Abstract
Regulation of bacterial gene expression at the post-transcriptional level has emerged as a major control mechanism, although not yet as well recognized as the mechanisms of control at the transcriptional level. In this article, we focus on regulated RNA decay in the control of gene expression in Gram-positive organisms, with a focus on Bacillus subtilis. Discovery of new ribonuclease activities in B. subtilis and other Gram-positive species, especially the dual-functioning RNase J1, which specifies both an endonuclease activity and the long-sought bacterial 5’-to-3’ exoribonuclease activity, has led to the recognition of intriguing mechanisms of gene regulation at the level of RNA decay.
Introduction
In eukaryotes, different modes of post-transcriptional regulation at the level of the RNA itself (i.e., not translational regulation) can occur. These include regulation of RNA splicing, polyadenylation, nuclear export, and decay. In prokaryotes, where splicing is rare, polyadenylation is not nearly as extensive, and there is no physical separation between the sites of RNA synthesis and RNA function, it is likely that RNA decay is a major form of post-transcriptional control. In this review, we discuss examples of regulated RNA decay in Gram-positive bacteria that are necessary for proper gene function. We describe four examples of this type of regulation from the model Gram-positive organism, Bacillus subtilis, and also cite examples from other Gram-positive organisms.
RNA decay in regulation of B. subtilis gene expression
glmS: metabolite-dependent mRNA decay
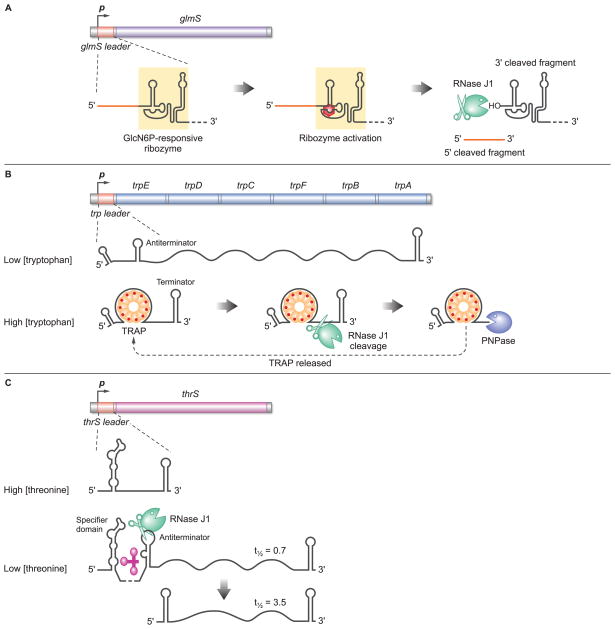
The B. subtilis glmS gene provides a prime illustration of gene expression control via regulated mRNA decay. Importantly, this mechanism involves RNase J1, a bi-functional enzyme found in many organisms that do not contain RNase E, the primary mRNA decay-initiating enzyme in Escherichia coli. RNase J1, an essential ribonuclease, is now known to have not only endonuclease activity but also the long-missing bacterial 5’-to-3’ exonuclease activity [1]. The 5’ exonuclease mode of RNase J1 is active on 5’-monophosphate and 5’-hydroxyl ends but is inactive on 5’-triphosphate ends [2–3]. (The closely related RNase J2 of B. subtilis is not essential and does not show significant 5’ exonuclease activity [4].)
The glmS gene codes for glucosamine-6-phosphate (GlcN6P) synthase and expression of the gene is inhibited by the product of the enzyme reaction, GlcN6P. It was known that binding of GlcN6P to the 5’ UTR of glmS mRNA results in decreased gene expression, thus putting glmS in the category of riboswitch genes [5]. However, unlike all other such genes, metabolite binding to the glmS UTR does not induce transcription termination or translational regulation but induces autocatalytic cleavage at a 5’-proximal site, leaving a downstream fragment with a 5’-hydroxyl end [6]. How this results in depression of gene expression was shown recently by Winkler and colleagues to be due to RNase J1-mediated degradation of the downstream cleavage product (Fig. 1A) [7]. Thus, glmS gene expression is regulated by a combination of metabolite-mediated ribozyme cleavage and exonucleolytic degradation. We can expect that other examples of this form of RNase-mediated regulation remain to be discovered. Connected to this, it is likely that the 5’ exonuclease activity of RNase J1 can act directly on the 5’ end of native transcripts, once the 5’ triphosphate is converted to a 5’-monophosphate by an RNA pyrophosphohydrolase (RppH) activity, similar to the one shown recently by Belasco and colleagues to exist in E. coli [8–9]. As such, regulation of an RppH-like activity, and subsequent RNase J1 5’ exonuclease activity, could be a mechanism by which mRNA half-life is controlled, similar to the regulation of the decapping step in eukaryotes [10–12].
Fig. 1.
Three forms of regulation by RNA decay in B subtilis. (A) Regulation of glmS mRNA stability. The 5'-leader region contains a ribozyme moiety, whose activity is induced by binding of GlcN6P (red hexagon), the product of the GlmS reaction. Ribozyme self cleavage exposes a 5'-hydroxyl group on the downstream fragment, which is a substrate for 5' exonuclease activity of RNase J1. (B) Regulation of trp operon transcription by trp leader RNA degradation. In conditions of low intracellular tryptophan, the entire operon is transcribed from a constitutive promoter. When tryptophan is abundant, excess tryptophan (red dots) binds to TRAP (orange spheres), which allows the 11-mer TRAP complex to bind to nascent trp transcript. TRAP binding favors formation of the terminator structure such that transcription ends before reaching the trp genes. This regulation depends on release of TRAP to allow for ongoing binding to nascent trp transcripts, which is accomplished by trp leader RNA degradation via RNase J1 endonuclease and PNPase 3' exonuclease activities. (Although RNase J1 has been shown to exist in a complex with RNase J2 [4], only RNase J1 is shown here for simplicity.) (C) Regulation of thrS mRNA stability. In conditions of high intracellular threonine, transcription from a constitutive promoter terminates in the thrS leader region. When threonine concentration is low, uncharged tRNAThr (pink) binds to the thrS 5'-UTR, resulting in antiterminator formation and transcription of the thrS gene. RNase J1 cleavage of the antiterminated transcript amplifies the positive effect of uncharged tRNAThr by generating a new 5' end that forms a protective stem-loop structure and confers five-fold increased stability.
trp operon: recycling of a regulatory RNA-binding protein
A more indirect mechanism for controlling gene expression by RNA decay is represented by the B. subtilis trp operon, which contains six ORFs that specify tryptophan synthesis enzymes. Transcription of the trp operon is regulated by a premature transcription termination decision, which is made in the process of transcribing the 140-nt trp leader region [13]. When the cell has ample amounts of tryptophan, excess tryptophan binds to a regulatory protein called TRAP (trp RNA-binding attenuation protein), which exists in the cell as an 11-mer complex. Binding of tryptophan to TRAP enables TRAP to bind to a series of 11 trinucleotide repeats in the central region of the trp leader RNA (Fig. 1B). This results in formation of a terminator structure and termination of trp operon transcription. Thus, the presence of excess tryptophan ensures repression of trp operon expression.
For this mechanism to work, TRAP must bind the nascent trp transcript as it is being synthesized. However, TRAP binds quite tightly to the trp leader sequence (Kd = 0.12 nM [14]), and the number of TRAP complexes per cell is estimated to be 200–400, not all of which would be activated by tryptophan and some of which would be bound to three other RNAs that also are regulated by TRAP [15]. In order to ensure ongoing termination of transcripts that initiate via the strong, constitutive trp promoter, it is critical that TRAP bound to the trp leader region be released quickly to maintain the free concentration of TRAP complex. This is achieved by rapid degradation of the trp leader region, which is initiated by RNase J1 endonuclease cleavage at around nt 100, followed by processive 3’-to-5’ exonuclease degradation by polynucleotide phosphorylase (PNPase) [3]. In a strain that is deleted for PNPase, TRAP is not recycled off trp leader region RNA and transcription of the trp operon becomes deregulated [16]. Accordingly, this is an example of regulation of expression that depends on degradation of an RNA to which a regulatory protein is bound.
Of relevance here is the recent surprising finding that, as shown in Caulobacter crescentus and E. coli, newly-synthesized RNA transcripts do not appear to diffuse throughout the cell, as had been previously assumed, but remain tethered to the site of transcription [17]. This would explain why genes encoding regulatory proteins that control operon gene expression are often located as part of, or nearby, the regulated operon. While the mtrB gene encoding TRAP is located only 7 kilobases upstream of the trp operon, the RNase J1 and PNPase genes are located hundreds of kilobases away from the trp operon. Thus, the cellular concentration of these ribonucleases must be such that either diffusion alone, or perhaps some directed complex, will allow regulation by RNA decay.
thrS: induced expression enhanced by mRNA stabilization
A third example of regulated expression that involves RNA stability is the B. subtilis thrS gene, encoding threonyl-tRNA synthetase, a member of the so-called T-box genes that respond to levels of uncharged cognate tRNA [18]. Binding of the uncharged tRNAThr to the highly structured thrS 5'-UTR promotes a conformational change that leads to formation of an antiterminator structure at the expense of a transcription attenuator (Fig. 1C) [19], in a premature transcription termination decision similar to that described for TRAP and the trp operon above. Curiously, under conditions of threonine starvation, when uncharged tRNAThr accumulates, the major read-through species found in the cell is not the full-length primary transcript, but an endonucleolytically cleaved thrS RNA bearing the transcription attenuator at its 5' end and a Rho-independent transcription terminator at its 3' end [20]. The cleavage efficiency is much greater under conditions of threonine starvation and the processed species is about 5-fold more stable than the primary transcript, suggesting that binding of uncharged tRNAThr has two effects: transcriptional read-through and regulated mRNA cleavage and stabilization that amplifies the antitermination effect. Although RNase J1 has been proposed as the main candidate for this endonucleolytic cleavage event, based on its ability to cleave at this site in vitro [21], that this enzyme is directly involved in the regulatory event and affects the amount of functional thrS mRNA produced under amino acid starvation conditions remains to be formally proven in vivo. It should be noted that B. subtilis contains another recently-discovered endonuclease, RNase Y, which has been implicated in the decay of many mRNAs [22–23]. RNase Y is thus another possible candidate for regulatory mechanisms that include endonucleolytic cleavage.
SR1 sRNA: curious effect of a small, regulatory RNA on mRNA stability
In the last decade, many small, regulatory RNAs (sRNAs) have been discovered in diverse species, and it has been suggested that a bacterial species may contain on average 200–300 sRNAs [24]. Two recent B. subtilis transcriptome studies have increased substantially the number of predicted sRNAs in this organism [25–26]. Most sRNAs that have been studied up to now base pair at or close to the translation initiation site of their target RNAs and have their primary effects on translation. Inhibition of translation in many cases has been shown to lead to a secondary effect on RNA stability, either because the RNA denuded of ribosomes is more vulnerable to nuclease attack or through direct recruitment of the degradation machinery through an interaction between RNase E and the RNA chaperone Hfq [27]. The Vogel lab was the first to show that RNA turnover could be the primary consequence of sRNA binding to its target, with the direct targeting of the S. typhimurium ompD mRNA for degradation upon binding of the small regulatory RNA MicC [28].
SR1 sRNA of B. subtilis is expressed in a sugar-dependent fashion, with expression being induced under gluconeogenic conditions [29]. It was shown a few years ago that SR1 sRNA can inhibit translation of ahrC, a transcriptional regulator of arginine catabolic operons [30]. Inhibition was due to base-pairing of SR1 sRNA to ahrC mRNA, which resulted in altered RNA structure downstream of the ribosome binding site [31]. Very recently [32], expression of SR1 sRNA was found by Brantl and colleagues to inhibit degradation of gapA operon mRNA, which encodes glycolytic enzymes (gap = glyceraldehyde-3-phosphate dehydrogenase). Surprisingly, the mechanism of SR1 action was independent of base-pairing, but instead required the small peptide encoded by SR1 sRNA, called SR1P. SR1P was found to interact with GapA protein itself, but it is not yet clear how stability of gapA mRNA is achieved. Stabilization may be a by-product of ribosome stalling that occurs when SR1P binds to nascent GapA, or SR1P may inhibit the putative ribonuclease activity of GapA [33]. Interestingly, the function of stabilization in this case is not to enhance expression, since the expression of gapA operon enzymes is low when SR1 expression is induced, i.e., under gluconeogenic conditions where glycolytic enzymes are not required. Rather, it has been hypothesized that stabilization of gapA mRNA via SR1P allows maintenance of some level of gapA mRNA in the cell, which can thus respond quickly to the appearance of glucose in the medium [32].
Regulated mRNA decay in other Gram-positive organisms
Small Regulatory RNAs
Although B. subtilis is the best developed Gram-positive organism in terms of our understanding of the mRNA turnover pathways and the enzymes involved, studies in other Gram-positive bacteria are beginning to hint at some interesting regulatory phenomena relating to mRNA stability. Two recent papers show how VR sRNA of Clostridium perfringens and FasX sRNA of Streptococcus pyogenes stabilize their target RNAs. A ~35 nt sequence near the 3' end of the VR RNA binds to the 5' UTR of its target, the collagenase gene colA, and provokes an endonucleolytic cleavage 2 nts downstream of the hybrid [34]. The cleavage reaction both frees up the ribosome binding site and results in sequestration of the 5' end of the transcript in a stable secondary structure. Although the primary stabilization effect appears to be due to increased translation, an effect of the secondary structure has not been ruled out. Transcription of the VR RNA is activated directly by the VirR/VirS two component system as part of the virulence pathway of C. perfringens.
The FasX sRNA of S. pyogenes also stabilizes its target, the streptokinase mRNA ska, but in this case by binding directly to its 5' end [35]. The 9-nt hybrid between FasX and the 5' end of ska is sufficient to stabilize the mRNA encoding this virulence factor. This was elegantly shown by the ability of the same sequence to stabilize the ska mRNA independently of FasX when fused in cis to the 5' end, whereas fusion of a scrambled sequence destabilized the ska mRNA despite the fact that FasX could still base-pair, just no longer at the 5' extremity. Since neither RNase Y nor PNPase were involved in this phenomenon, RNase J1 must be considered the most likely candidate. Interestingly, RNase Y (CvfA) does play a key role in growth medium-dependent expression of virulence factors in S. pyogenes [36]. However, since RNase Y expression is not directly affected by these conditions, it would appear that the effects are indirect.
Growth-phase dependent mRNA stability
A related phenomenon of growth-phase regulation of mRNA stability has been observed in Lactococcus lactis [37]. Although mRNAs stabilities had been measured previously at a global level in both E. coli and B. subtilis [38–40], this was the first study to compare mRNA half-lives at a global level in response to changes in growth conditions, specifically carbon source starvation. The mean half-life of mRNAs was dramatically increased from about 6 minutes in exponential phase to over 19 minutes under conditions of glucose starvation. At the level of individual transcripts, the median stability increased about 4-fold as the cells ran out of glucose, with short-lived transcripts in exponential phase showing the greatest levels of stabilization. This points to a global mechanism of mRNA turnover in L. lactis that responds to carbon source levels. It is not yet clear how this is achieved or which ribonucleases are involved.
A similar effect of growth phase has been observed for particular transcripts in group A Streptococci (GAS) [41]. Messenger RNAs in GAS have been divided into two categories, those that are unstable in both exponential and stationary phase (class I) and those that are highly stable in stationary phase (class II). RNAs in the latter class, e.g., sagA, sda and arcT, are highly enriched for transcripts involved in GAS virulence pathways. These RNAs are at least partially dependent on PNPase for their turnover. It was shown recently that, while class I mRNAs of GAS are typically turned over by either RNase J1 or RNase J2 (both essential in GAS), class II mRNAs are typically resistant to these enzymes for a period of time before finally succumbing to their activity [42]. It is not clear what causes this delay, but the suggestion has been made that RNase J1 and J2 are titrated by class I RNAs before becoming available to attack class II transcripts.
Other possible examples of regulated mRNA stability in Gram positives
There are a number of interesting cases to be found in Gram-positive bacteria that are potential examples of regulation at the level of mRNA stability and, although far from definitively proven, merit some discussion here. Enterococcus faecalis cells lacking RNase J2 are defective in pilin gene expression and biofilm formation [43]. RNase J2 is co-transcribed in a well-conserved operon with genes involved in the synthesis of peptidoglycan components meso-diaminopimelic acid (mDAP) and L-lysine, and therefore factors that regulate the expression of this operon (e.g., mDAP levels in E. coli or lysine levels in plants) would be expected to have an effect on pilin formation through the expression levels of RNase J2.
In E. coli, expression of three ribonuclease genes, rnc (RNase III), rne (RNase E), and pnp (PNPase), is known to be controlled by feedback regulation, i.e., gene product-mediated control of its own mRNA decay [44–49]. Not much is known, however, about autoregulation of the ribonuclease-encoding genes of B. subtilis or other Gram-positive bacteria. There is some suggestion of feedback regulation between the levels of RNase J1 and RNase J2 in GAS: the levels of RNase J1 increase under conditions of RNase J2 depletion [42]. The mechanism of such regulation is unknown, but since all known examples of autoregulation of RNase gene expression exploit enzyme activity and act at the level of mRNA stability, it would be no surprise if this were the case here. A clearer example of autoregulation is seen with the Streptomyces coelicolor absB gene, encoding RNase III [50]. The absB gene is found in an operon with two other genes in S. coelicolor. A point mutation causing decreased RNase III activity leads to about a three-fold increase in the stability of the full-length absB-containing transcript in vivo. In addition, RNase III cleaves this transcript at two sites, both in vitro and in vivo, one of which maps to a stem-loop structure in the absB ORF. Cleavage at this site is presumably responsible for the destabilization of the absB mRNA under conditions of RNase III excess.
RNase III has also been shown to affect antibiotic production and morphological differentiation in S. coelicolor. RNase III directly destabilizes the adpA mRNA, encoding a transcription activator involved in sporulation [51]. Interestingly, expression of AdpA leads to a decrease in RNase III levels through the induction of proteases that target it for degradation, creating an autoregulatory feed-back loop. Strains lacking RNase III show increased expression of sporulation genes and a concomitant decrease in antibiotic production. Since RNase III transcript levels decrease upon entry into stationary phase [52], just at the point where antibiotic production begins, it is likely that RNase III either destabilizes a transcript encoding a repressor of antibiotic production or stabilizes that of an activator. The details of this regulatory pathway remain to be determined.
Conclusion
Bringing examples of “unique” regulatory mechanisms to the public consciousness leads to discovery of such types of regulation for more genes and operons. The three examples of B. subtilis post-transcriptional regulation shown in Fig. 1 are quite different from each other, involving: (a) mRNA self cleavage to activate downstream decay; (b) RNA decay to release an RNA-binding protein; and (c) endonucleolytic cleavage to stabilize downstream mRNA. Variations on these themes are bound to be discovered in coming years. The complexity of bacterial gene regulation, with so many mechanisms that operate at the transcriptional level, is increased further by a growing number of examples of control at the level of the RNA itself.
Acknowledgments
Funding for C.C.’s laboratory was from the Agence Nationale de la Recherche (ANR project subtilRNA2). Funding for D.H.B.’s laboratory was from the National Institutes of Health (GM048804). We have tried to be as up-to-date as possible on forms of regulated mRNA decay in Gram-positive organisms. However, we acknowledge the distinct possibility that we have missed some examples, for which we apologize.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- **1.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5'-to-3' exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5' stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. Discovery of 5’-to-3’ exoribonuclease activity in bacteria. [DOI] [PubMed] [Google Scholar]
- **2.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. Structural and biochemical characterization of RNase J, showing a single catalytic site for both endonucleolytic and 5’ exonucleolytic activities. [DOI] [PubMed] [Google Scholar]
- 3.Deikus G, Condon C, Bechhofer DH. Role of Bacillus subtilis RNase J1 Endonuclease and 5'-Exonuclease Activities in trp Leader RNA Turnover. J Biol Chem. 2008;283:17158–17167. doi: 10.1074/jbc.M801461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *4.Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, de la Sierra-Gallay IL, Noirot P, Putzer H, Condon C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 2010;75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. In B. subtilis, the essential RNase J1 exists primarily in a complex with the non-essential RNase J12. Unlike RNase J1, RNase J2 shows no significant 5’ exonuclease activity in vitro. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- **7.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. Demonstration of mRNA destabilization by exposure of an RNase J1-sensitive 5’ end. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **9.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. Identification of the E. coli activity that converts a 5’ triphosphate end to a monophosphate end. [DOI] [PubMed] [Google Scholar]
- 10.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song MG, Kiledjian M. 3' Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 14.Baumann C, Otridge J, Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J Biol Chem. 1996;271:12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- 15.McCabe BC, Gollnick P. Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis. J Bacteriol. 2004;186:5157–5159. doi: 10.1128/JB.186.15.5157-5159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deikus G, Babitzke P, Bechhofer DH. Recycling of a regulatory protein by degradation of the RNA to which it binds. Proc Natl Acad Sci U S A. 2004;101:2747–2751. doi: 10.1073/pnas.0307343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. Demonstration that mRNAs (and ribosome that translate them) do not diffuse freely but are localized to the site of transcription. Important implications for regulation of gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 19.Putzer H, Condon C, Brechemier-Baey D, Brito R, Grunberg-Manago M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 2002;30:3026–3033. doi: 10.1093/nar/gkf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Condon C, Putzer H, Grunberg-Manago M. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1996;93:6992–6997. doi: 10.1073/pnas.93.14.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 29.Licht A, Preis S, Brantl S. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol Microbiol. 2005;58:189–206. doi: 10.1111/j.1365-2958.2005.04810.x. [DOI] [PubMed] [Google Scholar]
- 30.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 31.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–4346. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32.Gimpel M, Heidrich N, Mader U, Krugel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. Peptide encoded by SR1 sRNA involved in stabilization of an operon mRNA. [DOI] [PubMed] [Google Scholar]
- 33.Evguenieva-Hackenberg E, Schiltz E, Klug G. Dehydrogenases from all three domains of life cleave RNA. J Biol Chem. 2002;277:46145–46150. doi: 10.1074/jbc.M208717200. [DOI] [PubMed] [Google Scholar]
- *34.Obana N, Shirahama Y, Abe K, Nakamura K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5' leader sequence. Mol Microbiol. 2010;77:1416–1428. doi: 10.1111/j.1365-2958.2010.07258.x. mRNA stabilization by small RNAs in C. perfringens and S. pyogenes. [DOI] [PubMed] [Google Scholar]
- *35.Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. mRNA stabilization by small RNAs in C. perfringens and S. pyogenes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang SO, Caparon MG, Cho KH. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect Immun. 2010;78:2754–2767. doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redon E, Loubiere P, Cocaign-Bousquet M. Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J Biol Chem. 2005;280:36380–36385. doi: 10.1074/jbc.M506006200. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci U S A. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hambraeus G, von Wachenfeldt C, Hederstedt L. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol Genet Genomics. 2003;269:706–714. doi: 10.1007/s00438-003-0883-6. [DOI] [PubMed] [Google Scholar]
- 41.Barnett TC, Bugrysheva JV, Scott JR. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *42.Bugrysheva JV, Scott JR. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol Microbiol. 2010;75:731–743. doi: 10.1111/j.1365-2958.2009.07012.x. Both RNase J1 and RNase J2 are essential in S. pyogenes, and participate in decay of two classes of mRNA that differ significantly in half-life. [DOI] [PubMed] [Google Scholar]
- 43.Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol. 2010;192:5489–5498. doi: 10.1128/JB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain C, Belasco JG. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- 45.Schuck A, Diwa A, Belasco JG. RNase E autoregulates its synthesis in Escherichia coli by binding directly to a stem-loop in the rne 5' untranslated region. Mol Microbiol. 2009;72:470–478. doi: 10.1111/j.1365-2958.2009.06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardwell JC, Regnier P, Chen SM, Nakamura Y, Grunberg-Manago M, Court DL. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989;8:3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsunaga J, Simons EL, Simons RW. RNase III autoregulation: structure and function of rncO, the posttranscriptional “operator”. RNA. 1996;2:1228–1240. [PMC free article] [PubMed] [Google Scholar]
- 48.Jarrige AC, Mathy N, Portier C. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001;20:6845–6855. doi: 10.1093/emboj/20.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carzaniga T, Briani F, Zangrossi S, Merlino G, Marchi P, Deho G. Autogenous regulation of Escherichia coli polynucleotide phosphorylase expression revisited. J Bacteriol. 2009;191:1738–1748. doi: 10.1128/JB.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *50.Xu W, Huang J, Cohen SN. Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J Bacteriol. 2008;190:5526–5530. doi: 10.1128/JB.00558-08. Autoregulation of RNase III gene expression in S. coelicolor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu W, Huang J, Lin R, Shi J, Cohen SN. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol Microbiol. 2010;75:781–791. doi: 10.1111/j.1365-2958.2009.07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sello JK, Buttner MJ. The gene encoding RNase III in Streptomyces coelicolor is transcribed during exponential phase and is required for antibiotic production and for proper sporulation. J Bacteriol. 2008;190:4079–4083. doi: 10.1128/JB.01889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]