Abstract
Background
Corynebacterium pseudotuberculosis, a Gram-positive, facultative intracellular pathogen, is the etiologic agent of the disease known as caseous lymphadenitis (CL). CL mainly affects small ruminants, such as goats and sheep; it also causes infections in humans, though rarely. This species is distributed worldwide, but it has the most serious economic impact in Oceania, Africa and South America. Although C. pseudotuberculosis causes major health and productivity problems for livestock, little is known about the molecular basis of its pathogenicity.
Methodology and Findings
We characterized two C. pseudotuberculosis genomes (Cp1002, isolated from goats; and CpC231, isolated from sheep). Analysis of the predicted genomes showed high similarity in genomic architecture, gene content and genetic order. When C. pseudotuberculosis was compared with other Corynebacterium species, it became evident that this pathogenic species has lost numerous genes, resulting in one of the smallest genomes in the genus. Other differences that could be part of the adaptation to pathogenicity include a lower GC content, of about 52%, and a reduced gene repertoire. The C. pseudotuberculosis genome also includes seven putative pathogenicity islands, which contain several classical virulence factors, including genes for fimbrial subunits, adhesion factors, iron uptake and secreted toxins. Additionally, all of the virulence factors in the islands have characteristics that indicate horizontal transfer.
Conclusions
These particular genome characteristics of C. pseudotuberculosis, as well as its acquired virulence factors in pathogenicity islands, provide evidence of its lifestyle and of the pathogenicity pathways used by this pathogen in the infection process. All genomes cited in this study are available in the NCBI Genbank database (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers CP001809 and CP001829.
Introduction
Corynebacterium pseudotuberculosis is a facultative intracellular pathogen that mainly infects sheep and goats, causing the disease called caseous lymphadenitis (CL). This bacterium can also cause ulcerative lymphangitis in equines; superficial abscesses in bovines, pigs, deer and laboratory animals; arthritis and bursitis in ovines; pectoral abscesses in equines and, more rarely, in camels, caprines and deer [1]-[3]. In both disease manifestations, its main characteristic is abscessing of the lymph nodes [4]. Rare cases of human infection have also been reported [5], [6].
Despite the broad spectrum of hosts, the high incidence of CL reported from various countries, including Australia, New Zealand, South Africa, the United States of America, Canada and Brazil, mainly refers to small ruminants [7]-[11]. According to the World Animal Health Organization, among 201 countries that reported their sanitary situations, 64 declared the presence of animals with CL within their borders (OIE, 2009). The highest prevalence of CL has been reported in Brazil [12]. Pinheiro and colleagues (2000) reported 66.9% of animals with clinical signs of CL in the state of Ceará. In Minas Gerais state, a prevalence of 75.8% was reported for sheep [13] and 78.9% for goats [14]. In Australia, 61% of sheep flocks showed signs of infection [15]. In the USA, the prevalence ranges up to 43% [16]. Similar levels have been reported from the Canadian province of Quebec, with a prevalence of 21 to 36% [10]. In the United Kingdom, 45% of the producers that were polled reported abscesses in their sheep [9].
The high prevalence of CL in sheep and goats has made studies on ways to detect C. pseudotuberculosis in these hosts increasingly important; an efficient means to accomplish this would be a valuable tool for the control of this disease. Currently, there is no sufficiently sensitive and specific diagnostic test for subclinical CL. Diagnosis is currently achieved only by routine bacterial culture of purulent material collected from animals that have external abscesses, with subsequent biochemical identification of the isolates [17]. A few vaccines against CL are currently available, although they have not been licensed for use in many countries. Not all vaccines that have been developed for sheep are effective in goats. It is usually necessary to adjust vaccination programs to each animal host species [18].
Considering the current unfortunate status of CL prevalence in the world, especially in Brazil and Australia, there is a pressing need for more efficient alternatives for disease control that not only cure sick animals but also minimize or even prevent the onset of disease in herds. One of the major efforts to eradicate this disease involves the identification of genes that are related to the C. pseudotuberculosis pathogenicity and lifestyle. As an intracellular facultative pathogen, C. pseudotuberculosis exhibits several characteristics in its genome, such as gene loss, low GC content and a reduced genome [19] that differ from those of non-pathogenic Corynebacterium species. The finding of seven putative pathogenicity islands containing classical virulence elements, including genes for iron uptake, fimbrial subunits, insertional elements and secreted toxins [20], probably mostly acquired through horizontal transfer, contributes to our understanding of how this species causes disease. Comprehensive knowledge of an organism's genome facilitates an exhaustive search for candidates for virulence genes, vaccine and antimicrobial targets, and components that could be used in diagnostic procedures.
The information retrieved from a single genome is insufficient to provide an understanding of all C. pseudotuberculosis strains. Comparative genomics can shed light on the molecular attributes of a strain that affect its virulence, host specificity, dissemination potential and resistance to antimicrobial agents [21], [22]. Furthermore, comparison of entire genome sequences of strains belonging to the same species, but from different geographic, epidemiological, chronological and clinical backgrounds, as well as affecting different hosts, would be useful for determining the molecular basis of these differences. As part of an effort to provide means to control CL, we examined the genomes of two strains of C. pseudotuberculosis isolated from sheep and goats, respectively, and compared them to each other and to the genomes of two other strains already available in a public database [6], [23].
Results
Corynebacterium pseudotuberculosis genome
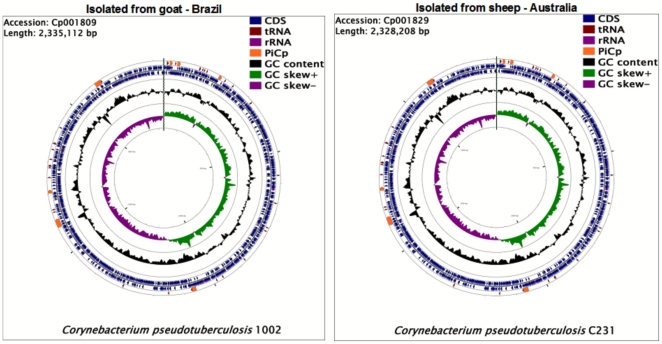
Overviews of the C. pseudotuberculosis genomes can be seen in Figure 1. The genomes are available in the NCBI GenBank database under accession numbers Cp1002:CP001809 and CpC231:CP001829.
Figure 1. The whole genome of Corynebacterium pseudotuberculosis.
Cp1002 strain isolated from a goat in Brazil and CpC231 strain isolated from sheep in Australia. Highlighted in yellow are the pathogenicity islands (PiCps) of C. pseudotubeculosis and its location in the genomes.
The two strains are very similar, with an amino acid similarity of at least 95% between their predicted proteins. In their genomic composition, the isolates were found to have the same mean i) GC content, ii) gene length, iii) operon composition and iv) gene density. However, some significant differences were observed in: i) genome size, ii) number of pseudogenes and iii) lineage-specific genes (Table 1).
Table 1. General features of the genomes of two Corynebacterium pseudotuberculosis strains.
| Genome feature | Cp1002 | CpC231 |
| Genome size (bp) | 2,335,112 | 2,328,208 |
| Gene number | 2111 | 2103 |
| Operon predicted number | 474 | 468 |
| Pseudogene number | 53 | 50 |
| tRNA number | 48 | 48 |
| rRNA operon | 4 | 4 |
| Gene mean length (bp) | 964 | 968 |
| Gene density (%) | 0.88 | 0.88 |
| Coding percentage | 84.9 | 85.4 |
| GC content (gene) (%) | 52.88 | 52.86 |
| GC content (genome) (%) | 52.19 | 52.19 |
| Lineage-specific genes | 52 | 49 |
Gene order in C. pseudotuberculosis
To determine whether synteny was maintained between the two C. pseudotuberculosis strains, we made a comparative analysis of global gene order. As expected, the two C. pseudotuberculosis strains showed high synteny conservation; approximately 97% of their genes were found to be conserved in the comparison between the two strains. Previous studies provide evidence of a high degree of conservation of gene order in four Corynebacterium genomes, C. diphtheriae, C. glutamicum, C. efficiens and C. jeikeium, showing only 10 gene-order breakpoints; rearrangement events during evolution in this species appear to be rare [24], [25]. We checked the validity of this conclusion by making a comparative analysis of the genomes of the two C. pseudotuberculosis strains against C. diphtheriae, the Corynebacterium species that is most closely related to C. pseudotuberculosis [26], [27].
Both C. pseudotuberculosis genomes showed a high degree of conservation in gene position, when compared to the C. diphtheriae genome, with few rearrangement points. This finding supports the hypothesis of a high degree of synteny conservation in this genus [25].
Pathogenicity islands (PAIs)
Pathogenicity islands in bacterial genomes can be characterized by looking for characteristics linked to horizontal gene transfer, such as differences in codon usage, G+C content, dinucleotide frequency, insertion sequences, and tRNA flanking regions, together with transposase coding genes, which are involved in incorporation of DNA by transformation, conjugation or bacteriophage infection [28].
Pathogenicity islands had not been reported for C. pseudotuberculosis; to date; we used a multi-pronged approach called PIPS (submitted article) to identify the putative PAIs of C. pseudotuberculosis. Seven regions with most or all of the characteristics of horizontally-acquired DNA were found in both strains, Cp1002 and CpC231: i) base composition and/or codon usage deviations, ii) tRNA flanking, and iii) transposase genes. These regions were not found in a non-pathogenic species belonging to the same genus, C. glutamicum, and were classified as putative pathogenicity islands in C. pseudotuberculosis (PiCp). PiCps encode for proteins involved in the ABC transport system, for glycosil transferase, a two-component system, the fag operon and phospholipase D Table 2 provides a list of some genes found in the PAIs, with their respective functions.
Table 2. Genes and proteins present in pathogenicity islands of the Corynebacterium pseudotuberculosis strain genomes.
| PAI | Cp1002 | CpC231 | Protein |
| tnp7109-9 | tnp7109-9 | Transposase for insertion sequence | |
| pld | pld | Phospholipase D precursor (PLD) | |
| PiCp 1 | fag C | fag C | ATP binding cytoplasmic membrane protein - FagC |
| fag B | fag B | Iron-enterobactin transporter - FagB | |
| fag A | fag A | Integral membrane protein - FagA | |
| fag D | fag D | Iron siderophore binding protein - FagD | |
| mgtE | mgtE | Mg2+ transporter mgtE | |
| malL | malL | Oligo-1,6-glucosidase | |
| PiCp 2 | tetA | tetA | Putative tetracycline-efflux transporter |
| cskE | cskE | Anti-sigma factor | |
| sigK | sigK | ECF family sigma factor K | |
| dipZ | dipZ | Integral membrane C-type cytochrome biogenesis protein DipZ | |
| potG | potG | Putrescine ABC transport system | |
| afuB | afuB | Putative transport system permease (iron) | |
| PiCp 3 | afuA | afuA | Iron (Fe3+) ABC superfamily ATP binding cassette transporter, binding protein |
| glpT | glpT | Glycerol-3-phosphate transporter | |
| phoB | phoB | Two-component regulatory protein | |
| lcoS | lcoS | Two-component sensor protein, sensor histidine kinase | |
| ciuA | ciuA | Putative iron transport system binding (secreted) protein | |
| ciuB | ciuB | Putative iron transport system membrane protein | |
| PiCp 4 | ciuC | ciuC | Putative iron transport system membrane protein |
| ciuD | ciuD | Putative iron ABC transport system | |
| ciuE | ciuE | Putative siderophore biosynthesis related protein | |
| σ70 | σ70 | Putative RNA polymerase sigma factor 70 | |
| Pseudogene | Pseudogene | Putative chromosome segregation ATPase | |
| PiCp 5 | hsdR | hsdR | Putative type III restriction-modification system |
| pfoS | pfoS | PfoR superfamily protein | |
| htaC | htaC | HtaA family protein | |
| guaB3 | guaB3 | Inosine 5-monophosphate dehydrogenase | |
| PiCp6 | pipA1 | pipB | Proline iminopeptidase |
| mfsD1 | mfsD1 | Major facilitator superfamily domain-containing protein 1 | |
| dcd | dcd | Deoxycytidine triphosphate deaminase | |
| udg | udg | UDP-glucose 6-dehydrogenase | |
| lysS1 | lysS1 | Lysyl-tRNA synthetase | |
| alaT | alaT | Aminotransferase AlaT | |
| ureA | ureA | Urease gamma subunit | |
| ureB | ureB | Urease beta subunit | |
| ureC | ureC | Putative urease subunit alpha | |
| PiCp 7 | ureE | ureE | Urease accessory protein |
| ureF | ureF | Urease accessory protein | |
| ureG | ureG | Urease accessory protein | |
| ureD | ureD | Urease accessory protein | |
| fepC2 | fepC2 | ABC superfamily ATP binding cassette transporter | |
| fecD | fecD1 | Iron(III) dicitrate transport system permease fecD | |
| phuC | phuC | Iron(III) dicitrate transport permease-like protein yusV | |
| arsR | arsR1 | ArsR-family transcription regulator |
Genetic composition of C. pseudotuberculosis Pathogenicity Islands
The genetic composition of PAIs can shed light on the lifestyle of pathogenic bacteria, since they include virulence genes that mediate mechanisms of adhesion, invasion, colonization, proliferation into the host and evasion of the immune system [29], [30]. In addition, PAIs are characterized as being unstable regions that can be affected by insertions and deletions, influencing bacterial adaptability to new environments and hosts [31]. Here follows descriptions of the most relevant genetic elements found in the C. pseudotuberculosis pathogenicity islands. For more information, see the list of these orthologous genes in other Corynebacterium species in the Table S1 (online supporting information).
PiCp 1
C. pseudotuberculosis PiCp 1 harbors key genes involved in virulence and pathogenicity; these include PLD, the major virulence factor of this organism, which plays a role in spreading through the host; the fag operon, responsible for extracellular iron acquisition and, consequently, for survival in hostile environments; and a transposase gene, probably responsible for insertion of the island into the C. pseudotuberculosis genome. The finding that C. ulcerans can produce phospholipase D protein [32] indicates acquisition of PiCp1 by both C. pseudotuberculosis and C. ulcerans.
PiCp 2
Gene mgtE of island 2 has Mg2+ influx activity [33]. In prokaryotes, Mg2+ has been identified as an important regulatory signal that is essential for virulence, since it is involved in thermal adaptation, protecting bacteria from heat shock caused by fever in warm-blooded mammals [34]. Translation of the mgtE gene is regulated by changes in cytosolic Mg2+ concentration; loss of MgtE reduces biofilm formation and motility in the pathogenic bacteria Aeromonas hydrophila [33].
The protein MalL (malL), a maltose-inducible α-glucosidase, hydrolyzes various disaccharides, such as maltose and isomaltose, which can serve as carbon and energy sources [35], [36].
The tetA gene codes for a tetracycline-efflux transporter protein that extrudes antibiotics from the cell and confers resistance to biofilm cells. The tetA gene is often carried by transmissible elements, such as plasmids, transposons, and integrons [37], thus explaining its presence in a PAI.
The sigK gene is an extracytoplasmic function sigma factor (sigma ECF) regulated by cskE, an anti-sigma factor. Another sigma ECF, sigK, mediates targeted alterations in bacterial transcription via transduction of extracellular signals. In M. tuberculosis, sigK regulates several genes (Rv2871, mpt83, dipZ, mpt70, Rv2876, and mpt53). Also, sigK mutations produce reduced quantities of the antigens MPT70 and MPT83 in vitro, and only induce strong expression during infection of macrophages [38]–[40].
PiCp2 also harbors a dipZ gene, which is regulated by sigK and seems to play a role in macrophage infection by M. tuberculosis, although its function is not clearly elucidated. DipZ is found as two separate proteins in most bacteria: CcdA and TlpA-like. Also, a full-length dipZ gene, found in the phylum Actinobacteria, is present exclusively in pathogenic bacteria (C. diphtheriae, C. jeikeium, M. avium, M. kansasii, M. marinum, M. ulcerans and M. tuberculosis) [40].
PiCp 3
potG gene, of the potFGHI operon, is a membrane-associated/ATP-binding protein that provides energy for putrescine (polyamine) uptake from the periplasmic space [41]. Although the potFGHI operon is a putrescine-specific transport system, potG is downregulated by another polyamine (spermine), which is produced only by eukaryotes. Carlson et al. (2009) demonstrated that transcription of the potG gene in Francisella tularensis decreases with high levels of spermine, while transcription of IS elements ISFtu1 and ISFtu2 increases in response to high levels of spermine in macrophages responding to bacterial infection. Also, many of the upregulated genes of F. tularensis (pseudogenes and transposase genes) are located near the IS elements in the chromosome [42].
The gene glpT belongs to the organophosphate:phosphate antiporter family of the major facilitator superfamily (MFS); it mediates transport of glycerol 3-phosphate (G3P) across the membrane in bacteria [43].
The PhoPR system regulates expression of various genes involved in metabolic, virulence and resistance processes in several intracellular bacterial pathogens [44]. Based on the information obtained from the complete genome sequence of C. pseudotuberculosis, we found that the PhoPR system is constituted of the phoP (714 bp) and phoR (1506 bp) genes, separated by a small 39-bp sequence, suggesting that these two genes are transcribed by a bicistronic operon. The size and organization of this system in C. pseudotuberculosis is similar to those of other Gram-positive bacteria [45]. Live bacteria attenuated via phoP inactivation are also promising vaccine candidates against tuberculosis. Several studies have reported the efficacy of attenuated mutant strains of M. tuberculosis as vaccines [46], [47]. Phylogenetic relationships within the class Actinobacteria strongly suggest correlation of the C. pseudotuberculosis PhoPR system with virulence mechanisms. The phoP gene is an important subject for regulation studies; and is also a probable vaccine candidate against CL.
PiCp4
The operon ciuABCDE (corynebacterium iron uptake) was described in C. diphtheriae as an iron transport and siderophore biosynthesis system. Proteins involved in iron acquisition are recognized as virulence factors, since they help pathogens to obtain iron from a host by using siderophores to strip iron from carrier proteins, such as transferrin, lactoferrin, and hemoglobin-haptoglobin [48,48].
PiCp5
Island 5 harbors a gene (pfoS) related to the pfoR superfamily. The pfoR gene was previously characterized as responsible for positive regulation of production of perfringolysin A (pfoA) and other toxins in Clostridium perfringens [50]. The virulence factors regulated by pfoR have not been totally elucidated. However, it is well known that deactivation of this gene inhibits hemolysis through negative regulation of several C. perfringens toxins. Clostridium perfringens harbors a phospholipase C gene (plc) that serves a function similar to that of phospholipase D [51]. Additionally, PiCp 5 contains a putative sigma 70 factor that is responsible for transporting the transcription machinery to specific promoters. Interestingly, the putative sigma 70 factor presents a nonsense mutation in C. pseudotuberculosis strain C231, which could be responsible for differential gene expression.
PiCp6
The pipA1 gene, which codes for a proline iminopeptidase, may have a role in pathogenesis, since it catalyses the removal of N-terminal proline residues from peptides; it also has a role in energy production [52]. In addition, a PIP-type protein is required for virulence of Xanthomonas campestris pv. campestris [53].
PiCp7
Island 7 harbors a urease operon that is also present in C. glutamicum; it is flanked, on both sides, by regions that are absent in the non-pathogenic C. glutamicum. This mosaicism is a common feature of pathogenicity islands [54]. The ure operon presents a codon usage deviation in C. glutamicum, as in C. pseudotuberculosis, indicating that this region is a putative genomic island in C. glutamicum.
The ure operon is responsible for nitrogen acquisition through hydrolysis of urea to carbamate and ammonia. Production of ammonia by uropathogenic and enteropathogenic bacteria causes cellular damage and compromises the action of the host's immune system [55]. Considering this fact, due to the intramacrophagic location of C. pseudotuberculosis and the finding of this operon in a non-pathogenic bacterial species, additional studies will be needed to elucidate how C. pseudotuberculosis obtains urea from the host and how this operon affects pathogenicity.
PiCp 7 also harbors a lysyl-tRNA synthetase (lysS), responsible for lysine incorporation into its respective transfer tRNA. The importance of lysS would normally make its location on a PAI inviable, since it is essential for cell metabolism. However, it is the only tRNA synthetase gene that is duplicated in the genome.
Protein classification of C. pseudotuberculosis in the biological process
Using the controlled vocabulary of functional terms proposed by the Gene Ontology (GO) Consortium for gene products classification [56], the predicted proteomes of the two genomes were analyzed according to the three organizing principles of gene ontology: cellular component, biological process and molecular function. The most abundantly represented categories are linked to metabolic processes in the two strains (cellular metabolic, biosynthetic, primary and macromolecule processes).
The gene products composition characterized using GO terminology suggests that C. pseudotuberculosis is a facultative intracellular pathogen. It is commonly found that pathogens specialized for an intracellular lifestyle have a high proportion of proteins linked to the above-mentioned processes. Moreover, the low proportion of proteins linked to the metabolism of secondary metabolites is an indication that C. pseudotuberculosis does not possess the metabolic machinery to deal with secondary metabolites, because they are supplied by the host.
Sub-cellular localization of C. pseudotuberculosis proteins
Prediction of the sub-cellular localization of C. pseudotuberculosis proteins was made by in silico analysis, using the SurfG+ tool [57]. Surfg+ is a pipeline for protein sub-cellular prediction, incorporating commonly used software for motif searches, including SignalP, LipoP and TMHMM, along with novel HMMSEARCH profiles to predict protein retention signals. Surfg+ starts by searching for retention signals, lipoproteins, SEC pathway export motifs and transmembrane motifs, roughly in this order. If none of these motifs are found in a protein sequence, then it is characterized as being cytoplasmic. A novel possibility introduced by Surfg+ is the ability to distinguish between integral membrane proteins versus PSE (potentially surface-exposed proteins). This is done by a parameter that determines the expected cell wall thickness, expressed in amino acids. Using published information or electron microscopy, it is possible to estimate cell wall thickness value for procaryotic organisms. C. pseudotuberculosis proteins were classified into four different sub-cellular locations: cytoplasmic, membrane, PSE (potentially surface exposed), or secreted. The C. pseudotuberculosis genomes were compared to those of other species of the genus, including C. diphtheriae, C. efficiens, C. glutamicum, C. jeikeium and C. urealyticum, also predicted by Surfg+, based on published cell wall thicknesses. Table 3 shows the number of predicted proteins in each sub-cellular location.
Table 3. Subcellular prediction of the protein locations derived from complete genomes of Corynebacterium species.
| Category/Species | Ce | CgB | CgK | CgR | Cj | Cd | Cu | Cp1002 | CpC231 | Total |
| Cytoplasm | 2,158 | 2,11 | 2,082 | 2,158 | 1,49 | 1,594 | 1,432 | 1,399 | 1,389 | 15,812 |
| Cytoplasm | 504 | 557 | 541 | 561 | 333 | 375 | 332 | 364 | 356 | 3,923 |
| PSE | 230 | 254 | 249 | 252 | 197 | 204 | 179 | 201 | 201 | 1,967 |
| Secreted | 102 | 136 | 121 | 109 | 100 | 99 | 79 | 95 | 107 | 948 |
| Total | 2,994 | 3,057 | 2,993 | 3,08 | 2,12 | 2,272 | 2,02 | 2,059 | 2,053 | 22,648 |
Ce: C. efficiens; CgB: C. glutamicum B; CgK: C. glutamicum K; CgR: C. glutamicum R; Cj: C. jeikeium; Cd: C. diphtheriae; Cu: C. urealyticum; Cp1002: C. pseudotuberculosis 1002; CpC231: C. pseudotuberculosis C231. PSE: potential surface exposure.
Comparison of the frequencies of subcellular occurrence of the C. pseudotuberculosis proteins and other Corynebacterium proteomes was made with Chi-square tests. The ratio between the four groups (cytoplasmic, membrane anchored, potentially exposed and secreted proteins) was found to be nearly constant among the Corynebacterium species. The proportions of the four protein categories cited above were similar to published data [58], [59]. Song and colleagues (2009) showed that approximately 30% of proteins secreted in gram-positive bacteria are exported through the Sec pathway. Few proteins (n = 27) were predicted to be secreted by the Tat pathway in Cp1002. About 2% of the proteins predicted to be secreted presented tertiary structures. In terms of proportions of secreted proteins, Cp1002 and CpC231 are at the higher end of the spectrum. They present 4.61 and 5.21%, respectively, predicted secreted proteins (Table 3).
Differences in metabolic pathways in the two strains of C. pseudotuberculosis
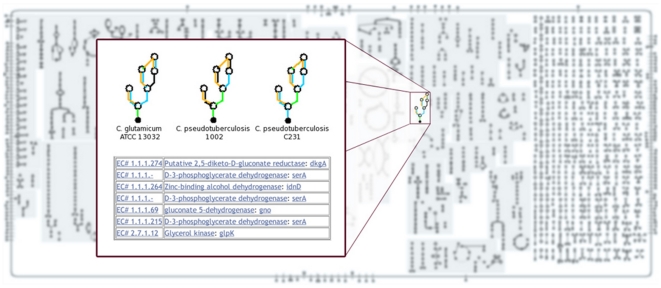
Automated reconstruction of the C. pseudotuberculosis Cp1002 metabolic pathways identified 156 pathways and 744 enzymatic reactions. As expected, quite similar results were encountered for strain CpC231: 154 pathways and 754 reactions (Table 4). Proteins of predicted functions that did not map to pathways, such as transport reactions, enzymes, transporters, and compounds, were also identified. The metabolic pathway database can be accessed online at http://corynecyc.cebio.org. This database enabled us to visualize and compare the metabolism of these two C. pseudotuberculosis strains (Figure 2).
Table 4. Comparative summary of the Corynebacterium pseudotuberculosis strain gene data types.
| Data Type | Cp1002 | CpC231 |
| Gene products | 2,059 | 2,053 |
| Pathways | 156 | 154 |
| Enzymatic Reactions | 744 | 754 |
| Transport Reactions | 8 | 4 |
| Polypeptides | 2,065 | 2,059 |
| Enzymes | 516 | 506 |
| Transporters | 10 | 10 |
| Compounds | 639 | 651 |
Figure 2. Corynebacterium glutamicum metabolic pathways overview.
C. glutamicum reactions are presented in blue and the reactions shared with C. pseudotuberculosis C231 and 1002 in red and green, respectively. By clicking on any compound or reaction, a window pops up showing details of each pathway. The fatty acid biosynthesis initiation pathway is the chosen example since computational evidence indicates it is not present only in strain C231.
We made a comparative analysis of transport reactions, pathways, compounds and proteins for C. pseudotuberculosis strains Cp1002 and CpC231 (Table 5). Despite the high similarity of the metabolic pathways, some differences were observed.
Table 5. Comparative summary of the number of pathways of Corynebacterium pseudotuberculosis strains Cp1002 and CpC231.
| Pathway Class | Cp1002 | CpC231 |
| - Pathway subclass | ||
| Biosynthesis | 105 | 104 |
| - Amine and Polyamine Biosynthesis | 5 | 3 |
| - Amino acid Biosynthesis | 25 | 26 |
| - Aminoacyl-tRNA Charging | 1 | 1 |
| - Aromatic Compound Biosynthesis | 1 | 1 |
| - Carbohydrate Biosynthesis | 10 | 7 |
| - Cell structure Biosynthesis | 4 | 4 |
| - Cofactor, Prosthetic Group, Electron Carrier Biosynthesis | 27 | 29 |
| - Fatty Acid and Lipid Biosynthesis | 8 | 7 |
| - Metabolic Regulator Biosynthesis | 1 | 2 |
| - Nucleoside and Nucleotide Biosynthesis | 12 | 12 |
| - Other Biosynthesis | 1 | 1 |
| - Secondary Metabolites Biosynthesis | 1 | 2 |
| Degradation/Utilization/Assimilation | 53 | 54 |
| - Alcohol Degradation | 2 | 1 |
| - Aldehyde Degradation | 1 | 1 |
| - Amine and Polyamine Degradation | 5 | 4 |
| - Amino Acid Degradation | 11 | 12 |
| - C1 Compound Utilization and Assimilation | 4 | 4 |
| - Carbohydrate Degradation | 7 | 7 |
| - Carboxylate Degradation | 5 | 4 |
| - Degradation/Utilization/Assimilation - Other | 5 | 5 |
| - Fatty Acid and Lipid Degradation | 3 | 2 |
| - Inorganic Nutrient Metabolism | 4 | 6 |
| - Nucleoside and Nucleotide Degradation and Recycling | 2 | 3 |
| - Secondary Metabolite Degradation | 5 | 5 |
| Generation of precursor metabolites and energy | 16 | 19 |
| Total | 163 | 164 |
The metabolic pathways in each of the two bacterial strains (Cp1002 and CpC231) were classified into several pathway classes; each pathway class was further broken down to show the distribution of pathways among the next-level subclasses. Analysis of the metabolism database of C. pseudotuberculosis strains Cp1002 and CpC231 revealed specific pathway differences between the two strains. Overall, CpC231 had 13 specific metabolic pathways not found in strain Cp1002, and the latter had 11 metabolic pathways not found in strain CpC231 (Table 6).
Table 6. Table listing the Corynebacterium pseudotuberculosis strain-specific pathways.
| Pathway Class | Cp1002 | CpC231 |
| Pathway Name | ||
| Biosynthesis - Amines and Polyamines Biosynthesis | ||
| choline degradation I | present | absent |
| glycine betaine biosynthesis I (Gram-negative bacteria) | present | absent |
| Biosynthesis - Amino acid Biosynthesis | ||
| citrulline-nitric oxide cycle | absent | present |
| Carbohydrates Biosynthesis | ||
| gluconeogenesis | present | absent |
| trehalose biosynthesis II | present | absent |
| trehalose biosynthesis III | present | absent |
| Biosynthesis - Cofactor, Prosthetic Group, and Electron Carrier Biosynthesis | ||
| adenosylcobalamin biosynthesis from cobyrinate a,c-diamide I | absent | present |
| heme biosynthesis from uroporphyrinogen I | present | absent |
| heme biosynthesis from uroporphyrinogen II | absent | present |
| siroheme biosynthesis | absent | present |
| Biosynthesis - Fatty Acid and Lipid Biosynthesis | ||
| biotin-carboxyl carrier protein | absent | present |
| cardiolipin biosynthesis I | present | absent |
| fatty acid biosynthesis initiation I | present | absent |
| Secondary Metabolite Biosynthesis | ||
| canavanine biosynthesis | absent | present |
| Biosynthesis - Metabolic Regulators Biosynthesis | ||
| citrulline-nitric oxide cycle | absent | present |
| Degradation - Alcohols Degradation | ||
| glycerol degradation II | present | absent |
| Degradation - Aldehyde Degradation | ||
| methylglyoxal degradation I | absent | present |
| methylglyoxal degradation III | present | absent |
| Degradation - Amine and Polyamine Degradation | ||
| choline degradation I | present | absent |
| Degradation - Amino Acid Degradation | ||
| 2-ketoglutarate dehydrogenase complex | absent | present |
| citrulline-nitric oxide cycle | absent | present |
| valine degradation I | present | absent |
| Degradation - Carboxylate Degradation | ||
| acetate formation from acetyl-CoA I | present | absent |
| Degradation - Fatty Acid and Lipids Degradation | ||
| triacylglycerol degradation | present | absent |
| Inorganic Nutrients Metabolism | ||
| nitrate reduction III (dissimilatory) | absent | present |
| nitrate reduction IV (dissimilatory) | absent | present |
| Degradation - Nucleoside and Nucleotide Degradation and Recycling | ||
| purine deoxyribonucleoside degradation | absent | present |
| Generation of precursor metabolites and energy | ||
| 2-ketoglutarate dehydrogenase complex | absent | present |
| nitrate reduction III (dissimilatory) | absent | present |
| nitrate reduction IV (dissimilatory) | absent | present |
Two amine and polyamine biosynthesis pathways, choline degradation I and glycine betaine biosynthesis I (Gram-negative bacteria), were found in strain Cp1002 but not in strain CpC231. Strain CpC231 was found to have an extra amino acid biosynthesis pathway, the citrulline-nitric oxide cycle. Strain Cp1002 was found to have three additional carbohydrate biosynthesis pathways: gluconeogenesis, trehalose biosynthesis II and trehalose biosynthesis III. Strain CpC231 showed three cofactor biosynthesis, prosthetic group and electron carrier pathways, corresponding to adenosylcobalamin biosynthesis from cobyrinate a,c-diamide I, heme biosynthesis from uroporphyrinogen II and siroheme biosynthesis. Strain Cp1002 showed only one unique cofactor biosynthesis pathway, heme biosynthesis from uroporphyrinogen I. Two extra pathways of fatty acid and lipid biosynthesis were found in strain Cp1002, cardiolipin biosynthesis I and fatty acid biosynthesis initiation I. Strain CpC231 showed only the biotin-carboxyl carrier protein. Among metabolic regulator biosynthesis genes, strain CpC231 showed the citrulline-nitric oxide cycle. Strain CpC231 also showed an extra pathway, the canavanine biosynthesis pathway, part of secondary metabolite biosynthesis.
Among degradation/utilization/assimilation pathways, strain Cp1002 showed an extra pathway: glycerol degradation II, for alcohol degradation, as well as choline degradation I for amine and polyamine degradation. Strain CpC231 was found to have two additional pathways, 2-ketoglutarate dehydrogenase complex and citrulline-nitric oxide cycle, for amino acid pathways; strain Cp1002 showed only one extra pathway, valine degradation I. Among carboxylate degradation pathways, involving fatty acid and lipid degradation, strain Cp1002 showed two extra pathways: one corresponding to acetate formation from acetyl-CoA I, and the second linked to triacylglycerol degradation. Two inorganic nutrient metabolism pathways were found in strain CpC231 but not in strain Cp1002: nitrate reduction III (dissimilatory) and nitrate reduction IV (dissimilatory), and a nucleoside and nucleotide degradation and purine deoxyribonucleoside recycling degradation pathway.
Finally, when we analyzed the generation of precursor metabolites and energy, strain CpC231 showed three extra pathways: 2-ketoglutarate dehydrogenase complex, nitrate reduction III (dissimilatory) and nitrate reduction IV (dissimilatory). The differences are presented in Table 6.
Metabolic pathways in C. pseudotuberculosis compared to other Corynebacterium species
The web interface enabled us to visually compare the metabolic pathways of strains Cp1002 and CpC231 reactions (Figure 2) with those of four other bacteria of the genus Corynebacterium: C. diphtheriae, C. efficiens, C. glutamicum, and C. jeikeium. Using these diagrams we were able to easily spot reactions present in C. pseudotuberculosis and absent in other Corynebacterium species.
A comparative analysis of reactions, pathways, compounds and proteins was also done for C. pseudotuberculosis and other closely-related bacteria in the same genus. The list of C. pseudotuberculosis specific pathways is shown in Table 7.
Table 7. List of Corynebacterium pseudotuberculosis specific metabolic pathways that were compared to those of closely-related bacteria, including C. diphtheriae, C. glutamicum, C. efficiens, and C. jeikeium.
| Pathway Class |
| Pathway Name |
| Biosynthesis - Amino acid Biosynthesis |
| Asparagine biosynthesis II |
| Lysine biosynthesis V |
| Biosynthesis - Metabolic Regulators Biosynthesis |
| Citrulline-nitric oxide cycle |
| Biosynthesis - Nucleoside and Nucleotide Biosynthesis |
| Salvage pathways of pyrimidine deoxyribonucleotides |
| Degradation - Alcohol Degradation |
| Glycerol degradation II |
| Degradation - Aldehyde Degradation |
| Methylglyoxal degradation III |
| Degradation - Amino Acid Degradation |
| Alanine degradation IV |
| Citrulline-nitric oxide cycle |
| Lysine degradation I |
| Degradation - C1 Compound Utilization and Assimilation |
| Reductive monocarboxylic acid cycle |
| Degradation - Carbohydrate Degradation |
| Chitobiose degradation |
| Degradation - Carboxylate Degradation |
| Conversion of succinate to propionate |
| Degradation - Fatty Acid and Lipid Degradation |
| Phospholipases |
| Inorganic Nutrients Metabolism |
| Ammonia oxidation I (aerobic) |
| Nitrate reduction IV (dissimilatory) |
| Degradation - Secondary Metabolite Degradation |
| D-glucarate degradation |
| Betanidin degradation |
| D-galactarate degradation |
| Generation of precursor metabolites and energy |
| Ammonia oxidation I (aerobic) |
We found that C. pseudotuberculosis has several pathways that are not found in other species of the genus Corynebacterium. However, little information is available about these pathways in Corynebacterium spp. We found no published information concerning the following pathways: asparagine biosynthesis II, citrulline-nitric oxide cycle (amino acid biosynthesis and degradation), pyrimidine deoxyribonucleotide salvage pathways, methylglyoxal degradation III, reductive monocarboxylic acid cycle, chitobiose degradation, conversion of succinate to propionate, ammonia oxidation I (aerobic), nitrate reduction IV (dissimilatory), D-glucarate degradation, betanidin degradation, D-galactarate degradation, and ammonia oxidation I (aerobic).
Some studies reported five pathways: lysine biosynthesis V, glycerol degradation II, alanine degradation IV, lysine degradation I and phospholipases. However, none of the studies, except for those concerning lysine degradation I and phospholipase pathways, involved C. pseudotuberculosis. Most of these studies were carried out with C. glutamicum.
Four papers concerning C. glutamicum were found for the lysine degradation I pathway [60]–[63]. Studies have focused on: acetohydroxyacid synthase, a novel target for improvement of L-lysine production [62], improvement of L-lysine formation by expression of the Escherichia coli pntAB genes [61], genetic and functional analysis of soluble oxaloacetate decarboxylase [63], and modeling and experimental design for metabolic flux analysis of lysine-producing Corynebacteria by mass spectrometry [64].
Six studies were found concerning the glycerol degradation II pathway, one performed with C. diphtheria [65] and four with C. glutamicum [66]–[69]. In the sixth study, made with C. glutamicum, we found information on the alanine degradation IV pathway [64].
Approximately 140 studies, of which 107 were made with C. glutamicum alone, dealt with the lysine degradation I pathway, in which cadaverine is biosynthesized from L-lysine. Cadaverine is reported to be essential for the integrity of the cell envelope and for normal growth of the organism, as well as for inhibiting porin-mediated outer membrane permeability, thereby protecting cells from acid stress [70], [71].
All studies of specific phospholipase pathways were carried out with C. pseudotuberculosis. Phospholipases hydrolyze phospholipids and are ubiquitous in all organisms. Several types of phospholipases were reported; phospholipase D is the best studied and has been considered a major virulence factor for C. pseudotuberculosis [72], [73]. In our analyses, none of the five bacteria of the genus Corynebacterium were found to have pathways belonging to the following subclasses: siderophore biosynthesis; chlorinated compound degradation; cofactor, prosthetic group, electron carrier, and hormone degradation. Clearly more biochemical studies are needed. Our current study brings new insight to relevant biochemical pathways that can be further explored experimentally.
We made a comparative summary of the metabolic pathways of C. pseudotuberculosis strains Cp1002 and CpC231 and C. glutamicum (Table 8). C. glutamicum has several metabolic pathways not found in C. pseudotuberculosis Cp1002 and/or in C. pseudotuberculosis CpC231. Overall, C. glutamicum has approximately 40 additional metabolic pathways.
Table 8. Comparative summary of Corynebacterium pseudotuberculosis strains Cp1002 and CpC231 and C. glutamicum pathways.
| Pathway Class | Cp1002 | CpC231 | C. glutamicum |
| - Pathway subclass | |||
| Biosynthesis | 105 | 104 | 131 |
| - Amine and Polyamine Biosynthesis | 5 | 3 | 3 |
| - Amino acid Biosynthesis | 25 | 26 | 29 |
| - Aminoacyl-tRNA Charging | 1 | 1 | 3 |
| - Aromatic Compound Biosynthesis | 1 | 1 | 1 |
| - Carbohydrate Biosynthesis | 10 | 7 | 9 |
| - Cell structure Biosynthesis | 4 | 4 | 4 |
| - Cofactor, Prosthetic Group, Electron Carrier Biosynthesis | 27 | 29 | 38 |
| - Fatty Acid and Lipids Biosynthesis | 8 | 7 | 14 |
| - Metabolic Regulator Biosynthesis | 1 | 2 | 1 |
| - Nucleoside and Nucleotide Biosynthesis | 12 | 12 | 10 |
| - Other Biosynthesis | 1 | 1 | 1 |
| - Secondary Metabolite Biosynthesis | 1 | 2 | 6 |
| Degradation/Utilization/Assimilation | 53 | 54 | 72 |
| - Alcohols Degradation | 2 | 1 | 2 |
| - Aldehyde Degradation | 1 | 1 | 1 |
| - Amine and Polyamine Degradation | 5 | 4 | 6 |
| - Amino Acid Degradation | 11 | 12 | 15 |
| - Aromatic Compound Degradation | 0 | 0 | 9 |
| - C1 Compound Utilization and Assimilation | 4 | 4 | 2 |
| - Carbohydrate Degradation | 7 | 7 | 10 |
| - Carboxylate Degradation | 5 | 4 | 6 |
| - Chlorinated Compound Degradation | 0 | 0 | 4 |
| - Degradation/Utilization/Assimilation - Other | 5 | 5 | 2 |
| - Fatty Acid and Lipid Degradation | 3 | 2 | 2 |
| - Inorganic Nutrient Metabolism | 4 | 6 | 9 |
| - Nucleoside and Nucleotide Degradation and Recycling | 2 | 3 | 1 |
| - Secondary Metabolite Degradation | 5 | 5 | 4 |
| Generation of precursor metabolites and energy | 16 | 19 | 25 |
| Total | 163 | 164 | 206 |
Among biosynthesis pathways, C. glutamicum showed around 30 extra pathways when compared to the two strains of C. pseudotuberculosis. These involve pathways of amino acid biosynthesis, aminoacyl-tRNA charging, cofactors, prosthetic groups, electron carrier biosynthesis, fatty acid and lipid biosynthesis and secondary metabolite biosynthesis. However, the two strains of C. pseudotuberculosis also have specific pathways that were not found in C. glutamicum, these being the pathways of amine and polyamine biosynthesis, carbohydrate biosynthesis and nucleoside and nucleotide biosynthesis.
Among the degradation/utilization/assimilation pathways, C. glutamicum presented around 20 extra pathways, when compared to C. pseudotuberculosis Cp 1002 and C. pseudotuberculosis CpC231. These specific pathways of C. glutamicum correspond to pathways of amine and polyamine degradation, amino acid degradation, aromatic compound degradation, carbohydrate degradation, carboxylate degradation, chlorinated compound degradation and the metabolism of inorganic nutrients. Again, the two strains of C. pseudotuberculosis also had specific pathways involving degradation/utilization/assimilation, fatty acid and lipid degradation and secondary metabolite degradation that were not found in C. glutamicum.
We found 25 pathways involving generation of precursor metabolites and energy in C. glutamicum, while C. pseudotuberculosis Cp1002 had only 16 and C. pseudotuberculosis CpC231 had 19.
Discussion
General aspects of the C. pseudotuberculosis genome
The C. pseudotuberculosis genome has proven to be one of the smallest genomes of the Corynebacterium genus sequenced so far, with Cp1002 being the smallest and Cp231 the fourth smallest, larger only than Cp1002, C. lipophiloflavum DSM 44291 (2,293,743 bp) and C. genitalium ATCC 33030 (2,319,774 bp); the latter two are both human pathogens. Corynebacterium pseudotuberculosis has a very small genetic repertoire, with considerable gene loss when compared to non-pathogenic species such as C. glutamicum and C. efficiens. When predicted proteomes were compared, C. pseudotuberculosis showed a loss of approximately 1,220 genes, in comparison with C. glutamicum. Classification of these proteins using GO terminology showed that the majority are linked to metabolic processes, such as cellular, primary, biosynthetic, macromolecule, nitrogen compound and oxidation reduction processes.
Other characteristics of the C. pseudotuberculosis genome include the lowest GC content in the Corynebacterium genus, this being 52% in both the goat and sheep strains, followed by C. diphtheriae with a GC content of 53%. This contrasts with C. urealyticum, which has a GC content of 64%. Furthermore, C. pseudotuberculosis has a higher number of predicted pseudogenes and a lower number of tRNAs, when compared to other species of the Corynebacterium genus for which genome sequences are available.
Merjeh et al. (2009) made a comparative analysis of 317 genomes of bacteria with different lifestyles (free-living, facultative intracellular and obligate intracellular). They found evidence that peculiar characteristics in bacterial genomes can drive the organisms to certain lifestyles. All characteristics cited in their work were identified in the C. pseudotuberculosis genomes. Lower GC content generally can occur due to gene loss, which is a means to contract the genome in response to a specialized environment. Moreover, presence of a higher number of pseudogenes could be evidence of bacterial mechanisms to generate non-functional genes and subsequent gene loss [19]. In addition, the high proportion of proteins linked to primary metabolism, and the small proportion of proteins related to secondary metabolism, is usually seen in facultative intracellular organisms. Taking these aspects of the genomic architecture of C. pseudotuberculosis into account, it can be affirmed that C. pseudotuberculosis has a facultative intracellular lifestyle.
High similarity in the genome architecture
Usually, pseudogenes are characterized as genes that have lost their function in the genome, due either to changes in the reading frame (frameshifts) or to a premature stop codon. Pseudogenes are common in prokaryotes; most have been linked to a sudden change in the environment of the pathogen, with simultaneous loss of metabolic and respiratory activities [74].
The high number of pseudogenes in these two strains of C. pseudotuberculosis (52 in Cp1002 and 50 pseudogenes in CpC231) suggest an evolutionary process involving a contracting genome in this species. An example of this is also seen in Mycobacterium leprae, which has a large number of pseudogenes (around 1,000). When we compare M. leprae to M. tuberculosis, the latter has both considerably fewer genes and a higher number of pseudogenes that can drive this gene loss.
Virulence factors acquired
Identification of pathogenicity islands (PAIs) in pathogenic bacteria is highly relevant for understanding the reasons behind different responses to vaccines and the biological mechanisms leading to genome plasticity. The biovars equi and ovis of C. pseudotuberculosis cause distinct diseases in their hosts; assessment of virulence genes could help identify genes involved in these host-specific differences.
Virulence genes, which are central to distinguishing pathogenic from non-pathogenic species, are present in PAIs in large numbers. Additionally, the fact that PAIs are a consequence of horizontal transfer events indicates that the virulence factors they contain can help increase the adaptability of strains to different host environments. This increase in adaptability is demonstrated by the finding of genes with functions associated with uptake of iron (fag operon), carbon (malL) and Mg2+ from the host, since this uptake improves survival under stress conditions, such as iron depletion, starvation and heat shock. Furthermore, PAIs of C. pseudotuberculosis present genes that respond to a macrophagic environment (potG, sigK and dipZ), which sheds new light on the mechanisms responsible for the intramacrophagic lifestyle of this organism.
Gene Sharing among C. pseudotuberculosis strains
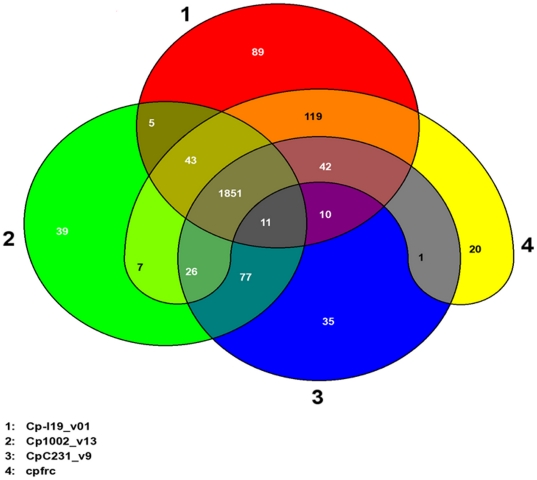
Considering the four available genomes of C. pseudotuberculosis strains (Cp1002, CpC231, and CpI19 pFRC41), we identified 1,851 whole genes shared among them (Figure 3).
Figure 3. Venn diagram illustrating the three genomic categories of four Corynebacterium pseudotuberculosis strains: core, accessory and extended genome.
Data obtained from the comparison of the predicted proteomes of four C. pseudotuberculosis speices in the EDGAR program (Blom et al., 2009). In red: Cp-I19; green: Cp1002; blue: CpC231 and yellow: CpFRC41. The remaining colors illustrate the shared genes among strains. The numbers within the forms indicate the number of shared genes.
This repertoire of genes is vast for this specie, since, among the four isolates the maximum number of genes is 2,377 (called the pangenome of the species). When we compare the number of genes shared by these four C. pseudotuberculosis strains with a study of 17 strains of the bacterium E. coli [75], we conclude that C. pseudotuberculosis has a greater proportion of shared genes. In isolates of E. coli, 2,220 genes constituted the core genome, less than half of the genes in this species, with a mean of 5,000 genes in each genome [75]. Other significant information that emerges from this data is that the C. pseudotuberculosis genomes are extremely similar, since we found no significant change in the composition of the repertoire of genes for this species after adding the two new strains (Figure 3).
Gene Sharing between C. pseudotuberculosis and other Corynebacterium species
Previous comparative studies of sequences of the rpoB gene of C. pseudotuberculosis and C. diphtheriae have suggested a close relationship between them [27], [76]. In our current study, we confirmed this close relationship with several types of evidence: i) a similar codon bias, ii) high similarity at the amino acid level and iii) conserved synteny. Synteny analysis of the genomes of the two C. pseudotuberculosis strains compared to C. diphtheriae indicates that these genomes are highly conserved; the gene position is conserved within the species. This observation reinforces the conclusions of previous research claiming conserved synteny in this genus, which indicated that few rearrangement events occurred during evolution [25].
Corynebacterium pseudotuberculosis shares more orthologous genes with C. glutamicum (1,345 genes), C. efficiens (1,330), C. diphtheriae (1,263 genes) and C. auricumucosum (1,273 genes); it shares only 1,030 genes with C. jeikeium and C. kroppenstedtii.
The larger number of genes shared between C. pseudotuberculosis, C. glutamicum and C. efficiens (72%), compared to other species (pathogenic species, 60%), may be a result not only of their close relationships, but also because a comparison is made among species with a larger gene repertoire, such as C. glutamicum and C. efficiens, which are non-pathogenic microorganisms, thus increasing the possibility of sharing genes.
Lineage-specific genes in C. pseudotuberculosis
Most of the lineage-specific genes are involved in processes of virulence, pathogenicity, drug resistance and response to certain types of stress. These factors can increase the adaptability of microorganisms to the niches they inhabit, but they are not indispensable to the survival of pathogens. Moreover, some copies of these genes can be acquired by horizontal transfer. These genes are not ORFans; they already have been characterized in other species. The terminology ‘lineage-specific’ portrays only some genes found among the four strains in our study; the same genes may be found in other species.
We found 49 lineage-specific genes in CpC231 and 52 in Cp1002. For most of them, we did not have a descriptive characterization of their products, and they were classified as hypothetical proteins. In addition, many of these identified genes, in both strains, encode membrane and secreted proteins and pseudogenes. On the other hand, some well-characterized proteins were found in the genome. One example is found in CpC231, which has the gene called pthA; this gene encodes an effector system of type III secretion and is related to bacterial growth and host cell lesions, as found in Xanthomonas campestris [77]. This gene may be a good target for understanding the development of C. pseudotuberculosis CpC231 inside the host and the necrosis seen in CL abscesses, where it plays the same role in this pathogen.
In Cp1002, a very interesting gene was found, tatA, which encodes a membrane protein translocase, involved in the secretion of proteins in their final conformation, through the inner membrane to the extracellular environment. This gene is interesting because it is independent of the Sec secretion system and is a unique copy among the strains, suggesting that Cp1002 may have other routes for secretion. Regarding the large number of hypothetical proteins found in this strain, it may harbor genes that came from horizontal transfer, including some from phylogenetically-distant organisms, for which genomic molecular characterization has not been made.
Finally, lineage-specific genes may be good tools for understanding the host-pathogen interaction and may be good targets for the development of computational tools for differentiation between these strains, for molecular epidemiology.
Biochemical properties of C. pseudotuberculosis
In the latest review of the biochemical properties of C. pseudotuberculosis [76], Dorella and colleagues gathered information concerning its metabolism, virulence and pathogenesis. They reported that the peptidoglycan in the cell wall is based on meso-DAP acid, and that arabinose and galactose are major cell-wall sugars. Our analyses predicted all of the reactions of the peptidoglycan biosynthesis II pathway; the meso-DAP acid compound was found as a product/substrate of the reaction catalyzed by UDP-N-acetylmuramyl tripeptide synthase (6.3.2.13). The complete pathway of UDP-galactose biosynthesis was also found; although there was no evidence of biosynthesis of arabinose, we detected a membrane transporter, known as arabinose efflux permease.
We also found short-chain mycolic acids; 10 variations of acids of this type were encountered, including 6-O-cis-keto-mycolyl-trehalose-6-phosphate, and 6-O-mycolyl-trehalose-6-phosphate. The two strains of C. pseudotuberculosis showed considerable fermentation ability, with several fermentation pathways, including glycolysis III, mixed acid fermentation and pyruvate fermentation to acetate IV, ethanol I and lactate.
Several sugar degradation pathways were also found in the two strains of C. pseudotuberculosis, including galactose, lactose, sucrose and L-and D-arabinose degradation. We confirmed that, as reported by Dorella et al. (2006), all these pathways produce acids and no gasses, generating large amounts of energy.
It was also previously reported that C. pseudotuberculosis is phospholipase D and catalase positive. Our analysis showed that both phospholipase D and catalase are involved in important processes. The main molecular functions of phospholipase D are phospholipase D activity, magnesium ion binding, NAPE-specific phospholipase D activity and sphingomyelin phosphodiesterase D activity. Catalase, which is produced by the cat gene, is involved in response to oxidative stress and oxidation reduction. Although two enzymes of the denitrification pathway (nitrate reduction I) were found, absence of the remaining enzymes is probably the determining factor for the inability of these strains to reduce nitrate to N2, as reported by Dorella et al. (2006).
We also detected iron acquisition genes (fag) A, B, C and D in both strains of C. pseudotuberculosis [78]. Genes fagA and fagB produce the integral membrane proteins FagA, an iron-enterobactin transporter, and FagBy; both have important roles, including ion, transmembrane, organic acid and protein transport. The ATP binding cytoplasmic membrane protein, FagC, produced by gene fagC, has two main molecular functions: ATP binding and ATPase activity. Finally, gene fagD produces the iron siderophore binding protein, FeAcquisition gene D, which has a role in iron ion transmembrane transport activity.
Computational reconstruction of the C. pseudotuberculosis pathways in our database not only allowed us to better visualize the metabolism of this bacterium, but also to compare it to closely related species. The main purpose of this analysis was to describe C. pseudotuberculosis metabolism by computational means, providing a predictive tool for “wet-lab” research.
Methods
Bacterial strains and growth conditions
Corynebacterium pseudotuberculosis 1002 biovar ovis (herein referred to as Cp1002) is a wild strain, isolated from a caprine host in Brazil. Corynebacterium pseudotuberculosis C231 biovar ovis (herein referred to as CpC231) is also a wild strain, isolated from an ovine host in Australia. Both strains were confirmed to be C. pseudotuberculosis by routine biochemical tests (API CORYNE, Biomerieux, Marcy l'Etoile, France). These strains were maintained in brain-heart-infusion broth (BHI – HiMedia Laboratories Pvt. Ltda, India) at 37°C, under rotation.
Preparation of high molecular weight DNA
Chromosomal DNA extraction was performed as follows: 50 mL of 48–72 h cultures of the two strains were centrifuged at 4°C and 2000 x g for 20 min. Cell pellets were re-suspended in 1 mL Tris/EDTA/NaCl [10 mM Tris/HCl (pH 7.0), 10 mM EDTA (pH 8.0), and 300 mM NaCl] and centrifuged again under the same conditions. Supernatants were discarded, and the pellets were re-suspended in 1 mL TE/lysozyme [25 mM Tris/HCl (pH 8.0), 10 mM EDTA (pH 8.0), 10 mM NaCl, and 10 mg lysozyme/mL]. Samples were then incubated at 37°C for 30 min. Thirty milliliters of 30% (w/v) sodium N-lauroyl-sarcosine (Sarcosyl) were added to each sample and the mixtures were incubated for 20 min at 65°C, followed by incubation for 5 min at 4°C. DNA was purified using phenol/chloroform/isoamyl alcohol (25∶24∶1) and precipitated with ethanol. DNA concentrations were determined spectrophotometrically, and the DNA was visualized in ethidium bromide-stained 0.7% agarose gels.
Construction of Corynebacterium pseudotuberculosis genomic libraries and Sanger sequencing
For the shotgun strategy used to sequence C. pseudotuberculosis 1002, four small fragment libraries were constructed using the TOPO Shotgun cloning kit and the pCR4 Blunt-TOPO vector (Invitrogen), according to the manufacturer's instructions. Sanger sequencing was carried out using the Minas Gerais Genome Network (http://rgmg.cpqrr.fiocruz.br). A total of 6,144 forward and reverse reads were produced using the DYEnamic Dye Terminator kit and run in a Megabace 1000 automated sequencer (GE Healthcare).
Genome Sequencing
Cp1002 was sequenced using both Sanger and pyrosequencing technologies. Pyrosequencing was carried out using 454 Life Sciences (Branford, CT). A total of 397,147 high quality reads and 86,154,153 high quality bases were obtained, which translates into approximately 31-fold coverage. The average length of the sequences was 253 bases. The sequences were delivered after quality filtering and preassembly with the Newbler assembler (454 Life Sciences).
CpC231 was sequenced with a Roche-454 FLX sequencer at the Australian Animal Health Laboratory, Geelong, Australia. A total of 347,361 reads generated 80,336,550 bases, giving 34-fold coverage of the genome. De novo assembly of the filtered sequence data was carried out using the Newbler software. This assembly produced 10 large contigs in four scaffolds. The remaining gaps in the genomic sequence were closed by PCR walking and Sanger sequencing of the resulting fragments.
Treatment and assembly data
The raw Sanger data obtained from sequencing were processed using the Phred-Phrap-Consed package [75]. Possible contaminants (plasmid DNA, sequences with similarity to vectors and other contaminants) were discarded using the Cross_match program (www.phrap.org). The quality value used in the base-calling program was Q = 40 (Probability of incorrect base call 1 in 10,000/base call accuracy 99.99%). An assembly using Phrap parameters (Force Level: 40 and Gap Length: 10,000) was carried out.
The 454 data were processed using the Newbler assembler (454 Life Sciences), and the final genomic consensus sequence was obtained using the Phrap algorithm.
Genome annotation
The annotation procedures involved the use of several algorithms in a multi-step process. Structural annotation was performed using the following software: FgenesB: gene predictor (www.softberry.com); RNAmmer: rRNA predictor [79]; tRNA-scan-SE: tRNA predictor [80]; and Tandem Repeat Finder: repetitive DNA predictor (tandem.bu.edu/trf/trf.html). Functional annotation was performed by similarity analyses, using public databases and InterProScan analysis [81]. Manual annotation was performed using Artemis [82].
Identification and confirmation of putative pseudogenes in the genome was carried out using Consed. Manual analysis was performed based on the Phred quality of each base in the frameshift area. This analysis enabled the identification of erroneous insertions or deletions of bases in the genome information produced by the sequencing process, and it avoided identification of false-positive pseudogenes.
Predictions of the cellular locations of Corynebacterium proteins were made using the program SurfG Plus (version 1.0), with a minimum protein size of 73 amino acids. Classification of predicted proteins in functional categories was made using the BLAST2GO program (www.blast2go.org). The cutoff value used was 10−6 (http://www.blast2go.org/).
In silico Identification of Pathogenicity Islands
In order to accurately identify and classify putative Pathogenicity Islands (PAIs) in the corynebacterial genomes, we developed a combined computational approach using several in-house scripts to integrate the prediction of diverse algorithms and databases, namely: Colombo-SIGIHMM [83], Artemis [82], tRNAscan-SE [80]; EMBOSS-geecee [84], ACT: the Artemis Comparison Tool [85], and mVIRdb [86].
In silico metabolic pathway construction
The two main data sources used for reconstructing the C. pseudotuberculosis metabolic pathways were the genome sequence file in FASTA format and the genome annotation file in GBK format. Metabolic pathways databases for strains 1002 and C231 were created using the Pathway tools 13 software, developed by SRI International [87]. The Pathway tools software contains algorithms that predict metabolic pathways of an organism from its genome by comparison to a reference pathways database known as MetaCyc [88]. Construction of a metabolic pathways database was done using BioCyc [89], in order to compare the different bacteria, C. diphtheriae NCTC 13129, C. efficiens YS-314, C. glutamicum ATCC 13032, and C. jeikeium K411, to the deduced C. pseudotuberculosis pathways.
Comparative analysis of Corynebacterium pseudotuberculosis strains
Comparative analyses were made for the two C. pseudotuberculosis strains. Similarity analyses of the two genomes were made using the BLAST - NCBI [90], [91] and InterProScan databases. The Mauve algorithm (gel.ahabs.wisc.edu/mauve) and the ACT tool were used to identify whether blocks had undergone gene rearrangements or remained preserved. The Plotter program of the MUMMer 3.22 package (mummer.sourceforge.net) was used for synteny analysis.
Supporting Information
Orthologous genes present inside PAIs regions of C. pseudotuberculosis and their counterparts in other Corynebacterium species.
(DOC)
Acknowledgments
The authors acknowledge the scientific support of two genomics networks: Rede Paraense de Genômica e Proteômica and Rede Genoma de Minas Gerais. In addition, the authors thank for valuable contribution of all Public Institutions involved and their respective co-authors during the development of the present work.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project received financial support from the following grants: FAPEMIG (Fundação de Amparo à pesquisa do Estado de Minas Gerais) (CBB-1181/0 and REDE-186/08 to Guilherme Oliveira), NIH (National Institutes of Health) - Fogarty (TW007012 to Guilherme Oliveira). Guilherme Oliveira and Vasco Azevedo are CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) fellows. The work also received support from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ayers JL. Caseous lymphadenitis in goat and sheep: review of diagnosis, pathogenesis, and immunity. JAVMA. 1977;171:1251–1254. [PubMed] [Google Scholar]
- 2.Brown CC, Olander HJ, Alves SF. Synergistic hemolysis-inhibition titers associated with caseous lymphadenitis in a slaughterhouse survey of goats and sheep in northeastern Brazil. Can J Vet Res. 1987;51:46–49. [PMC free article] [PubMed] [Google Scholar]
- 3.Merchant IA, Packer RA. The genus Corynebacterium. In: Merchant IA, Packer RA, editors. Veterinary Bacteriology and Virology. USA: The Iowa State University Press; 1967. pp. 425–440. [Google Scholar]
- 4.Piontkowski MD, Shivvers DW. Evaluation of a commercially available vaccine against Corynebacterium pseudotuberculosis for use in sheep. J Am Vet Med Assoc. 1998;212:1765–1768. [PubMed] [Google Scholar]
- 5.Join-Lambert OF, Ouache M, Canioni D, Beretti JL, Blanche S, et al. Corynebacterium pseudotuberculosis necrotizing lymphadenitis in a twelve-year old patient. Pediatr Infect Dis J. 2006;25(9):848–851. doi: 10.1097/01.inf.0000234071.93044.77. [DOI] [PubMed] [Google Scholar]
- 6.Trost E, Ott L, Schneider J, Schroder J, Jaenicke S, et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics. 2010;11(1):728–745. doi: 10.1186/1471-2164-11-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor KM, Quirie MM, Baird G, Donachie W. Characterization of united kingdom isolates of Corynebacterium pseudotuberculosis using pulsed-field gel electrophoresis. J Clin.Microbiol. 2000;38:2633–2637. doi: 10.1128/jcm.38.7.2633-2637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben Saïd MS, Ben Maitigue H, Benzarti M, Messadi L, Rejeb A, et al. Epidemiological and clinical studies of ovine caseous lymphadenitis. Arch Inst Pasteur Tunis. 2002;79:51–57. [PubMed] [Google Scholar]
- 9.Binns SH, Bailey M, Green LE. Postal survey of ovine caseous lymphadenitis in the United Kingdom between 1990 and 1999. Vet Rec. 2002;150:263–268. doi: 10.1136/vr.150.9.263. [DOI] [PubMed] [Google Scholar]
- 10.Arsenault J, Girard C, Dubreuil P, Daignault D, Galarneau JR, et al. Prevalence of and carcass condemnation from maedi-visna, paratuberculosis and caseous lymphadenitis in culled sheep from Quebec, Canada. Prev Vet Med. 2003;59:67–81. doi: 10.1016/s0167-5877(03)00060-6. [DOI] [PubMed] [Google Scholar]
- 11.Paton MW, Walker SB, Rose IR, Watt GF. Prevalence of caseous lymphadenitis and usage of caseous lymphadenitis vaccines in sheep flocks. Aust Vet J. 2003;81:91–95. doi: 10.1111/j.1751-0813.2003.tb11443.x. [DOI] [PubMed] [Google Scholar]
- 12.Pinheiro RR, Gouveia AMG, Alves FSF, Haddad JP. Aspectos epidemiológicos da caprinocultura cearense. Arquivo Brasileiro de Medicina Veterinária e Zootecnia. 2000;52:534–543. [Google Scholar]
- 13.Guimarães AS, Seyffert N, Portela RWD, Meyer R, Carmo FB, et al. Caseous lymphadenitis in sheep flocks of the state of Minas Gerais, Brazil: prevalence and management surveys. Small Ruminants Research. 2009;87(1):86–91. [Google Scholar]
- 14.Seyffert N, Guimarães AS, Pacheco LGC, Portela RW, Bastos BL, et al. High seroprevalence of caseous lymphadenitis in brazilian goat herds revealed by Corynebacterium pseudotuberculosis secreted proteins-based ELISA. Res Vet Sci. 2010;88:50–55. doi: 10.1016/j.rvsc.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Eggleton DG, Middleton HD, Doidge CV, Minty DW. Immunisation against ovine caseous lymphadenitis: comparison of Corynebacterium pseudotuberculosis vaccines with and without bacterial cells. Aust Vet J. 1991;68:317–319. doi: 10.1111/j.1751-0813.1991.tb03085.x. [DOI] [PubMed] [Google Scholar]
- 16.Stoops SG, Renshaw HW, Thilsted JP. Ovine caseous lymphadenitis: disease prevalence, lesion distribution, and thoracic manifestations in a population of mature culled sheep from western United States. Am J Vet Res. 1984;45(3):557–61. [PubMed] [Google Scholar]
- 17.Ribeiro MG, Júnior JGD, Paes AC, Barbosa PG, Júnior GN, et al. Punção aspirativa com agulha fina no diagnóstico de Corynebacterium pseudotuberculosis na linfadenite caseosa caprina. Arq Inst Biol. 2001;68:23–28. [Google Scholar]
- 18.Dorella FA, Estevam EM, Pacheco LGC, Guimarães CT, Lana UGP, et al. In vivo insertional mutagenesis in Corynebacterium pseudotuberculosis: an efficient means to identify DNA sequences encoding exported proteins. Appl Environ Microbiol. 2006;72:7368–7372. doi: 10.1128/AEM.00294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 2009;4:13–37. doi: 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb SAR, Karleh CM. Bench-to-bedside review: Bacterial virulence and subversion of host defences. Critical Care. 2008;12:234–241. doi: 10.1186/cc7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobrindt U, Hentschel U, Kaper JB, Hacker J. Genome plasticity in pathogenic and nonpathogenic enterobacteria. Curr Top Microbiol Immunol. 2002;264:157–175. [PubMed] [Google Scholar]
- 22.Hall BG, Ehrlich GD, Hu FZ. Pan-genome analysis provides much higher strain typing resolution than multi-locus sequence typing. Microbiology. 2010;156(4):1060–8. doi: 10.1099/mic.0.035188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva A, Schneider MPC, Cerdeira L, Barbosa MS, Ramos RTJ, et al. Complete genome sequence of Corynebacterium pseudotuberculosis I19, a strain isolated from a cow in Israel with bovine mastitis. J Bacteriol. 2011;193(1):323–324. doi: 10.1128/JB.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura Y, Nishio Y, Ikeo K, Gojobori T. The genome stability in Corynebacterium species due to lack of the recombinational repair system. Gene. 2003;317:149–155. doi: 10.1016/s0378-1119(03)00653-x. [DOI] [PubMed] [Google Scholar]
- 25.Tauch A, Kaiser O, Hain T, Goesmann A, Weisshaar B, et al. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J Bacteriol. 2005;187:4671–4682. doi: 10.1128/JB.187.13.4671-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khamis A, Raoult D, La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004;42(9):3925–31. doi: 10.1128/JCM.42.9.3925-3931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol. 2005;43:1934–1936. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 29.Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, et al. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U.S.A. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumann W. Thermosensors in eubacteria: role and evolution. J Biosci. 2007;32:549–557. doi: 10.1007/s12038-007-0054-8. [DOI] [PubMed] [Google Scholar]
- 31.Hentschel U, Hacker J. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 2001;3:545–548. doi: 10.1016/s1286-4579(01)01410-1. [DOI] [PubMed] [Google Scholar]
- 32.McNamara PJ, Cuevas WA, Songer JG. Toxic phospholipases D of Corynebacterium pseudotuberculosis, C. ulcerans and Arcanobacterium haemolyticum: cloning and sequence homology. Gene. 1995;156:113–118. doi: 10.1016/0378-1119(95)00002-n. [DOI] [PubMed] [Google Scholar]
- 33.Moomaw AS, Maguire ME. The unique nature of Mg2+ channels. Physiology (Bethesda) 2008;23:275–285. doi: 10.1152/physiol.00019.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connor K, Fletcher SA, Csonka LN. Increased expression of Mg(2+) transport proteins enhances the survival of Salmonella enterica at high temperature. Proc Natl Acad Sci U.S.A. 2009;106:17522–17527. doi: 10.1073/pnas.0906160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schönert S, Buder T, Dahl MK. Identification and enzymatic characterization of the maltose-inducible alpha-glucosidase mall (sucrase-isomaltase-maltase) of Bacillus subtilis. J Bacteriol. 1998;180:2574–2578. doi: 10.1128/jb.180.9.2574-2578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto H, Serizawa M, Thompson J, Sekiguchi J. Regulation of the glv operon in Bacillus subtilis: yfia (glvR) is a positive regulator of the operon that is repressed through ccpA and cre. Bacteriol. 2001;183:5110–5121. doi: 10.1128/JB.183.17.5110-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May T, Ito A, Okabe S. Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob Agents Chemother. 2009;53:4628–4639. doi: 10.1128/AAC.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saïd-Salim B, Mostowy S, Kristof AS, Behr MA. Mutations in Mycobacterium tuberculosis RV0444c, the gene encoding anti-sigK, explain high level expression of mpb70 and mpb83 in Mycobacterium bovis. Mol Microbiol. 2006;62:1251–1263. doi: 10.1111/j.1365-2958.2006.05455.x. [DOI] [PubMed] [Google Scholar]
- 40.Veyrier F, Saïd-Salim B, Behr MA. Evolution of the mycobacterial sigK regulon. J Bacteriol. 2008;190:1891–1899. doi: 10.1128/JB.01452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vassylyev DG, Tomitori H, Kashiwagi K, Morikawa K, Igarashi K. Crystal structure and mutational analysis of the Escherichia coli putrescine receptor: structural basis for substrate specificity. J Biol Chem. 1998;273:17604–17609. doi: 10.1074/jbc.273.28.17604. [DOI] [PubMed] [Google Scholar]
- 42.Carlson PEJ, Horzempa J, O'Dee DM, Robinson CM, Neophytou P, et al. Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J Bacteriol. 2009;191:6855–6864. doi: 10.1128/JB.00995-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enkavi G, Tajkhorshid E. Simulation of spontaneous substrate binding revealing the binding pathway and mechanism and initial conformational response of glpT. Biochemistry. 2010;49:1105–1114. doi: 10.1021/bi901412a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez E, Samper S, Bordas Y, Guilhot C, Gicquel B, et al. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol. 2001;41(1):179–87. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 45.Soto CY, Menéndez MC, Pérez E, Samper S, Gómez AB, et al. IS6110 Mediates Increased Transcription of the phoP Virulence Gene in a Multidrug-Resistant Clinical Isolate Responsible for Tuberculosis Outbreaks. J Clin Microb. 2004;42(1):212–219. doi: 10.1128/JCM.42.1.212-219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aguilar D, Infante E, Martin C, Gormley E, Gicquel G, Pando RH. Immunological responses and protective immunity against tuberculosis conferred by vaccination of Balb/C mice with the attenuated Mycobacterium tuberculosis (phoP) SO2 strain. Clin Exper Immunol. 2006;147:330–338. doi: 10.1111/j.1365-2249.2006.03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernández-Pando R, et al. PhoP: A Missing Piece in the Intricate Puzzle of Mycobacterium tuberculosis Virulence. PLoS ONE. 2008;3(10):1–11. doi: 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carson SD, Klebba PE, Newton SM, Sparling PF. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J Bacteriol. 1999;181:2895–2901. doi: 10.1128/jb.181.9.2895-2901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunkle CA, Schmitt MP. Analysis of a dtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J Bacteriol. 2005;187:422–433. doi: 10.1128/JB.187.2.422-433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu T, Okabe A, Minami J, Hayashi H. An upstream regulatory sequence stimulates expression of the perfringolysin o gene of Clostridium perfringens. Infect Immun. 1991;59:137–142. doi: 10.1128/iai.59.1.137-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urbina P, Flores-Díaz M, Alape-Girón A, Alonso A, Goni FM. Phospholipase C and sphingomyelinase activities of the Clostridium perfringens alpha-toxin. Chem Phys Lipids. 2009;159:51–57. doi: 10.1016/j.chemphyslip.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Selby T, Allaker RP, Dymock D. Characterization and expression of adjacent proline iminopeptidase and aspartase genes from Eikenella corrodens. Oral Microbiol Immunol. 2003;18:256–259. doi: 10.1034/j.1399-302x.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang L, Jia Y, Wang L, Fang R. A proline iminopeptidase gene upregulated in planta by a luxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris. Mol Microbiol. 2007;65:121–136. doi: 10.1111/j.1365-2958.2007.05775.x. [DOI] [PubMed] [Google Scholar]
- 54.Böltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol. 2002;184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burne RA, Chen YY. Bacterial ureases in infectious diseases. Microbes Infect. 2000;2:533–542. doi: 10.1016/s1286-4579(00)00312-9. [DOI] [PubMed] [Google Scholar]
- 56.Huntley RP, Binns D, Dimmer E, Barrell D, O'Donavan C, et al. QuickGO: a user tutorial for the web-based Gene Ontology browser. Database. 2009;10:1–19. doi: 10.1093/database/bap010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barinov A, Loux V, Hammani A, Nicolas P, Langella P, et al. Prediction of surface exposed proteins in Streptococcus pyogenes, with a potential application to other gram-positive bacteria. Proteomics. 2009;9:61–73. doi: 10.1002/pmic.200800195. [DOI] [PubMed] [Google Scholar]
- 58.Song C, Kumar A, Saleh M. Bioinformatic comparison of bacterial secretomes. Genomics Proteomics Bioinformatics. 2009;7:37–46. doi: 10.1016/S1672-0229(08)60031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wooldridge L, Lissina A, Cole DK, van den Berg HA, Price DA, Sewell AK. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology. 2009;126:147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wittmann C, Kiefer P, Zelder O. Metabolic Fluxes in Corynebacterium glutamicum during Lysine Production with Sucrose as Carbon Source. Applied and Environmental Microbiology. 2004;70:7277–7287. doi: 10.1128/AEM.70.12.7277-7287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabus A. Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl Microbiol Biotechnol. 2007;75:47–53. doi: 10.1007/s00253-006-0804-9. [DOI] [PubMed] [Google Scholar]
- 62.Blombach B, Arndt A, Auchter M, Eikmanns BJ. L-Valine production during growth of pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum in the presence of ethanol or by inactivation of the transcriptional regulator SugR. Appl Environ Microbiol. 2009;75:1197–1200. doi: 10.1128/AEM.02351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klaffl S, Eikmanns BJ. Genetic and Functional Analysis of the Soluble Oxaloacetate Decarboxylase from Corynebacterium glutamicum. . Journal of Bacteriology. 2010;192:2604–2612. doi: 10.1128/JB.01678-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wittmann C, Heinzle E. Modeling and experimental design for metabolic flux analysis of lysine-producing Corynebacteria by mass spectrometry. Metab Eng. 2001;3(2):173–91. doi: 10.1006/mben.2000.0178. [DOI] [PubMed] [Google Scholar]
- 65.Parche S, Thomae AW, Schlicht M, Titgemeyer F. Corynebacterium diphtheriae: a PTS View to the Genome. J Mol Microbiol Biotechnol. 2001;3(3):415–422. [PubMed] [Google Scholar]
- 66.Rübenhagen R, Rönsch H, Jung H, Krämer R, Morbach S. Osmosensor and osmoregulator properties of the betaine carrier betP from Corynebacterium glutamicum in proteoliposomes. J Biol Chem. 2000;275:735–741. doi: 10.1074/jbc.275.2.735. [DOI] [PubMed] [Google Scholar]
- 67.Rittmann D, Schaffer S, Wendisch VF, Sahm H. Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch Microbiol. 2003;180:285–292. doi: 10.1007/s00203-003-0588-6. [DOI] [PubMed] [Google Scholar]
- 68.Kiefer P, Heinzle E, Zelder O, Wittmann Z. Comparative Metabolic Flux Analysis of Lysine-Producing Corynebacterium glutamicum Cultured on Glucose or Fructose. Applied and Environmental Microbiology. 2004;70:229–239. doi: 10.1128/AEM.70.1.229-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rumbold K, Buijsen HJJ, Overkamp KM, Groenestijn JW, Punt PJ, Werf MJ. Microbial production host selection for converting second-generation feedstocks into bioproducts. Microbial Cell Factories. 2009;8:1–11. doi: 10.1186/1475-2859-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casalino M, Prosseda G, Barbagallo M, Lacobino A, Ceccarini P, et al. Interference of the cadC regulator in the arginine-dependent acid resistance system of Shigella and enteroinvasive E. coli. Int J Med Microbiol. 2010;300(5):289–95. doi: 10.1016/j.ijmm.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 71.Alvarez-Ordóñez A, Fernández A, Bernardo A, López M. Arginine and lysine decarboxylases and the acid tolerance response of Salmonella typhimurium. Int J Food Microbiol. 2010;136:278–282. doi: 10.1016/j.ijfoodmicro.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 72.Hodgson AL, Carter K, Tachedjian M, Krywult J, Corner LA, et al. Efficacy of an ovine caseous lymphadenitis vaccine formulated using a genetically inactive form of the Corynebacterium pseudotuberculosis Phospholipase D. Vaccine. 1999;17:802–808. doi: 10.1016/s0264-410x(98)00264-3. [DOI] [PubMed] [Google Scholar]
- 73.D'Afonseca V, Moraes PM, Dorella FA, Pacheco LGC, Meyer R, et al. A description of genes of Corynebacterium pseudotuberculosis useful in diagnostics and vaccine applications. Genet Mol Res. 2008;7:252–260. doi: 10.4238/vol7-1gmr438. [DOI] [PubMed] [Google Scholar]
- 74.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 75.Rasko DA, Rosovitz MJ, Garry SA, Emmanuel FM, Fricke WF, et al. The pan-genome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. Journal of Bacteriology. 2008;190(20):6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dorella FA, Pacheco LGC, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res. 2006;37:201–218. doi: 10.1051/vetres:2005056. [DOI] [PubMed] [Google Scholar]
- 77.Shiotani H, Yoshioka T, Yamamoto M, Matsumoto R. Susceptibility to citrus canker caused by Xanthomonas axonopodis pv. citri depends on the nuclear genome of the host plant. J Gen Plant Pathol. 2008;74:133–137. [Google Scholar]
- 78.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 79.Lagesen K, Hallin P, Rødland EA, Staerfeldt H, Rognes T, et al. Rnammer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lowe TM, Eddy SR. Trnascan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zdobnov EM, Apweiler R. Interproscan--an integration platform for the signature-recognition methods in INTERPRO. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 82.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 83.Waack S, Keller O, Asper R, Brodag T, Damm C, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden markov models. BMC Bioinformatics. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rice P, Longden I, Bleasby A. Emboss: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 85.Carver TJ, Rutherford KM, Berriman M, Rajandream M, Barrell BG, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 86.Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, et al. Mvirdb--a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 2007;35:D391–394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karp PD, Paley S, Romero P. The pathway tools software. Bioinformatics. 2002;18(1):S225–32. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 88.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2008;36:D623–31. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caspi R, Karp PD. Using the metacyc pathway database and the biocyc database collection. Curr Protoc Bioinformatics Chapter. 2007;1:Unit1.17. doi: 10.1002/0471250953.bi0117s20. [DOI] [PubMed] [Google Scholar]
- 90.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 91.Krauthammer M, Rzhetsky A, Morozov P, Friedman C. Using BLAST for identifying gene and protein names in journal articles. Gene. 2000;259:245–252. doi: 10.1016/s0378-1119(00)00431-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Orthologous genes present inside PAIs regions of C. pseudotuberculosis and their counterparts in other Corynebacterium species.
(DOC)