Abstract
PURPOSE
Emerging evidence suggests that aberrant expression of oncogenes contributes to development of lung malignancy. The thyroid transcription factor 1 (TITF-1) gene functions as a lineage survival gene abnormally expressed in a significant fraction of NSCLCs, in particular lung adenocarcinomas.
EXPERIMENTAL DESIGN
To better characterize TITF-1 abnormality: patterns in NSCLC, we studied TITF-1’s gene copy number using fluorescent in situ hybridization (FISH) and quantitative PCR, as well as its protein expression by immunohistochemistry analysis in a tissue microarray comprised of surgically resected NSCLC (N=321) including 204 adenocarcinomas and 117 squamous cell carcinomas (SCCs). TITF-1 copy number and protein expression were correlated with patients’ clinicopathologic characteristics, and in a subset of adenocarcinomas with EGFR and KRAS mutation status.
RESULTS
We found that increased TITF-1 protein expression was prevalent in lung adenocarcinomas only and was significantly associated with female gender (p<0.001), never smokers (p=0.004), presence of EGFR mutations (p=0.05) and better overall survival (all stages, p=0.0478. stages I and II, p=0.002). TITF-1 copy number gain (CBG) was detected by FISH analysis in both adenocarcinomas (18.9%; high CNG, 8.3%) and SCCs (20.1%; high CNG, 3.0%), and correlated significantly with the protein product (p=0.004) and presence of KRAS mutations (p=0.008) in lung adenocarcinomas. Moreover, multivariate analysis revealed that TITF-1 copy number gain was an independent predictor of poor survival of NSCLC (p=0.039).
CONCLUSIONS
Our integrative study demonstrates that the protein versus genomic expression patterns of TITF-1 have opposing roles in lung cancer prognosis and may occur preferentially in different subsets of NSCLC patients with distinct oncogene mutations.
Keywords: NSCLC, TITF-1, gene copy gain, lineage-specific oncogenes
Introduction
It is estimated that lung cancer is the leading cause of cancer-related deaths in the United States (1). The majority of diagnosed lung cancers are non-small cell lung cancers (NSCLCs) which include two major histological subtypes; lung adenocarcinomas and SCCs (2). Lung adenocarcinomas and SCCs appear to develop progressively by different pathogenic phases; SCCs typically develop near the central airways whereas lung adenocarcinomas occur predominantly in the lung periphery (3). Therefore, it is plausible to assume that characterization of molecular and biological markers for better understanding the similarities and differences in the development of the different subtypes of NSCLC will favorably impact the clinical management of this deadly disease (4).
It has been suggested that lineage-specific genes, that play important roles in normal developmental processes such as organogenesis or tissue homeostasis and remain to be expressed or become amplified during an acquired pathological condition, are crucial for maintenance of the disease state (5, 6). Moreover, lineage-oncogenes have been shown to be important for mediating the prosurvival properties of cancer cells of different histopathological characteristics; for example adenocarcinomas versus SCCs (5). In addition, tumor cells are addicted to aberrant and growth-promoting cell signaling mediated by lineage-specific oncogenes (7), e.g. presence of the BCR-ABL fusion oncoprotein in chronic myelogenous leukemia (CML) (8), mutations in the KIT oncogene in gastrointestinal stromal tumors (GISTs) (9) and mutations in the epidermal growth factor receptor (EGFR) in lung adenocarcinomas (10–12).
NK2 homebox 1 (NKX2-1) otherwise known as thyroid transcription factor 1 (TITF-1) is a homeodomain-containing transactivating factor predominantly expressed in the terminal lung bronchioles and lung periphery in the developing and adult mouse (13–15). In addition, TITF-1 is crucial for branching morphogenesis during normal lung development (14–17) and transactivates the expression of the surfactant proteins (SPs) such as SPs-A, -B and –C which are in turn typically expressed in the Clara cells and are important for the differentiation of alveolar type II pneumocyte cells in the peripheral lung (18–20). More recently, TITF-1 expression and function have been shown to be important in the etiology of congenital pulmonary disease (21) and NSCLC (22–24). TITF-1 is part of the 14q13.3 cytoband locus which is amplified in a significant fraction of lung tumors (22, 24). In addition, knockdown of TITF-1 expression by RNA interference results in lung adenocarcinoma cell growth inhibition and apoptosis demonstrating a lineage-specific dependency of lung adenocarcinomas on TITF-1 (22–24). In contrast to the expected pro-survival properties of a cell-lineage oncogene, TITF-1 protein expression by immunohistochemistry is a marker of favorable prognosis in NSCLC patients (25–28) and is associated with good prognosis in early stage (stage-I) lung adenocarcinoma patients (29). More recently, amplification of the TITF-1 gene has been identified in lung squamous cell carcinomas (28) despite lack of protein expression; however the relevance of this copy number gain to the clinical outcome of NSCLC patients is still unclear, although Weir et al reported no significant difference in patient survival between adenocarcinoma tumors with or without amplification of TITF-1 (24).
In this study, we sought to investigate protein and copy number levels of the TITF-1 gene in NSCLC patients and correlate, in parallel, both levels of analyses with the clinicopathological and molecular features of the patients. We demonstrate the significant and close association of TITF-1 protein with the protein expression of all three surfactant proteins tested. In addition, we find a significant association of TITF-1 protein expression and copy gain with favorable and poor prognosis, respectively, in lung adenocarcinoma patients, despite a significant positive correlation between the gene’s copy number and protein expression. Lastly, we unravel distinct significant correlations between TITF-1 protein expression and DNA copy gain with mutations in the lung adenocarcinoma-prevalent EGFR and KRAS oncogenes.
Methods
Human lung tissues and tissue microarray (TMA)
All human tissues were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at the M.D. Anderson Cancer Center (Houston, TX). For each tissue sample, the percentage of malignant tissue was calculated and the cellular composition of specimens was determined by histological examination (I.I.W.) following Hematoxylin-Eosin (H&E) staining.
Specimens resected from NSCLC patients with stages I–IV disease according to the revised International System for Staging Lung Cancer (30) who had no prior chemotherapy or radiotherapy were used for TMA analysis by immunohistochemistry. Clinicopathological characteristics of the patients are summarized in Supplementary Table 1. Patients who had smoked at least 100 cigarettes in their lifetime were defined as smokers. Samples were fixed in formalin, embedded in paraffin, stained with H&E, and reviewed by an experienced pathologist (I.I.W.). The 321 tissue specimens collected from 321 patients included 117 SCCs and 204 adenocarcinomas. All tumors and lesions were classified according to the World Health Organization (WHO) 2004 criteria as previously described (31). The TMAs were prepared with a manual tissue arrayer (Advanced Tissue Arrayer ATA100, Chemicon International, Temecula, CA) using 1-mm-diameter cores in triplicate for tumors. Sections were then determined if they were suitable for analysis of TITF-1 protein expression by immunohistochemistry and TITF-1 amplification by FISH analysis. Of the entire TMA set, 179 and 170 adenocarcinomas were used for immunohistochemistry and FISH analyses, respectively. In addition, 117 and 99 lung SCC sections were used for immunohistochemistry and FISH analyses, respectively. Moreover, 151 lung adenocarcinomas were subsequently analyzed for correlation of TITF-1 expression by immunohistochemistry and FISH analyses. Furthermore, data on EGFR and KRAS mutational status was available and performed on 178 and 120 lung adenocarcinoma sections, respectively.
Immunohistochemistry analysis
Tissue section slides were baked at 56°C overnight, then deparaffinized in xylene and rehydrated through a graded series of ethanol concentrations. Antigen retrieval was performed using a decloaker for analysis of SP-B expression and a steamer (PH=9, 20 minutes) for assessment of TITF-1 protein expression. Intrinsic peroxidase activity was blocked by 3% hydrogen peroxide in H2O2 for 15 min and 5% goat serum (Sigma) solution was used for blocking nonspecific antibody binding by incubating at room temperature for 60 min. Slides were then incubated at room temperature for 1 hour with primary antibodies raised against TITF-1 (dilution 1:100, clone 8G7G3/1, Cell Marque, Rocklin, CA) , SP-A (dilution 1:150, clone PE10, Thermo Scientific, Fremont, CA), SP-B (dilution 1:150, clone SPB02, Thermo Scientific) and SP-C (dilution 1:300, Chemicon, Billerica, MA). After three washes in Tris-buffered saline with Tween-20 (TBST) for 5 minutes each, slides were incubated with Dako Envision+ Dual Link for 30 minutes at room temperature. Following three additional washes in TBST, slides were incubated with Dako chromogen substrate for 5 min and were counterstained with hemotoxylin for 5 min. Slides were read under microscope. Two pathologists (X.T. and I.W.) examined both the intensity and extent of immunostaining by light microscopy using a ×20 magnification objective. Cytoplasmic and nuclear expression were quantified using a four-value intensity score (0, none; 1+, weak; 2+, moderate; and 3+, strong) and the percentage (0–100%) of the extent of reactivity. A final expression score was obtained by multiplying the intensity and reactivity extension values (range, 0–300).
FISH analysis
TITF-1 DNA was originally provided by Dr. Wam Lam (British Columbia Cancer Research Center, Vancouver, BC, Canada). The DNA was labeled with Spectrum Red conjugated dUTP (Abbott Laboratories, Abbott Park, IL) using the Vysis Nick translation kit, according to the manufacturer’s instructions. A chr14 control probe (14q11) was prepared using the BAC clone RP11-324B11 and labeled with Spectrum Green conjugated dUTP using the same procedure as described above for the TITF-1 probe. The FISH assay was performed on the TMAs using a standard protocol (32). Briefly, slides were incubated for 4 hours at 56°C, deparaffinized in Citri-Solv (Thermo Fisher Scientific, Waltham, MA) and rehydrated. The slides were then incubated in 2x saline-sodium citrate (SSC) buffer at 75°C for 10–13 min, digested in 0.25mg/ml Proteinase K/2×SSC at 45°C for 10–13 minutes, washed in 2x SSC for 5 min and dehydrated. Following application of the probe mixtures, samples were denaturated for 15 min at 80°C and incubated at 37°C for 48 hours, after which post-hybridization washes were performed with 2×SSC/0.3%NP40 (pH 7.0–7.5) at 72 °C for 2 min. Slides were then washed in 2×SSC for 2 min, dehydrated and chromatin was counterstained with DAPI (0.3 µg/ml in Vectashield mounting medium, Vector Laboratories, Burlingame, CA). Analysis was performed in epifluorescence microscope using single interference filters sets for green (Fluorescein isothiocyanate, FITC), red (Texas Red) and blue (DAPI) as well as dual (red/green) band pass filters. Monochromatic images were captured and merged using the CytoVision workstation (Genetix). The quality of the preparation and the intensity of the fluorescence signal were variable per slide and per tissue core. The assay was repeated once for each array in order to obtain results in higher number of cores. Specimens displaying a gene signal number per cell of 4 or greater were considered to exhibit copy number gain (4–10 signals, low copy gain; greater than 10 signals, high level of copy number gain). The maximum value among the three cores was considered to represent each case.
DNA extraction and quantitative PCR (qPCR)
NSCLC tissues (n=82, 53 adenocarcinomas and 29 SCCs) were dissected from formalin-fixed paraffin embedded (FFPE) Hematoxylin-stained tissue sections using manual microdissection under stereomicroscope to ensure that tumor cell proportions are greater than 70% for subsequent DNA extraction. Tumor DNA was extracted using the PicoPure DNA extraction Kit (Arcturus, Mt View, CA) according to the manufacturer’s instructions. Two µl of DNA was added to a 20 µl final volume reaction mixture consisting of 10 µl Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) and 0.5 µmol/l of each of forward and reverse primers spanning the TITF-1 gene (forward, 5’- gctgtgcgtttgtcgcttac-3’; reverse, 5’- ccatgccgctcatgttca-3’). β-actin gene was used as an endogenous reference gene. TaqMan® Control Human Genomic DNA (Applied Biosystems, Foster City, CA) was amplified as a standard control for calibration. All samples and standard DNA reactions were carried out in triplicates. qPCR was performed using an ABI 7300 Real Time PCR System Sequence (Applied Biosystems) at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The quantity of the target genes were normalized using the level of the β-actin gene, and expressed as relative quantities (RQ) compared with the value of the Human Genomic DNA. RQ equal or larger than 2 was considered as gene copy gain.
EGFR and KRAS mutational status
Exons 18 and 21 of EGFR and exons 1 and 2 of KRAS were PCR-amplified using DNA extracted from microdissected tumor cells, as previously described (33). Each PCR was done using HotStarTaq Master Mix (Qiagen) for 40 cycles at 94°C for 30 s, at annealing temperature for 30 s, and 72°C for 30 s, followed by a 7-min extension at 72°C. All primer sequence and annealing temperatures are list in Supplementary Table 2. PCR products were directly sequenced using the Applied Biosystems PRISM dye terminator cycle sequencing method (Perkin-Elmer Corp.). Sequence variants were confirmed by independent PCR amplifications from at least two independent microdissections and in both directions.
Statistical analyses
Data were summarized using descriptive statistics and frequency tabulation. Associations between categorical variables were assessed via cross-tabulation, chi-squared tests and Fisher’s exact tests. Differences in continuous markers between groups were tested using the rank-based non-parametric Wilcoxon-rank sum test or Kruskal-Wallis tests. Survival rates were calculated using the Kaplan-Meier method for estimation of survival probability. Univariate and multicovariate Cox proportional hazard models were applied to assess the effects of covariates on overall and recurrence free survival. All computations were performed in Statistical analysis Software (SAS) (Cary, NC) and S-plus 7.0 (TIBCO software inc., Palo Alto, CA).
Results
Immunohistochemical expression of TITF-1, SP-A, SP-B and SP-C proteins in NSCLC
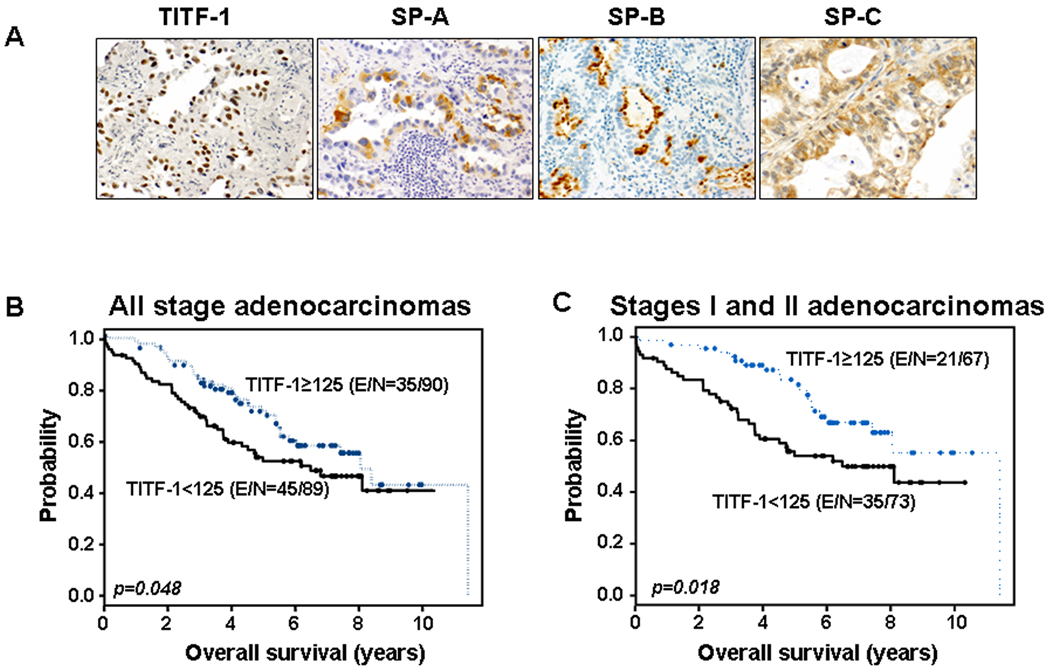
We assessed the protein expression of TITF-1 and the surfactant proteins, SP-A, SP-B and SP-C which are typically transactivated by TITF-1 (18–20), in a series of lung adenocarcinomas (n=179) and SCCs (n=117) by immunohistochemistry. TITF-1 protein expression was mainly nuclear (Figure 1A), prevalent in lung adenocarcinomas (68.7% with TITF-1 protein greater than 0) and absent in SCCs (all zero TITF-1 protein) (p<0.0001) (Supplementary Table 3). Similarly, the immunohistochemical protein expression (greater than a score of zero) of SP-A and SP-B was mainly cytoplasmic (Figure 1A) and only evident in lung adenocarcinomas (19.8% and 16.4%, respectively) and was absent in SCC (p<0.0001) (Supplementary Table 3). In contrast, SP-C protein was expressed in both NSCLC subtypes as 84.5% of lung adenocarcinomas and 93.6% of SCCs exhibited an SP-C expression score of greater than zero (Supplementary Table 3). A significant positive correlation was found between the expression of any of the three surfactant proteins analyzed and TITF-1 expression when lung adenocarcinoma patients were stratified by absence or presence of TITF-1 immunoreactivity (Table 1).
Figure 1. TITF-1 immunohistochemical expression in NSCLC.
A. Representative photomicrographs of the immunohistochemical expression of TITF-1, SP-A, SP-B and SP-C. Kaplan-Meier survival probability plots of all stages (B) and stages I and II only (C) lung adenocarcinoma patients stratified by TITF-1 protein expression. P-values were obtained by the log-rank test. E/N, censored events/total number of cases.
Table 1.
Correlation of the immunohistochemical protein expression of TITF-1 and the SP-A, SP-B and SP-C surfactant proteins in lung adenocarcinomas.
| Covariate (N) | Covariate expression (N) | TITF-1 negative [N (%)] | TITF-1 positive [N (%)]* | p-value |
|---|---|---|---|---|
| SP-A (171) | negative (135) | 49 (36) | 86 (64) | 0.004 |
| positive (36) | 4 (11) | 32 (89) | ||
| SP-B (168) | negative (137) | 47 (34) | 90 (66) | 0.002 |
| positive (31) | 2 (7) | 29 (94) | ||
| SP-C (171) | negative (23) | 14 (61) | 9 (39) | 0.0005 |
| positive (148) | 37 (25) | 111 (75) |
P-values were obtained by the Fisher’s exact test.
TITF-1 immunoreactivity greater than zero was considered positive.
We then correlated TITF-1 protein expression with other clinicopathological features besides histology (Table 2). TITF-1 protein expression was significantly associated with female gender (p=0.0001), never- compared to ever-smokers (p<0.001) and never compared to former or current smokers (p=0.0004) (Table 2).
Table 2.
Association of TITF-1 protein expression with NSCLC histology, gender, tobacco history and smoking habits.
| Covariate | Covariate type (N) | TITF-1 negative [N (%)] |
TITF-1 positive [N (%)] |
p-value |
|---|---|---|---|---|
| Gender | Female (148) | 65 (43.9) | 83 (56.1) | <0.0001 |
| Male (135) | 95 (70.4) | 40 (29.6) | ||
| Tobacco history | Never (53) | 17 (32.1) | 36 (67.9) | 0.0001 |
| Ever (229) | 142 (62) | 87 (38) | ||
| Smoking habit | Never (53) | 17 (32.1) | 36 (67.9) | 0.0004 |
| Former (140) | 86 (61.4) | 54 (38.6) | ||
| Current (89) | 56 (62.9) | 33 (37.1) | ||
| Histology | Adenocarcinoma (179) | 56 (31.3) | 123 (68.7) | <0.0001 |
| SCC (104) | 104 (100) | 0 (0) |
P-values were obtained by the Fisher’s exact test.
We next asked whether TITF-1 protein expression exhibits prognostic properties in lung adenocarcinomas, the NSCLC subtype where it is predominantly expressed. Lung adenocarcinoma patients with higher than the median expression of TITF-1 protein (greater than or equal to 125) exhibited favorable survival compared to patients with lower expression (less than total score of 125) (p=0.0478 of the log-rank test) with a hazard ratio (HR) of 0.639 (Figure 1B). Similar results were obtained when stages I and II lung adenocarcinoma patients (n=140) were analyzed alone using the same cut-off for TITF-1 protein expression (p=0.01 of the log-rank test, HR=0.485) (Figure 1C). In contrast to those findings, no significant association was found between TITF-1 protein expression and recurrence-free survival in lung adenocarcinoma patients (data not shown). In addition, multivariate Cox proportional hazard regression analysis revealed that TITF-1 protein expression was not an independent predictor of overall or recurrence free survival in lung adenocarcinoma (data not shown).
TITF-1 gene amplification in NSCLC and association with clinicopathological features
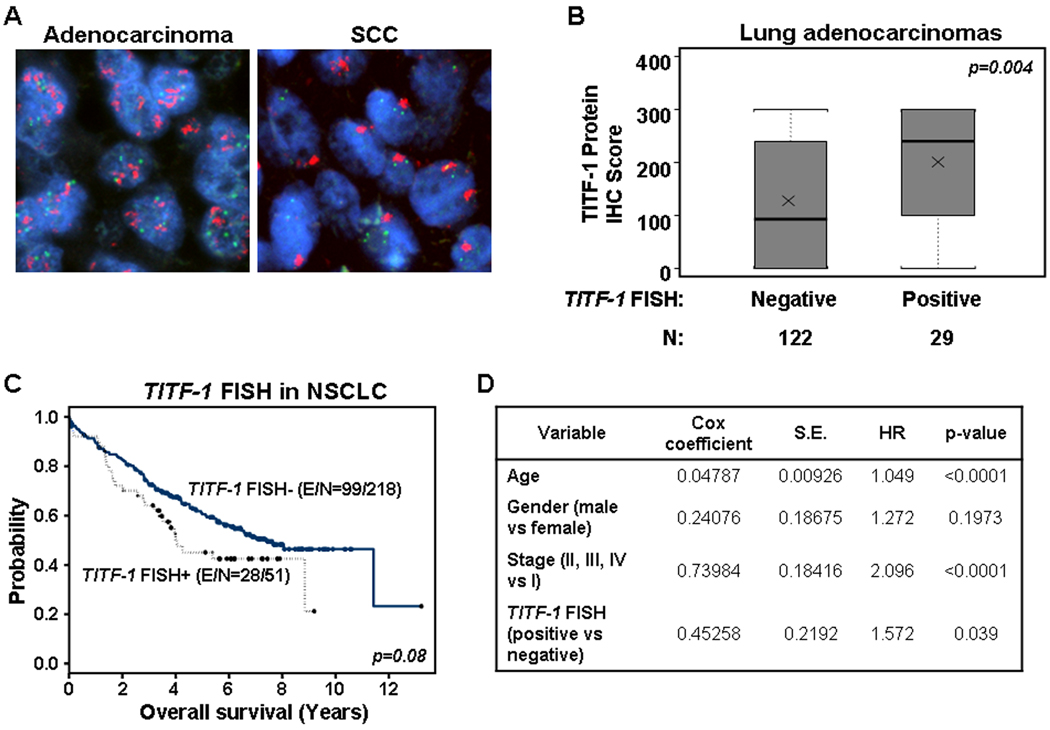
We next assessed the gene dosage levels of the TITF-1 gene in NSCLC patients by two different methodologies, copy number gain and amplification by FISH (170 lung adenocarcinomas and 99 SCCs) and copy gain by qPCR in 46 lung adenocarcinomas and 36 SCCs. TITF-1 copy number gain by FISH was found in 18.9% of lung adenocarcinomas and in 20.2% of lung SCCs (Figure 2A and Supplementary Table 4). A high level of TITF-1 copy number gain (greater than ten copies) was found in 8.3% and 3.0% of lung adenocarcinomas and SCCs, respectively (Supplementary Table 4). Two lung adenocarcinoma cases and one case of lung SCC were found to have greater than 20 signals per cell; the high level of TITF-1 copy gain by FISH detected in the SCC case is depicted in Supplementary Figure 1. We then determined to correlate TITF-1 protein with its DNA copy gain in lung adenocarcinomas since both variables were assessed and found in these set of patients. A significant positive correlation was found between the TITF-1 protein expression (as a continuous score variable) with the presence of TITF-1 copy gain assessed by FISH (p=0.004) (Figure 2B) in lung adenocarcinomas. The mean and median of the TITF-1 protein score, assessed by immunohistochemical analysis, were 200.69 ± 115.73 and 240 (min, 0; max, 300), respectively, in TITF-1 FISH positive lung adenocarcinoma patients (n=29) and 127.1 ± 122.61 and 93 (min, 0; max, 300), respectively in TITF-1 FISH negative adenocarcinoma specimens (n=122).
Figure 2. TITF-1 copy number gain in NSCLC.
A. Representative photomicrographs (magnification ×1000) of lung adenocarcinoma (left) and SCC (right) cells with copy number gain of TITF-1. B. Statistically significant differences in TITF-1 protein score between lung adenocarcinomas with and without TITF-1 copy gain by FISH. p-value was determined by the non-parametric Wilcoxon-rank sum test. C. Kaplan-Meier curves of the overall survival of NSCLC patients (n=269) based on the presence or absence of TITF-1 copy number gain by FISH. D. Cox proportional multivariate hazard model of overall survival of NSCLC patients. S.E., standard error; HR, hazard ratio.
We also assessed TITF-1 copy number gain by qPCR in a subset of patients that we had analyzed by FISH (n=82, 46 lung adenocarcinomas and 36 SCCs). A similar percentage of lung adenocarcinomas (18.4%) and SCCs (18.2%) exhibited greater than three copies of TITF-1 analyzed by qPCR (Supplementary Table 5). Similar to the FISH analysis, a slightly higher percentage of lung adenocarcinomas (10.2%) displayed a higher ratio (≥5) of TITF-1 copy gain than that of SCCs (4.6%, Supplementary Table 5). Moreover, we found a significant correlation between levels of TITF-1 copy gain by FISH and qPCR analyses in both lung adenocarcinomas (p=0.0037) and lung SCCs (p=0.048). The mean and median levels of relative TITF-1 copy number by quantitative PCR in TITF-1 FISH positive lung adenocarcinomas (n=11) were 5.12 ± 5.55 and 3.41 (min, 0.53; max, 21), compared to 2.49 ± 4.31 and 1.41 (min, 0.48; max, 24.22) in FISH negative adenocarcinomas (n=35). In addition, the mean and median levels of TITF-1 expression assessed by quantitative PCR in TITF-1 FISH positive SCCs (n=9) were 13.61 ± 24.16 and 2.23 (min, 0.82; max, 74.93), compared to 1.64 ± 1.18 and 1.28 (min, 0.56; max, 4.8) in TITF-1 FISH negative SCCs.
We next investigated the role of TITF-1 copy number gain by FISH in prognosis of both lung adenocarcinoma and SCC patients since we detected TITF-1 gain in both subtypes of NSCLC. NSCLC patients with TITF-1 copy number gain (low and high) by FISH (n=51, 33 adenocarcinomas and 18 SCCs) displayed a trend for poorer survival (p=0.08) compared to NSCLC patients lacking TITF-1 copy number gain (n=218, 137 adenocarcinomas and 81 SCCs). However, multivariate Cox proportional hazard regression analysis revealed that copy gain of the TITF-1 gene by FISH was an independent predictor of poor survival in NSCLC alone with age (p<0.0001), and stage-I disease (p<0.0001) (Figure 2D). Similar results were obtained when NSCLC patients were dichotomized based on presence or absence of high level of TITF-1 copy gain (greater than 10 signals; data not shown).
Association of TITF-1 expression with EGFR and KRAS mutations in lung adenocarcinoma
Molecular abnormalities in the expression or function of KRAS and EGFR contribute to tumor development and progression and therefore serve as crucial targets for molecularly driven target-specific therapies (2, 34). We had previously investigated the prevalence of EGFR and KRAS mutations in our clinical sample sets and correlated the extent of the mutations of those two oncogenes with patients’ clinicopathological features (33, 35). However, the relationship of TITF-1 abnormality with the mutational pattern of oncogenes prevalent in lung adenocarcinoma, e.g. EGFR and KRAS, is not well understood. Therefore, we sought to correlate the presence of mutations in the KRAS and EGFR oncogenes with DNA copy gain and protein levels of the TITF-1 gene. Interestingly, the protein levels and gene dosage extent of TITF-1 exhibited different correlation patterns. TITF-1 protein expression as a continuous variable displayed border-line significant positive correlation with presence of EGFR mutations (p=0.05) (Supplementary Figure 2). The mean and median levels of TITF-1 immunohistochemical protein in EGFR mutant lung adenocarcinomas (n=22) were 183.94 ± 124.58 and 225 (min, 0; max, 300), compared to 124.62 ± 121.06 and 100 (min, 0; max, 300) in EGFR wild type adenocarcinomas (n=137). No significant differences were found in TITF-1 protein expression between KRAS mutant (n=10) (mean, 126.67 ± 127.57; median, 75) and wild type (n=94) (mean, 112.78 ± 117.68; median, 66.67) lung adenocarcinomas. On the other hand, TITF-1 copy gain by FISH displayed a significant positive correlation with presence of KRAS mutations as 55% or six out of eleven KRAS mutant patients exhibited TITF-1 copy number gain compared to 15.9% of patients with wild type KRAS (14 out of 88 patients) (p=0.008 of the Fisher’s exact test) (Table 3). Moreover and in accordance with analysis of TITF-1 protein expression as a continuous variable, 64% of lung adenocarcinoma patients with mutant EGFR exhibited a TITF-1 protein score greater than or equal to 200 compared to 33% of patients with wild type EGFR (p=0.006) (Table 3). These new findings further demonstrate the dissimilarities between the extent of protein and DNA expression levels of TITF-1 in lung adenocarcinomas with EGFR versus KRAS mutations.
Table 3.
Significant positive associations of TITF-1 copy number and TITF-1 protein expression with mutations in KRAS and EGFR, respectively.
| TITF-1 FISH | TITF-1 IHC ≥ 200* | ||||
|---|---|---|---|---|---|
| Covariate | Covariate type | N positive/Total (%) | p-value | N positive/Total (%) | p-value |
| EGFR | mutant | 5/16 (31) | 0.338 | 14/22 (64) | 0.006 |
| wild type | 27/133 (20) | 45/137 (33) | |||
| KRAS | mutant | 6/11 (55) | 0.008 | 3/10 (30) | 0.933 |
| wild type | 14/88 (16) | 27/94 (29) | |||
P-values were obtained by the Fisher’s exact test.
TITF-1 immunoreactivity greater than or equal to 200 was considered positive.
Discussion
In this study, we investigated the protein and copy number gain of the TITF-1 cell-lineage gene in a tissue microarray comprised mainly of lung adenocarcinomas and squamous cell carcinomas and in correlation with molecular and clinicopathological features. We found that immunohistochemical protein expression of TITF-1 is prevalent in lung adenocarcinomas and absent in SCCs, correlates positively and significantly with the protein expression of the SP-A, SP-B and SP-C surfactant proteins and with female gender and never smoking status. Moreover, copy gain of TITF-1 DNA, assessed by FISH, was observed in both adenocarcinomas and SCCs and correlated positively and significantly with TITF-1 protein in lung adenocarcinomas as well as copy gain in NSCLC assessed by qPCR. Interestingly, while higher expression of TITF-1 protein was predictive of favorable prognosis in lung adenocarcinomas, TITF-1 copy number gain was an indicator of poor survival in NSCLC. Lastly, TITF-1 protein and DNA copy gain exhibited distinct correlations with oncogene mutation status in lung adenocarcinomas as the former correlated significantly with EGFR mutations, whereas the latter was associated with mutations in KRAS. The opposing prognostic, clinicopathological and molecular characteristics of TITF-1 protein expression and copy number gain suggest that different subsets of NSCLC patients may express TITF-1 through different cellular mechanisms.
Our findings on the association of higher levels of TITF-1 protein expression with favorable prognosis and overall survival are in accordance with previous findings by independent groups (25–29). In this study, we further investigated TITF-1 protein expression in clinical samples of NSCLC by correlating its expression with that of three surfactant proteins, SP-A, SP-B and SP-C. We unraveled significant associations of TITF-1 protein expression with that of the three surfactant proteins. It is noteworthy that staining patterns of TITF-1 in the developing and adult lung are very similar to those observed for the three surfactant proteins (18–20, 36). Moreover, TITF-1 is known to facilitate the differentiation of alveolar type II cells, in part, by transactivating the expression of the three surfactant genes following direct binding on their promoter regions (18–20, 36). It is worthy to mention that the combined use of Napsin A, an aspartic proteinase involved in the maturation of SP-B, and thyroid transcription factor-1 results in improved sensitivity and specificity for identifying pulmonary adenocarcinoma in primary lung tumors and in metastatic settings (37). Here, we provide new evidence of positive association of TITF-1 protein and that of the three major surfactant proteins in clinical specimens of human lung adenocarcinoma. These findings raise the possibility of the combined use of TITF-1 and surfactant protein expression in the diagnosis of pulmonary adenocarcinomas as the usage of TITF-1 with any of the SPs may increase the number of adenocarcinoma cases with positive expression of either marker.
We tested the association of TITF-1 protein expression with prognosis in lung adenocarcinomas because we only noted the protein in this subtype of NSCLC. In contrast, we analyzed the prognostic capacity of TITF-1 copy number gain in lung adenocarcinomas and SCCs as we detected increased TITF-1 gene dosage in both subtypes of NSCLC. It is worthwhile to note, that Kwei et al previously demonstrated amplification of TITF-1 assessed by comparative genomic hybridization (CGH) in both lung adenocarcinomas and SCCs, albeit at a lower frequency (11% and 3%, respectively) (22). In the present study, we detected TITF-1 both low and high copy number gain in 20.1% of lung SCCs assessed. Specifically, 17.2% of lung SCCs exhibited low level of amplification (4 to 10 copies, data not shown) and 3% (three cases) displayed high level of TITF-1 copy number gain (greater than ten copies, Supplementary Table 4), which is similar to the rate of amplification found in lung SCC in the previously reported CGH study by Kwei et al (22). Interestingly, we also found that one case of lung SCC harbored greater than 20 copies of TITF-1 (Supplementary Figure 1). It is worthwhile to note that a recent study by Bass et al using single nucleotide polymorphism (SNP) arrays showed the absence of TITF-1 amplification in a set of lung SCCs (n=47) (5). In our study and as mentioned before only three out of a total of 99 lung SCCs were found to display more than greater than 10 signals of TITF-1 per cell which we considered to harbor high copy gain of the gene rather than amplification. Due to the lack normalization to DNA content, a shortcoming in our FISH analysis, it is not clear whether specimens displaying high level of TITF-1 copy number gain actually harbor amplified TITF-1.
We found a significant positive correlation between TITF-1 protein expression and DNA copy number levels assessed by FISH analysis in lung adenocarcinomas. Previously, Perner et al demonstrated a similar positive correlation between the protein products and the DNA copy gain levels of the TITF-1 gene (28). Moreover, we further demonstrated that, unlike the association of TITF-1 protein with overall survival of lung adenocarcinoma patients, TITF-1 copy gain by FISH analysis appears to be a marker of poor prognosis in NSCLC. We also attempted to investigate the survival of lung adenocarcinoma patients alone with TITF-1 copy number gain stratified by presence or absence of TITF-1 protein expression. Lung adenocarcinoma patients with positive amplification of TITF-1 displayed worse survival than adenocarcinoma patients with both copy gain and protein expression (data not shown), demonstrating the association of TITF-1 copy number gain and TITF-1 protein expression with poor and good prognosis, respectively, in the lung adenocarcinoma population alone. The better survival correlations with TITF-1 protein and worse outcomes associated with TITF-1 copy gain are in close agreement with those reported earlier by Barletta et al (38). It is uncertain why protein levels and DNA copy gain of TITF-1 have opposing associations with prognosis of NSCLC patients including lung adenocarcinomas alone. However, it is worthwhile to mention that oncogene copy number gain is a feature of cell-lineage genes that elicit prosurvival functions important for the population of cells expressing the oncogene or for the pathological condition, i.e. cancer, whose cells are driven by amplification of the oncogene (5, 6). In this context, it is not surprising that NSCLC patients exhibiting copy number gain of the TITF-1 gene display poor survival and these findings no longer become counterintuitive. Moreover, it is plausible to suggest that TITF-1 copy number gain may be more reliable than protein expression for studying cell-lineage patterns of expression in NSCLC. It is noteworthy that TITF-1 protein was found to be elevated in primary lung adenocarcinoma compared to matched metastatic lesions in the brain (39). This is in accordance with our findings and the results of previous reports by other groups (25–29) on the utility of TITF-1 protein expression as a marker of favorable prognosis in lung cancer. Interestingly, Tanaka et al reported that TITF-1 gene dosage, which we demonstrated in this study to be a marker of dismal prognosis in NSCLC, was higher in metastatic sites compared to primary lung tumors (23). Therefore, our current findings and previous reports by others suggest that lung adenocarcinoma cells with either elevated TITF-1 protein expression or copy gain of the TITF-1 gene may originate from different cell populations with dissimilar biological properties and effects on patient clinical outcome. Congruent with this probable thought, is our novel finding that TITF-1 protein expression and TITF-1 gene copy number were associated with mutations in different oncogenes in lung adenocarcinomas. Our current study shed light on positive associations between TITF-1 gene dosage and protein expression level with mutations in the KRAS and EGFR oncogenes, respectively. It is noteworthy, that over-expression of HRAS inhibits the mRNA and protein levels of TITF-1 as well as its transcriptional factor function (40). Thus, it is plausible to suggest that expression of TITF-1 is favored at the DNA level in lung adenocarcinomas with mutant KRAS. Since, mutations in EGFR and KRAS occur almost mutually exclusively in lung adenocarcinomas (2), our results suggest that elevated expression of TITF-1 may be variably controlled (copy gain versus transcription) in distinct subsets of lung adenocarcinoma patients. Moreover, the association of TITF-1 protein, but not copy gain, with EGFR mutations is in accordance with our finding on the increased levels of TITF-1 protein in never-smoker compared to ever-smoker lung cancer patients.
It has been suggested that other genes within the TITF-1 amplicon (14q13.3) may facilitate, cooperate with, or even enhance the prosurvival biological properties of TITF-1 (22, 24, 41, 42). Kwei et al and Weir et al reported the location of several genes within the TITF-1 amplicon including the paired box transcriptional factor family member PAX9 and NKX2.8 (22, 24). It is worthy to note that Kendal et al demonstrated that co-amplified TITF-1, PAX9 and NKX2.8 exhibit oncogenic cooperation and cell prosurvival and proliferative properties (42). Over-expression of both TITF-1 and NKX2.8 simultaneously in BEAS-2B immortalized human bronchial epithelial cells elicited the highest increase in cell colony growth compared to single-gene transfected cells (42). Moreover, pathway genes signatures that overlap downstream of both TITF-1 and NKX2.8 defined lung adenocarcinoma patients with most dismal prognosis compared to signatures downstream of either transcriptional factor alone (41). It is therefore tempting to speculate that co-amplification or copy gain of NKX2.8 and/or PAX9 together with TITF-1 may contribute to the observed poor survival patterns observed in NSCLC patients exhibiting copy gain of the TITF-1 gene compared to patients with exhibiting only elevated expression of the protein product of this cell-lineage gene. It is also plausible that TITF-1 copy gain and its correlation with important biological outcomes in NSCLC may only be a surrogate marker, e.g. in SCCs, of another molecular defect in a gene nearby or within the 14q13.3 amplicon, e.g. NKX2.8 or PAX9. It is worthwhile to mention that NKX2.8 displays dissimilar patterns of expression relative to those of TITF-1 in the developing and adult mouse lung (43). Therefore, it is possible that expression of the NKX2.8 protein or its copy number gain may subdivide patients exhibiting positive expression of TITF-1 protein into subsets with different clinical outcomes. Moreover, lung SCC patients with copy number gain of the TITF-1 gene, as shown in this study and previously (28, 38), may display positive expression of the NKX2.8 protein and that transactivation of TITF-1 may be inhibited in lung SCCs and not in adenocarcinomas by yet unknown mechanisms.
In summary and herein, we further highlight the cell-lineage abnormality pattern of TITF-1 in human NSCLC and its correlation at the protein level with that of surfactant proteins in clinical specimens of lung adenocarcinoma. Moreover, we demonstrate that TITF-1 protein and gene dosage are associated with discordant clinical outcomes. We also noted in lung SCCs TITF-1 copy number gain in the absence of protein expression which warrants future studies. Furthermore, we highlight previously unknown associations of TITF-1 gene dosage and protein expression with differing oncogene mutation patterns suggesting that lung adenocarcinomas exhibiting elevated protein or amplification of the gene may be further divided into different cell-lineage subsets that can be subject to distinct treatment strategies.
Statement of Translational Relevance
Thyroid transcription factor 1 (TITF-1) is a transactivating factor with important functions in the normal peripheral lung and that exhibits dysregulated expression in NSCLC and amplification in a significant fraction of lung tumors typical of a lineage-specific oncogene. We tested the hypothesis that the abnormal expression of TITF-1 at the protein and DNA levels may occur in different subsets of patients with distinct clinicopathological and unique molecular features. We demonstrated significant association of TITF-1 protein expression and copy number gain with favorable and poor prognosis, respectively, in NSCLC patients. Importantly, we unraveled distinct correlations between TITF-1 protein expression and copy number gain of the gene with mutations in the EGFR and KRAS oncogenes, respectively, in lung adenocarcinoma. Our findings, suggest that lung adenocarcinomas exhibiting elevated protein or copy gain of TITF-1 may be of different cell-lineage populations with distinct oncogene mutation patterns, and therefore subject to dissimilar anti-cancer therapies.
Supplementary Material
Acknowledgements
This study was supported in part by grants from the Department of Defense (W81XWH-04-1-0142 W.K.H. and I.I.W.) and by the Specialized Program of Research Excellence in Lung Cancer Grants P50CA70907 (A.F.G. and I.I.W.) and P50CA58187 (M.V.G.) as well as Cancer Center Support Grant CA-16672 from the National Cancer Institute.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wistuba II, Gazdar AF. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1:331–348. doi: 10.1146/annurev.pathol.1.110304.100103. [DOI] [PubMed] [Google Scholar]
- 4.Wistuba II. Genetics of preneoplasia: lessons from lung cancer. Curr Mol Med. 2007;7:3–14. doi: 10.2174/156652407779940468. [DOI] [PubMed] [Google Scholar]
- 5.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 8.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 9.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 10.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 11.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev Biol. 2008;317:296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 14.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- 15.Yuan B, Li C, Kimura S, Engelhardt RT, Smith BR, Minoo P. Inhibition of distal lung morphogenesis in Nkx2.1(−/−) embryos. Dev Dyn. 2000;217:180–190. doi: 10.1002/(SICI)1097-0177(200002)217:2<180::AID-DVDY5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Bingle CD. Thyroid transcription factor-1. Int J Biochem Cell Biol. 1997;29:1471–1473. doi: 10.1016/s1357-2725(97)00007-1. [DOI] [PubMed] [Google Scholar]
- 17.Kimura S, Hara Y, Pineau T, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 18.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda K, Clark JC, Shaw-White JR, Stahlman MT, Boutell CJ, Whitsett JA. Gene structure and expression of human thyroid transcription factor-1 in respiratory epithelial cells. J Biol Chem. 1995;270:8108–8114. doi: 10.1074/jbc.270.14.8108. [DOI] [PubMed] [Google Scholar]
- 20.Minoo P, Hu L, Xing Y, et al. Physical and functional interactions between homeodomain NKX2.1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol Cell Biol. 2007;27:2155–2165. doi: 10.1128/MCB.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krude H, Schutz B, Biebermann H, et al. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J Clin Invest. 2002;109:475–480. doi: 10.1172/JCI14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 24.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barlesi F, Pinot D, Legoffic A, et al. Positive thyroid transcription factor 1 staining strongly correlates with survival of patients with adenocarcinoma of the lung. Br J Cancer. 2005;93:450–452. doi: 10.1038/sj.bjc.6602717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berghmans T, Mascaux C, Martin B, Ninane V, Sculier JP. Prognostic role of thyroid transcription factor-1 in stage III non-small cell lung cancer. Lung Cancer. 2006;52:219–224. doi: 10.1016/j.lungcan.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Berghmans T, Paesmans M, Mascaux C, et al. Thyroid transcription factor 1--a new prognostic factor in lung cancer: a meta-analysis. Ann Oncol. 2006;17:1673–1676. doi: 10.1093/annonc/mdl287. [DOI] [PubMed] [Google Scholar]
- 28.Perner S, Wagner PL, Soltermann A, et al. TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J Pathol. 2009;217:65–72. doi: 10.1002/path.2443. [DOI] [PubMed] [Google Scholar]
- 29.Anagnostou VK, Syrigos KN, Bepler G, Homer RJ, Rimm DL. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J Clin Oncol. 2009;27:271–278. doi: 10.1200/JCO.2008.17.0043. [DOI] [PubMed] [Google Scholar]
- 30.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 31.Behrens C, Feng L, Kadara H, et al. Expression of interleukin-1 receptor-associated kinase-1 in non-small cell lung carcinoma and preneoplastic lesions. Clin Cancer Res. 16:34–44. doi: 10.1158/1078-0432.CCR-09-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang X, Varella-Garcia M, Xavier AC, et al. Epidermal growth factor receptor abnormalities in the pathogenesis and progression of lung adenocarcinomas. Cancer Prev Res (Phila Pa) 2008;1:192–200. doi: 10.1158/1940-6207.CAPR-08-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117:2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 36.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 37.Bishop JA, Sharma R, Illei PB. Napsin A and thyroid transcription factor-1 expression in carcinomas of the lung, breast, pancreas, colon, kidney, thyroid, and malignant mesothelioma. Hum Pathol. 41:20–25. doi: 10.1016/j.humpath.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 38.Barletta JA, Perner S, Iafrate AJ, et al. Clinical significance of TTF-1 protein expression and TTF-1 gene amplification in lung adenocarcinoma. J Cell Mol Med. 2009;13:1977–1986. doi: 10.1111/j.1582-4934.2008.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald JM, Pelloski CE, Ledoux A, et al. Elevated phospho-S6 expression is associated with metastasis in adenocarcinoma of the lung. Clin Cancer Res. 2008;14:7832–7837. doi: 10.1158/1078-0432.CCR-08-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vita G, Bauer L, da Costa VM, et al. Dose-dependent inhibition of thyroid differentiation by RAS oncogenes. Mol Endocrinol. 2005;19:76–89. doi: 10.1210/me.2004-0172. [DOI] [PubMed] [Google Scholar]
- 41.Hsu DS, Acharya CR, Balakumaran BS, et al. Characterizing the developmental pathways TTF-1, NKX2-8, and PAX9 in lung cancer. Proc Natl Acad Sci U S A. 2009;106:5312–5317. doi: 10.1073/pnas.0900827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kendall J, Liu Q, Bakleh A, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian J, Mahmood R, Hnasko R, Locker J. Loss of Nkx2.8 deregulates progenitor cells in the large airways and leads to dysplasia. Cancer Res. 2006;66:10399–10407. doi: 10.1158/0008-5472.CAN-06-1564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.