the failure of modern species concepts. Trends Ecol Evol. 2006;21(8):447. doi: 10.1016/j.tree.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Mansfeld A. Urwald-Dokumente. Vier Jahre unter den Crossfluss-Negern Kameruns. 1. Berlin: Reimer; 1908. [Google Scholar]
- Vigilant L, Hofreiter M, Siedel H, Boesch C. Paternity and relatedness in wild chimpanzee communities. Proc Natl Acad Sci USA. 2001;98(23):12890–12895. doi: 10.1073/pnas.231320498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arandjelovic M, Guschanski K, Schubert G, Harris TR, Thalmann O, Siedel H, Vigilant L. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol Ecol Resour. 2009;9(1):28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- Arandjelovic M, Head J, Kühl H, Boesch C, Robbins MM, Maisels F, Vigilant L. Effective non-invasive genetic monitoring of multiple wild western gorilla groups. Biological Conservation. 2010;143(7):1780–1791. doi: 10.1016/j.biocon.2010.04.030. [DOI] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- Leberg PL. Estimating allelic richness: Effects of sample size and bottlenecks. Mol Ecol. 2002;11(11):2445–2449. doi: 10.1046/j.1365-294X.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- Cockerham CC, Weir BS. Estimation of gene flow from F-statistics. Evolution. 1993;47(3):855–863. doi: 10.2307/2410189. [DOI] [PubMed] [Google Scholar]
- Beaumont MA. In: Handbook of Statistical Genetics. 3. Balding DJ, Bishop JM, Cannings C, editor. Vol. 1. London: John Wiley & Sons, Ltd; 2007. Conservation Genetics; pp. 1021–1066. full_text. [Google Scholar]
- Wang J. A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet Res. 2001;78:243–257. doi: 10.1017/S0016672301005286. [DOI] [PubMed] [Google Scholar]
- Nei M, Tajima F. Genetic drift and estimation of effective population size. Genetics. 1981;98(3):625–640. doi: 10.1093/genetics/98.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics. 1989;121(2):379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Williamson EG, Thompson EA. Monte Carlo evaluation of the likelihood for N(e) from temporally spaced samples. Genetics. 2000;156(4):2109–2118. doi: 10.1093/genetics/156.4.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Smith NGC, Ellegren H. Microsatellite evolution inferred from human- chimpanzee genomic sequence alignments. Proc Natl Acad Sci USA. 2002;99(13):8748–8753. doi: 10.1073/pnas.122067599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA. Serial SimCoal: A population genetics model for data from multiple populations and points in time. Bioinformatics. 2005;21(8):1733–1734. doi: 10.1093/bioinformatics/bti154. [DOI] [PubMed] [Google Scholar]
- Laval G, Excoffier L. SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics. 2004;20(15):2485–2487. doi: 10.1093/bioinformatics/bth264. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian Computation in population genetics. Genetics. 2002;162(4):2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger C, Wegmann D. Bayesian Computation and Model Selection Without Likelihoods. Genetics. 2010;184(1):243–252. doi: 10.1534/genetics.109.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information. Here we provide additional information regarding laboratory methods and data analyses.