SUMMARY
Cotton is an important cash crop worldwide and serves as a significant source of fiber, feed, foodstuff, oil and biofuel products. Considerable effort in genetics and genomics has been expended to increase sustainable yield and quality through molecular breeding and genetic engineering of new cotton cultivars. With the effort of whole genome sequencing of cotton, it is essential to develop molecular tools and resources for large-scale analysis of gene functions at the genome-wide level. We have successfully established an Agrobacterium-mediated virus-induced gene silencing (VIGS) assay in several cotton cultivars with different genetic backgrounds. The genes of interest were potently and readily silenced within 2 weeks after inoculation at the seedling stage. Importantly, we showed that silencing GhNDR1 and GhMKK2 compromised cotton resistance to the infection by Verticillium dahliae, a fungal pathogen causing Verticillium wilt. Furthermore, we established a cotton protoplast system for transient gene expression to study gene functions by a gain-of-function approach. The viable protoplasts were isolated from green cotyledons, etiolated cotyledons, and true leaves, and responded to a wide range of pathogen elicitors and phytohormones. Remarkably, cotton plants possess conserved, but also distinct MAP kinase activation with Arabidopsis upon bacterial elicitor flagellin perception. Thus, we demonstrated that GhNDR1 and GhMKK2 are required for Verticillium resistance in cotton using gene silencing assays, and established the high throughput loss-of-function and gain-of-function assays for functional genomic studies in cotton.
Keywords: Gossypium hirsutum, pathogen elicitors, protoplast transient gene expression assay, resistance, Verticillium dahliae, virus-induced gene silence (VIGS)
INTRODUCTION
Cotton (Gossypium spp.) is an important crop around the world, which sustains one of the world’s largest industries (textiles) and serves as a significant source of feed, foodstuff, oil and biofuel production (Sunilkumar et al., 2006). With about 50 species in the Gossypium genus, four are cultivated in agriculture, including two allotetraploids (G. hirsutum and G. barbadense) and two diploids (G. herbaceum and G. arboreum) (Wendel, 2000, Zhang et al., 2008a). G. hirsutum, also known as Upland cotton, produces more than 95% of the annual cotton crop worldwide (Chen et al., 2007). In addition to its economic importance, cotton also provides a model for studying genome size evolution and polyploidization (Grover et al., 2004, Zhang et al., 2008a). Cotton fiber, a single and greatly elongated cell from the epidermal layer of the ovule, is a unique system for the study of plant cell elongation and cell wall and cellulose biosynthesis (Kim & Triplett, 2001).
Considerable effort has been expended to increase sustainable yield and quality of cotton through improved plant cultivars, cultivation practices and disease control. The recent advances in cotton genetics and genomics have generated significant excitement in the molecular breeding of new cotton cultivars. Genetic engineering of novel genes into cotton provides an alternative and robust approach of genome improvement. Cotton has led the way for commercialization of genetically-engineered plants, taking advantage that it is not a food crop. Sequencing and decoding of the cotton genome has been initiated by worldwide cotton genome scientists (Chen et al. 2007). The D genome species G. raimondii was prioritized for the whole-genome shotgun sequence because of its smallest genome size among Gossypium species (880Mb for the haploid). Recently, a draft physical map of G. raimondii was assembled, providing the foundation for BAC-based sequencing and validating whole-genome shotgun sequences (Lin et al. 2010). With the availability of cotton whole-genome sequence, it is essential to develop molecular tools and resources for large-scale analysis of gene functions at the genome-wide level. Ultimately, understanding the molecular mechanisms of gene function and regulation will lead to cotton improvement by genetic engineering and molecular breeding.
Various large-scale approaches, including chemical mutagenesis, T-DNA and transposon-based insertional mutant populations have been developed for forward genetic studies (Robinson & Parkin, 2009, Ostergaard & Yanofsky, 2004, Martienssen, 1998). Large collections of mutant populations have greatly facilitated the study of gene functions at the genome-wide level for certain model plant species such as Arabidopsis thaliana because of ease transformation (Chory et al., 2000). Although these technologies are being developed in cotton to understand the features that make cotton unique (Paterson, 2010), currently, they have not been widely and successfully deployed in cotton functional genomics due to the technical challenge of transformation, long growth cycle and large genome size. In addition, these functional genomic approaches have certain drawbacks, such as large populations required for disrupting a gene of interest, gene targeting bias and complication with multiple mutations or insertions, lack of obvious phenotypes due to the presence of large gene families, gene duplication and polyploidy, and lethality caused by complete loss-of-functions (Borevitz & Ecker, 2004). Virus-induced gene silencing (VIGS) offers an alternative and complementary approach for large-scale functional analysis of individual genes by knocking down the expression of endogenous genes (Burch-Smith et al., 2004, Dinesh-Kumar et al., 2003, Lu et al., 2003b). The technique was developed based on the RNA-mediated post-transcriptional gene silencing (PTGS) that functions as an antivirus defense system in plants and other organisms (Hamilton & Baulcombe, 1999). This rapid and efficient approach avoids plant transformation and overcomes functional redundancy (Burch-Smith et al., 2004, Becker & Lange, 2010). The power of VIGS as a tool for high-throughput functional genomics was further accelerated with the Agrobacterium tumefaciens-mediated transient assays. Seedlings or roots could be syringe or vacuum-infiltrated, agrodrench-inoculated or sprayed with the Agrobacterium culture carrying the viral vector containing the gene of interest to degrade the endogenous genes, making it feasible to inoculate a large number of plants within a short time (Burch-Smith et al., 2004, Ryu et al., 2004). Recently, a bombardment-based geminivirus-mediated gene silencing vector was developed from cotton leaf crumple virus and shown to be able to silence two endogenous genes, Magnesium chelatase subunit I (ChlI) and phytoene desaturase gene (PDS) in cotton (Tuttle et al., 2008). In that experiment, the microprojectile coated with viral DNA was delivered into plants by a particle acceleration unit attached to a high-pressure helium tank.
To further decipher gene functions and dissect signal transduction pathways at the cellular, biochemical and molecular levels, various transient gain-of-function assays, for instance, protoplast transient expression, have been developed in different plants. The protoplasts freshly isolated from plant tissues retain their cell identity and differentiated state and display physiological and cell-autonomous responses to a broad spectrum of signals, including plant hormones, metabolites, environmental cues, and pathogen-derived elicitors (Sheen, 2001, He et al., 2007, Yoo et al., 2007). Protoplasts can be used as a vehicle to transiently express or silence genes for high throughput screen and systemic characterization of candidate genes. Notably, the specific responses triggered by expression of the transfected genes can be assayed within a day or two, which is particularly attractive for crop plants with a long growth cycle and recalcitrant for stable transgenic plants. Cotton protoplasts have been isolated from callus, cotyledons, hypocotyls, leaves and cell suspension cultures since the 1970s for plant regeneration studies (Bhojwani et al., 1977). However, it has not been used as a tool for gene discovery and functional studies.
Verticillium wilt, one of the most devastating plant diseases worldwide, is caused by the soil-borne fungi Verticillium spp (Fradin & Thomma, 2006). Verticillium wilt in cotton, caused typically by V. dahliae, is a major concern for cotton producers triggering severe yield losses each year. This pathogen is particularly difficult to control in cotton as the hyphae reside in the woody vascular tissues and is thus protected from fungicides. In addition, V. dahliae survives in soil for many years due to its extremely persistent resting structures called microsclerotia (Fradin & Thomma, 2006). The genetic and molecular mechanisms underlying cotton resistance to Verticillium infection are poorly understood. Recently, the effort has been made to develop mapping populations for identifying the quantitative trait loci (QTL) responsible for resistance in cotton to V. dahliae (Yang et al., 2008, Bolek et al., 2005). In tomato, the Ve locus has been bred into most tomato cultivars for resistance to Verticillium wilt. Positional cloning identified two closely linked genes, Ve1 and Ve2, both of which encode receptor-like proteins with an extracellular leucine-rich repeat domain (Kawchuk et al., 2001). Arabidopsis is also a host to V. dahliae. Genetic analysis identified a dominant locus, V. dahliae-tolerance 1 (VET1), in ecotype C-24, conferring the increased tolerance to V. dahliae infection (Veronese et al., 2003). Interestingly, it seems that gene silencing machinery is also required for resistance in Arabidopsis to Verticillium wilt, since several components in RNA silencing pathways were found to alter defense to V. dahliae (Ellendorff et al., 2009). By using a VIGS approach, EDS1 (Enhanced Disease Susceptibility1), NDR1 (Non-race-specific Disease Resistance1), BAK1 (BRI1-Associated Kinase1) and MEK2 (MKK2, MAP kinase kinase 2), were shown to be required for resistance in tomato to V. dahliae (Fradin et al., 2009). However, it remains enigmatic whether a similar mechanism exists in cotton that exhibits resistance to V. dahliae infection.
In this study, we successfully established an Agrobacterium-mediated VIGS assay on cotton seedlings for efficient silencing of cotton genes in different cotton cultivars. Silencing of GhNDR1 or GhMKK2 in a commercial cotton cultivar compromised its resistance to V. dahliae infection. We further developed a protoplast transient transfection assay for studying cotton gene functions by gain-of-function approach. We showed that cotton protoplasts responded to various pathogen elicitors and phytohormones. The loss-of-function and gain-of-function assays established in this study lay important foundations for functional genomics studies of cotton genes.
RESULTS
Silencing of the cotton CLA1 gene by Agrobacterium-mediated VIGS
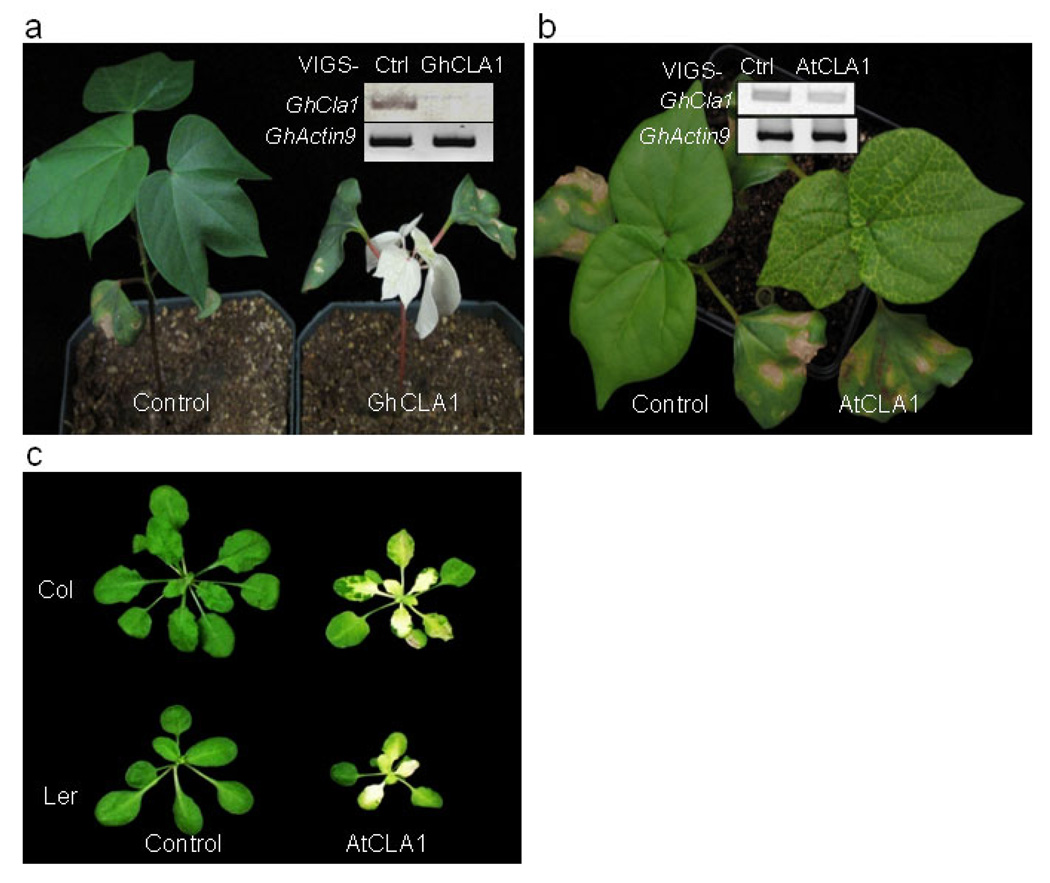
VIGS has been proven to be a powerful tool for gene function studies and functional genomics in various higher eudicots including Solanaceae, Arabidopsis, cucurbit and legume species, and monocot crop species like barley, rice, wheat and maize (Becker & Lange, 2010). Among several viral VIGS vectors, tobacco rattle virus (TRV) invades a wide range of hosts and is able to spread vigorously throughout the entire plant yet producing mild symptoms in the hosts (Burch-Smith et al., 2004). In an initial experiment to develop an Agrobacterium-mediated VIGS system in cotton, we silenced the expression of endogenous Cloroplastos alterados 1 gene (CLA1) with TRV-based VIGS vector. In Arabidopsis, CLA1 gene (At4g15560) encodes 1-deoxyxylulose 5-phosphate synthase, the first enzyme of the 2-C-methyl-D-erythritol-4-phosphate pathway involved in chloroplast development (Estevez et al., 2000). The CLA1 gene is highly conserved in different plant species and disruption of CLA1 by T-DNA insertion in Arabidopsis resulted in an albino phenotype (Mandel et al., 1996). We used AtCLA1 gene as a query to blast against the Gossypium unigenes database at http://www.cottondb.org/blast/blast.html. We identified a cotton CLA1 gene (GhCLA1) sharing 81% identities with AtCLA1 at the nucleotide level. GhCLA1 possesses 80% identity and 85% similarity with AtCLA1 at the amino acid level (Figure S1). We PCR amplified a 500bp fragment of GhCLA1 from cotton cDNA and inserted it into an improved pTRV-RNA2 (pTRV2) VIGS vector, pYL156 (Liu et al., 2002a, Liu et al., 2002b). A mixture of Agrobacterium cultures containing pTRV-RNA1 (pTRV1) and pTRV2-GhCLA1 in a 1:1 ratio were hand-inoculated into the underside of the cotyledons of 2-week-old cotton plants cultivar Deltapine 90 with a needleless syringe. To facilitate infiltration, a few holes were gently punched on the underside of the cotyledons with a syringe needle (gauge 22G11/2). The plants were kept at 25°C. As a control, the same number of cotton plants were infiltrated with the mixture of Agrobacterium culture containing pTRV1 and pTRV2 vector. Initiation of photobleaching phenotype in the GhCLA1-agrobacteria infiltrated plants was observed on the newly emerging leaves approximately 10 days post-infiltration. One month later, 100% of GhCLA1-agrobacteria infiltrated plants exhibited a strong photobleaching phenotype, which was uniformly distributed on the entire true leaves (Figure 1a, Table S1). The stems of GhCLA1-agrobacteria infiltrated plants turned pink or purple, and the plants became severely stunted by the late growth stage (six-week-old) likely due to the strong inhibition of chlorophyll biosynthesis. The plants infiltrated with the pTRV2 empty vector did not display any photobleaching phenotype and grew normally (Figure 1a).
Figure 1. Agrobacterium-mediated VIGS in cotton.
a. The photobleaching phenotype of cotton leaves triggered by VIGS-GhCLA1. Agrobacterium culture carrying pTRV1 and pTRV2-GhCLA1 was infiltrated into two fully expanded cotyledons of two-week-old seedlings of cotton cultivar Deltapine90. pTRV2-GFP was used as a vector control. The insert shows the expression of GhCLA1 in control and silenced plants by RT-PCR analysis. GhActin9 was used as a control. b. The photobleaching phenotype triggered by VIGS-AtCLA1 in cotton. The insert shows the expression of GhCLA1 in control and silenced plants by RT-PCR analysis. c. The photobleaching phenotype triggered by VIGS-AtCLA1 in Arabidopsis Col-0 and Ler-0. The pictures were taken at 3 weeks (a and b) and 5 weeks (c) after Agrobacterium infiltration. The experiments were repeated five times with similar results.
Silencing of GhCLA1 by a heterologous gene fragment from Arabidopsis thaliana
TRV VIGS vector carrying phytoene desaturase gene (PDS) orthologs from a wide range of plant species in Solanaceous family has been shown to silence NbPDS in the VIGS-efficient plant N. benthamiana (Senthil-Kumar et al., 2007). To test whether a heterologous gene sequence from a different family is able to trigger VIGS in cotton, we infiltrated the cotton cotyledons (cultivar Deltapine 90) with AtCLA1 from Arabidopsis thaliana. The photobleaching phenotype in AtCLA1-agrobacteria infiltrated cotton plants was initially observed 14-days after infiltration in true leaves, but to a lesser extent than that in GhCLA1-agrobacteria infiltrated plants (Fig. 1b and Table S1). The photobleaching was limited to the veins of true leaves in AtCLA1-agobacteria infiltrated plants, compared to whole leaves in GhCLA1-agrobacteria infiltrated plants. Nevertheless, the heterologous gene sequence of AtCLA1 gene from Arabidopsis thaliana in TRV VIGS vector is able to suppress the expression of its cotton ortholog, suggesting cotton plants are readily amenable to VIGS. As expected, infiltration of AtCLA1-agrobacteria, but not the vector control, in Arabidopsis induced the silencing of endogenous CLA1 and caused the photobleaching phenotype in both ecotypes Col-0 and Ler-0 (Fig. 1c). However, infiltration of GhCLA1-agrobacteria in Arabidopsis Col-0 did not induce photobleaching phenotype (data not shown).
Highly efficient VIGS-mediated gene silencing in different cotton cultivars
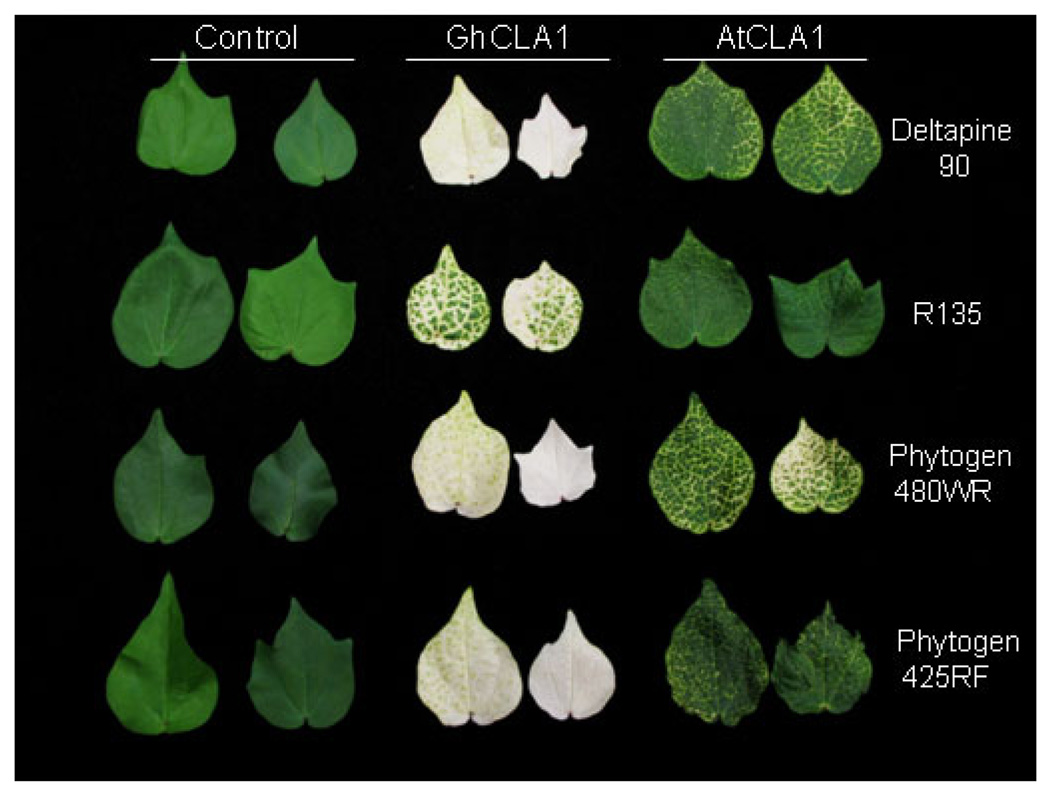
For many crops, the generation of stable transgenic plants is largely limited to a few genetic backgrounds. In cotton transformation, a persistent challenge has been the genotype-specificity of cotton somatic embryogenesis, which is the most efficient transformation method (Trolinder, 2009; Paterson, 2010). Cotton transformation is limited to highly embryogenic cultivars, such as Coker 312 and Coker 5110 (Trolinder, 2009; Paterson, 2010). To utilize VIGS as a tool for cotton gene function studies and functional genomics, silencing genes readily in different genotypes would be desirable. To this end, we tested the VIGS efficiency on a wide range of cotton breeding lines or commercial cultivars that were at one time grown in Texas, USA, including Deltapine 90, R135, Phytogen 480WR, Phytogen 425RF, Fibermax 832, and PSC355. Similar with cultivar Deltapine 90 (Fig. 1), the photobleaching phenotype was observed in all these cultivars infiltrated with GhCLA1-agrobacteria within 2 weeks after inoculation (Fig. 2 and Table S1). One month later, the photobleaching phenotype had uniformly spread to the entire leaves, and the silencing efficiency on all cultivars tested was equally high and consistently reached 100% with multiple independent biological trials (Fig. S2 and Table S1). The photobleaching phenotype was incomplete, with some green color remaining, on line R135 compared with other lines although the silencing efficiency was also 100% four weeks after infiltration. Similarly, infiltration of AtCLA1-agrobacteria triggered the silencing of GhCLA1 and caused photobleaching phenotype in most cotton cultivars except R135 (Fig. S3). Taken together, these data suggest that TRV VIGS vector could trigger potent silencing of endogenous genes in cotton, and cotton plants are readily amenable to VIGS experiments for gene loss-of-function studies.
Figure 2. Agrobacterium-mediated VIGS in a diverse array of cotton cultivars.
Two fully expanded cotyledons of different cotton genotypes were infiltrated with Agrobacterium culture containing pTRV1 and pTRV2, pTRV2-GhCLA1 or pTRV2-AtCLA1. The pictures were taken at 5 weeks after Agrobacterium infiltration. The experiments were repeated three times with similar results.
GhNDR1 and GhMKK2 are required for resistance against Verticillium dahliae in cotton
VIGS has been successfully used to decipher gene function in numerous cellular and developmental processes in plants, particularly plant disease resistance. For example, VIGS has been used in tobacco and tomato to study disease resistance gene (R)-mediated resistance to bacteria and virus (Liu et al., 2002a, Jin et al., 2002, Ekengren et al., 2003). We sought to explore the VIGS approach to study the genetic requirement of resistance in cotton to V. dahliae by silencing orthologs of several genes known to be involved in disease resistance in other plant species. In Arabidopsis, NPR1 (Non-expressor of Pathogenesis-Related genes-1), plays an essential role in plant systemic acquired resistance. Constitutive expression of NPR1 in several plant species, including cotton, confers enhanced resistance to various bacterial and fungal pathogens (Cao et al., 1998, Quilis et al., 2008, Parkhi et al., 2010). Cotton NPR1 (GhNPR1) was induced by infection of Fusarium oxysporum pv. vasinfectum and Xanthomonas axonopodis pv. malvacearum, suggesting potential involvement in cotton disease resistance (Zhang et al., 2008b). In tomato, Ve1 locus, encoding a receptor-like protein, provides resistance against V. dahliae (Kawchuk et al., 2001, Fradin & Thomma, 2006). Arabidopsis NDR1 (Non race-specific disease resistance 1) is involved in plant race-specific resistance to infection by bacterial and fungal pathogens (Century et al., 1997). MKK2 encodes a MAP kinase kinase involved in plant responses to various biotic and abiotic stresses (Qiu et al., 2008). In tomato, Ve1, MKK2, and NDR1 have been suggested as required for resistance against race 1 of V. dahliae (Fradin et al., 2009). Race 1 of V. dahliae would likely be the “nondefoliating” type of the fungus in cotton. We examined the functional requirement of cotton GhVe1, GhNPR1, GhNDR1, and GhMKK2 genes in cotton resistance to V. dahliae King isolate, a “defoliating” type and aggressive cotton pathogen.
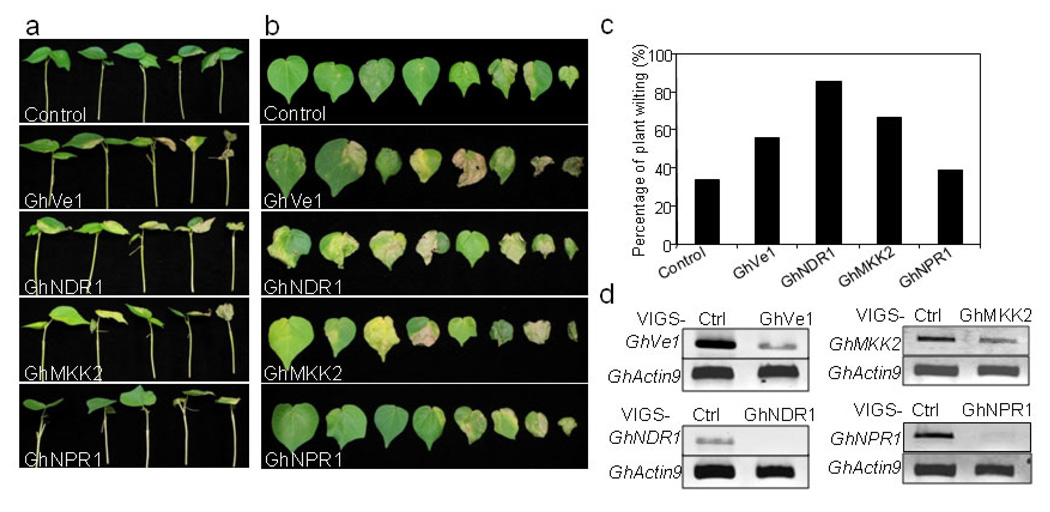
We designed the VIGS-GhNPR1 primers according to published GhNPR1 sequence (Zhang et al., 2008b). Full-length tomato Ve1, Arabidopsis NPR1, NDR1 and MKK2 sequences were used as a query to blast search against the Gossypium unigenes database, to identify their cotton orthologs. We further designed the primers to amplify the relatively conserved regions of cotton orthologs and cloned them into VIGS pTRV2 vector. The Agrobacterium culture carrying the recombinant pTRV vectors was hand-infiltrated into cotton cotyledons of cultivar Fibermax (FM) 9160B2F (Bayer Cropsciences, Lubbock, TX). At least eight plants were infiltrated for each construct, pTRV-GhVe1, GhNDR1, GhMKK2 and GhNPR1. Two weeks after Agrobacterium infiltration, when the plants infiltrated with GhCLA1 displayed photobleaching phenotype, we hand-inoculated a spore suspension of V. dahliae into the stems of cotton seedlings with a 25-G syringe at 1×106 conidia/ml. Three weeks later, wilting of the true leaves was scored. FM 9160B2F was selected as a partially resistant genotype because it exhibited lower incidence of Verticillium wilt and less defoliation in six field studies compared to most other cultivars (Wheeler and Woodward, 2010). For vector control inoculated-seedlings, about 30% of plants exhibited a weak partial leaf wilting phenotype (Fig. 3). Significantly, silencing of GhVe1, GhNDR1 or GhMKK2 enhanced plant susceptibility to V. dahliae, and plants exhibited more severe wilting phenotype than the vector control plants (Fig. 3a, 3b and 3c). In particular, more than 80% of GhNDR1 VIGSed plants were severely infected by V. dahliae, which caused all the leaves to wilt in some cases. More than 65% of GhMKK2 VIGSed plants also showed strong wilting symptoms on leaves. Interestingly, silencing of GhNPR1 did not seem to compromise cotton resistance to V. dahliae (Fig. 3a, 3b and 3c). However, we do not exclude the possibility of existence of other functional NPR1 homologs in cotton that was not silenced by this GhNPR1 construct. The silencing of these genes was further confirmed by RT-PCR analysis with RNAs isolated from leaves or stems (Fig. 3d and S4). Together, our VIGS assays identified GhNDR1, GhMKK2, and GhVe1 as important components in cotton resistance to V. dahliae infection.
Figure 3. Silencing of GhVe1, GhNDR1 and GhMKK2 enhanced plant susceptibility to V. dahliae infection.
The cotton cultivar FM9160B2F was first silenced with individual genes by VIGS, and then inoculated with V. dahliae (isolate King) suspension spores at the concentration of 1 ×106/ml. Whole plant (a) and detached leaves (b) were pictured at 18 days post V. dahliae infection. c. The percentage of plants that showed Verticillium wilting phenotype at 18 days post infection. The disease rate was scored based on 18~21 plants. d. The gene expression of GhVe1, GhNDR1, GhMKK2 and GhNPR1 in silenced and control plant leaves by RT-PCR analysis. GhActin9 was used as a control. The experiments were repeated three times with similar results.
Cotton mesophyll protoplasts as a vehicle to express transfected genes for functional studies
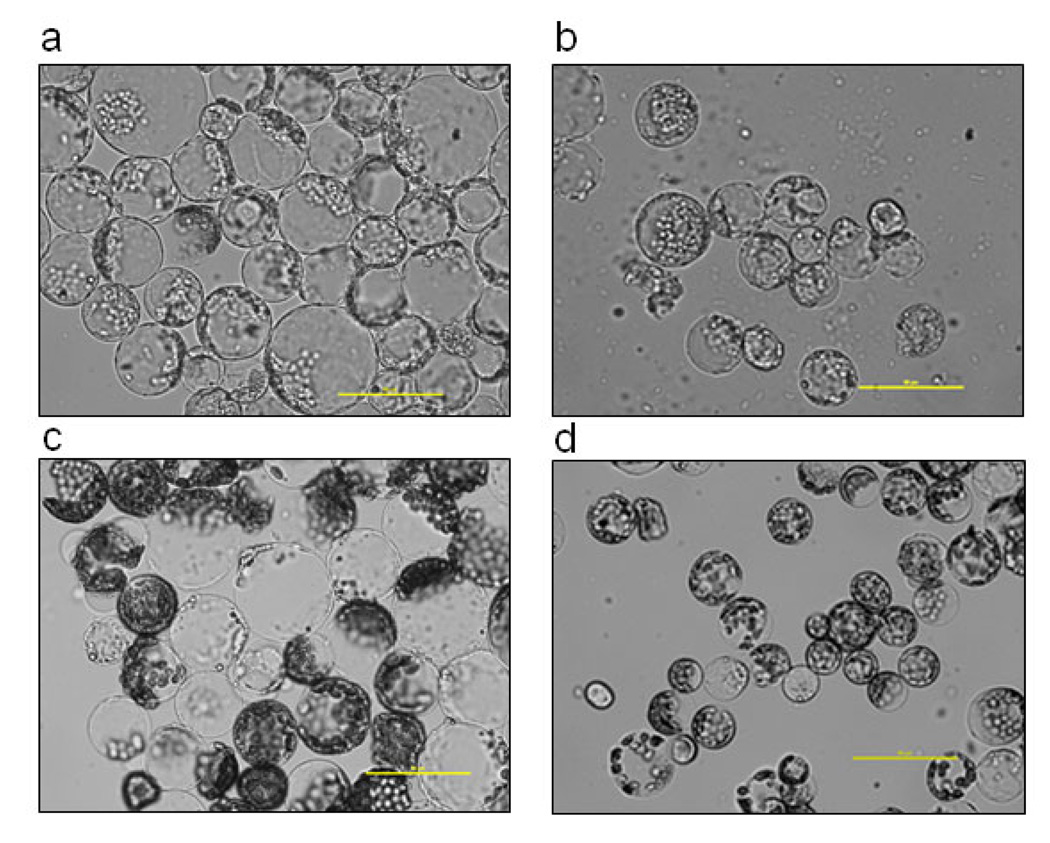
Protoplast transient assays have proven to be useful in studying gene functions and dissecting signal transduction pathways without the need to generate transgenic plants. We initiated studies to isolate cotton protoplasts for transient gene expression assays. We first compared the protoplast yield and quality isolated from leaves or cotyledons at one-, two- or three-week-old stage under either 12/12 hour light/dark cycle or dark (etiolated) growth conditions for two different cotton cultivars, Cropland Genetics 3035RF and FM9160B2F (Table 1). The protoplasts isolated from green tissues were uniformly spherical and contained green chloroplasts, whereas the protoplasts from etiolated tissues were pale green and contained less chloroplasts. In general, the protoplasts from cotyledons were bigger in size than those from true leaves (Figure 4; Table 1). The isolation procedure was optimized with different incubation times of either 3 hr or 12 hr after a 30 min vacuum treatment (see Experimental Procedures for the detail protocol). As summarized in Table 1, the 12 hr incubation yielded more protoplasts than the 3 hr incubation based on three independent experiments for both green and etiolated tissues. In general, green cotyledons, etiolated cotyledons, and true leaves produced comparable amount of protoplasts (Table 1). There was also no apparent difference in protoplast production between two cultivars.
Table 1.
The efficiency of cotton protoplast isolation.
| Cultivar | Tissue | Age (week) |
Protoplast yield /ml enzyme solution |
Incubation time |
Protoplast size |
|---|---|---|---|---|---|
| Cropland Genetics 3035RF | Green cotyledon | 1 | 3 ×105 | 12 hr | Medium |
| Cropland Genetics 3035RF | Green cotyledon | 2 | 7.5 ×105 | 12 hr | Big |
| Cropland Genetics 3035RF | Green cotyledon | 2 | 0.64 ×105 | 3 h | Big |
| Cropland Genetics 3035RF | True leaf | 3 | 2.34 ×105 | 12 hr | Small |
| Cropland Genetics 3035RF | Etiolated cotyledon | 1 | 8.4 ×105 | 12 hr | Small |
| Cropland Genetics 3035RF | Etiolated cotyledon | 2 | 0.04 ×105 | 3 h | Small |
| FM9160B2F | Green cotyledon | 1 | 3.4 ×105 | 12 hr | Medium |
| FM9160B2F | Green cotyledon | 2 | 4 ×105 | 12 hr | Big |
| FM9160B2F | Green cotyledon | 2 | 1.16 ×105 | 3 hr | Big |
| FM9160B2F | True leaf | 3 | 13.2 ×105 | 12 hr | Small |
| FM9160B2F | Etiolated cotyledon | 1 | 5.4 ×105 | 12 hr | Small |
| FM9160B2F | Etiolated cotyledon | 2 | 0.08 ×105 | 3 hr | Small |
Figure 4. The protoplasts isolated from different tissues of cotton plants of cultivar FM9160B2F.
The protoplasts were isolated from (a) 2-week-old green cotyledon, (b) 2-week-old etiolated cotyledon, (c) 3-week-old green cotyledon and (d) 3-week-old true leaf. The pictures are representative of three independent repeats.
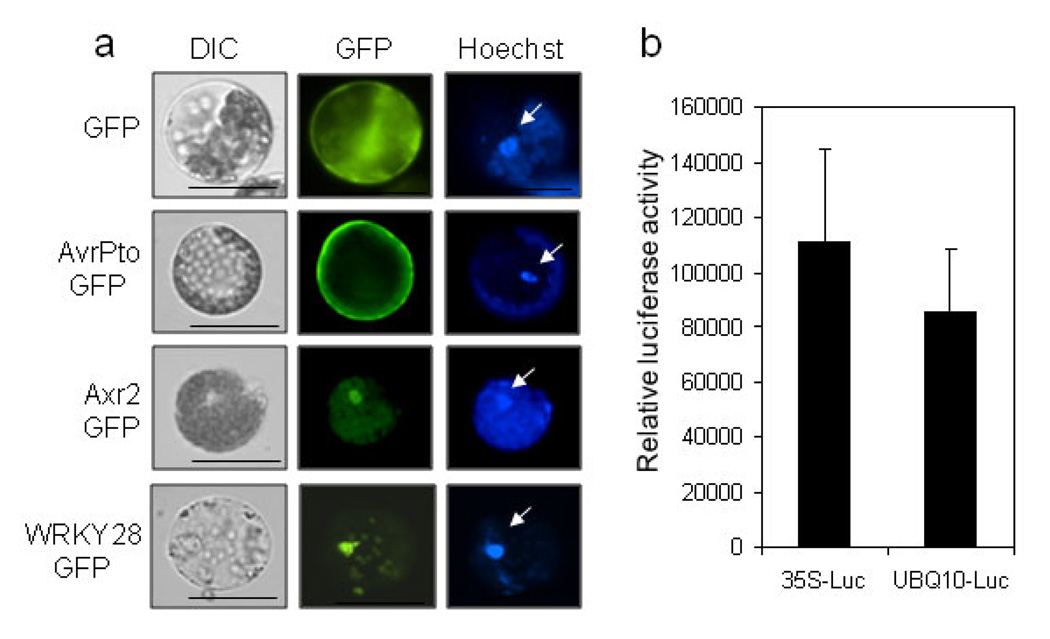
We further examined the feasibility of expressing exogenous genes in cotton protoplasts. PEG-mediated DNA transfection was carried out to introduce various reporter genes including green fluorescence protein (GFP), β-glucuronidase (GUS) and luciferase (LUC), into cotton protoplasts isolated from 2-week-old green cotyledons. GFP reporter is a vital marker for protein subcellular localization. The cotton protoplasts transfected with GFP under the control of constitutive CaMV35S promoter showed very strong fluorescence signal, and the signal was uniformly and diffusively distributed in the cytosol and nucleus (Figure 5a). Bacterial protein AvrPto is localized to plasma membrane in Arabidopsis and tobacco. Similarly, AvrPto-GFP was predominantly observed in the plasma membrane in cotton protoplasts (Figure 5a). AXR2-GFP or WRKY28-GFP, two transcription factors fused with GFP, exhibited strong nuclear localization in cotton (Figure 5a). Taken together, the distinct subcellular localization of different GFP fusion proteins in cotton protoplasts demonstrated that cotton protoplasts are suitable for protein subcellular localization studies.
Figure 5. Transient expression of exogenous genes in cotton protoplasts.
a. Subcellular localization of GFP proteins in cotton protoplasts. The protoplasts were transfected with indicated constructs and incubated for 12 hr before GFP observation. Arrows indicate the Hoechst 33342 staining of nucleus, confirming the nuclear localization of Axr2-GFP and WRKY28-GFP. b. Expression of luciferase reporter gene in cotton protoplasts. The protoplasts were transfected with 35S-LUC or UBQ-LUC, and the luciferase activity was detected 6 hr after transfection by a plate reader. The protoplasts were isolated from the cotyledons of cultivar FM9160B2F. The experiments were repeated three times with similar results. The data in (b) are shown as means ± standard errors from three independent biological replicates.
LUC reporters under the control of different promoters were well established to readily and quantitatively monitor gene transcriptional level. We transfected the LUC gene under the control of either the 35S promoter or UBQ (ubiquitin) promoter into cotton protoplasts. Both 35S-LUC and UBQ-LUC transfected protoplasts exhibited about 100,000 fold higher luciferase activity than the empty vector control (Figure 5b). We also attempted to transfect 35S-GUS and UBQ-GUS reporters into cotton protoplasts. However, cotton protoplasts exhibited a very high background level of GUS activity, which made it difficult to detect the activity of transfected genes (data not shown).
Cotton protoplasts respond to multiple pathogen elicitors and activate convergent MAP kinase signaling
Plant defense against pathogen infection is triggered by detecting a collection of pathogen elicitors, termed pathogen-associated molecular patterns (PAMPs), through cell surface pattern recognition receptors (PRRs) (Boller & He, 2009). Upon PAMP perception, plants activate complex defense responses, including MAP kinase activation, reactive oxygen species (ROS) production, and induction of an array of defense-related genes (Boller & He, 2009). We tested whether cotton protoplasts can be used as a tool to study signal transduction in plant defense by detecting the MAPK activation upon PAMP perception. Several common elicitors, including bacterial flg22 (a 22 amino acid peptide from bacterial flagellin), elf18 (an 18 amino acid peptide from bacterial elongation factor-Tu), HrpZ1 harpin, peptidoglycan (PGN), lipopolysaccharide (LPS), fungal chitin and oomycete NPP1, were tested for the activation of MAPK in cotton protoplasts. We also included VdNEP, a necrosis- and ethylene-inducing protein, from V. dahliae (Wang et al., 2004).
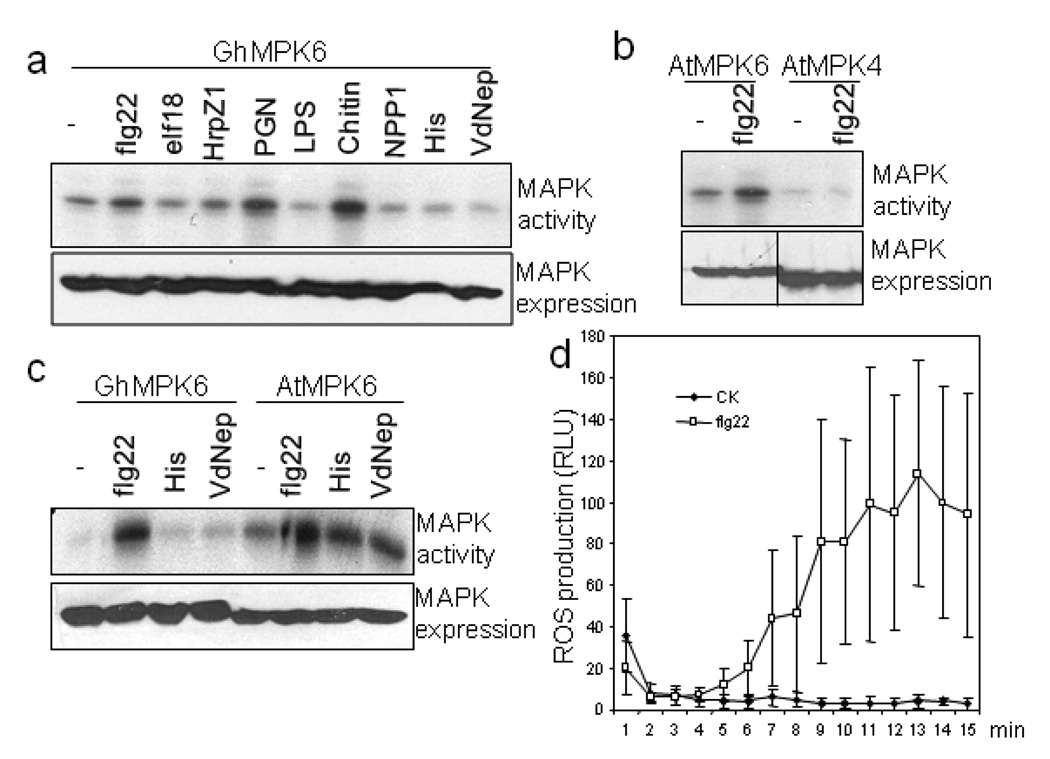
Arabidopsis MPK3, MPK4 and MPK6, and their orthologs in other species, were strongly activated by these elicitors (Rodriguez et al., 2010). We cloned full length cotton MPK6 (GhMPK6) with an HA (human influenza hemagglutinin) epitope tag. We transiently expressed GhMPK6-HA in cotton protoplasts and treated the protoplasts with different elicitors. An immunocomplex kinase assay with MBP as a substrate was used to detect MAPK activation. Strikingly, treatment of flg22, PGN or Chitin activated GhMPK6 (Fig. 6a). Chitin showed the strongest activation of GhMPK6 among these elicitors. We did not detect the activation of GhMPK6 by elf18, LPS, NPP1 and VdNEP (Fig. 6a). GhMPK6 was well expressed in protoplasts as detected by anti-HA Western blot (Fig. 6a, lower panel). We also transiently expressed Arabidopsis AtMPK3 and AtMPK4 in cotton protoplasts and treated with flg22. Interestingly, flg22 activated AtMPK6, but not AtMPK4 in cotton cells (Fig. 6b) while flg22 activated AtMPK3, AtMPK4 and AtMPK6 in Arabidopsis (Rodriguez et al., 2010), indicating the conserved but also distinct MAPK signaling triggered by flagellin in different plant species. Similarly, when we expressed GhMPK6 in Arabidopsis protoplasts, flg22, but not VdNEP, activated GhMPK6 (Fig. 6c). The activation level of GhMPK6 by flg22, as well as its expression level, was comparable with AtMPK6 in Arabidopsis cells. Together, the data suggest that MPK6 signaling is conserved between cotton and Arabidopsis, whereas MPK4 signaling is likely divergent between these two species.
Figure 6. Cotton protoplasts respond to multiple PAMPs and elicit convergent defense signaling.
a. flg22, PGN and Chitin activate GhMPK6 in cotton protoplasts. The protoplasts were transfected with GhMPK6, and incubated for 6 hr before treatment with different PAMPs for another 10 min. The concentrations of individual PAMPs are flg22, 1 uM; elf18, 1 uM; HrpZ1 harpin, 100 nM; PGN, 50 µg/ml; chitin, 50 µg/ml; LPS, 50 µg/ml; NPP1, 20 nM; VdNEP, 20 nM. His is the control of His-VdNEP. The top panel shows the MAPK activity detected by phosphorylation of MBP substrate, and the bottom panel is GhMPK6 expression detected by Western blot with an anti-HA antibody. b. flg22 activated AtMPK6, but not AtMPK4, in cotton protoplasts. c. flg22 activated GhMPK6 in Arabidopsis protoplasts. “-“ is the H2O control for flg22 and His is the control of His-VdNEP. d. ROS burst in cotton leaves triggered by flg22 treatment. The experiments were repeated three times with similar results. The data in (d) are shown as means ± standard errors from ten independent biological replicates.
Flagellin can induce immune responses in a wide range of plants, including tomato, tobacco, and potato (Felix et al., 1999). To further confirm that cotton plants have a functional perception system for flg22, we tested the production of ROS in cotton seedlings infiltrated with flg22. As shown in Fig. 6d, flg22 treatment induced ROS production compared to H2O control treatment, suggesting cotton plants possess flagellin perception machinery. In the future, it will be interesting to determine whether the cotton ortholog of Arabidopsis flagellin receptor FLS2 functions as flagellin receptor, and the mechanism that flg22 is unable to activate MPK4 in cotton.
Cotton protoplasts respond to multiple phytohormones
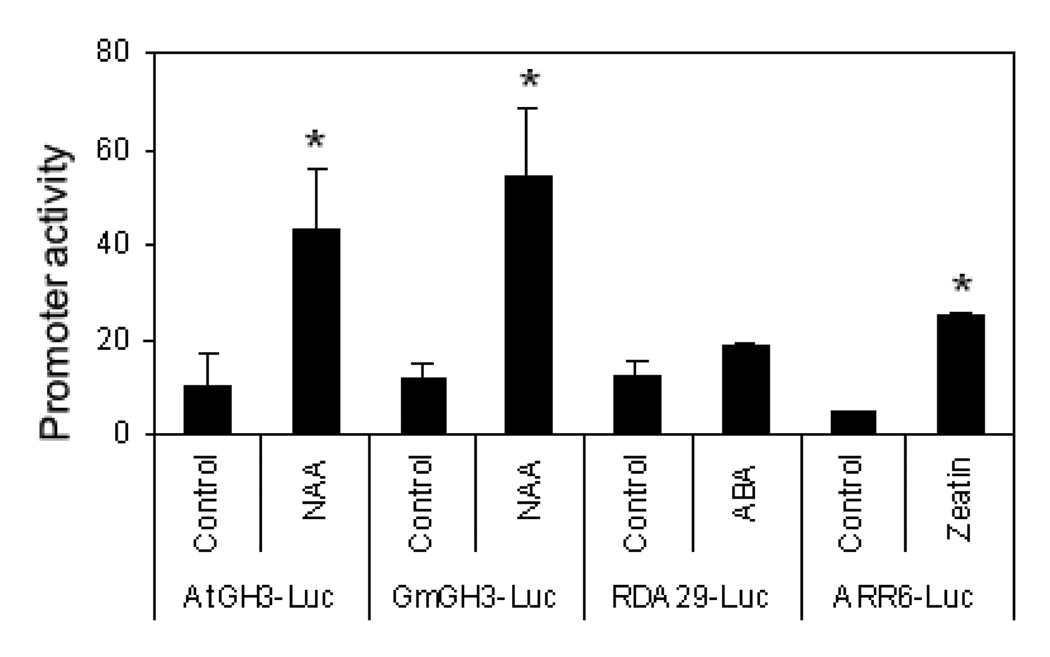
Freshly isolated protoplasts also respond to multiple phytohormones that activate specific transcriptome changes (Sheen, 2001). To examine whether cotton protoplasts could be used to study growth hormone regulated responses, we transiently expressed hormone marker genes and determined the reporter gene activity upon hormone treatment. As shown in Fig. 7, auxin (NAA) treatment strongly activated GH3-LUC reporter genes from both Arabidopsis (AtGH3-LUC) and soybean (GmGH3-LUC). Cytokinin (Zeatin) activated its reporter ARR6-LUC. Although ABA treatment also weakly induced RD29A-LUC reporter in multiple independent experiments, this induction was not statistically significant. Nevertheless, the reporter-mediated cotton protoplast transient transfection assay could potentially provide a tool to study growth hormone regulation and signaling in cotton. Future development of the cotton gene promoter reporters in response to different hormones will likely potentiate their responsiveness to the corresponding signals.
Figure 7. Cotton protoplasts respond to growth hormones.
Cotton protoplasts were transfected with specific marker genes for different plant hormones, then treated with 1 uM of NAA, 100 uM ABA or 1 uM Zeatin for 3 hr. The experiments were repeated four times with similar results. The data are shown as means ± standard errors from three independent biological replicates. * indicates a significant difference with p<0.05 when compared with data from control based on the results of an unpaired Student’s t-test.
DISCUSSION
As an important crop plant, the molecular biology and functional genomics of cotton are lagging behind other crop plants, such as rice, wheat and maize, largely due to limited molecular tools and resources. Here, we established two powerful transient assays for rapid gene discovery and gene function studies in cotton in a relatively short time frame. The Agrobacterium-mediated VIGS is able to readily silence the genes of interest for loss-of-function assays, and protoplast transient transfection provides a tool to express the genes of interest for gain-of-function studies. More importantly, unlike cotton transformation, these two transient assays are not limited to a few selective cotton cultivars. We consistently obtained 100% of gene silencing efficiency of GhGLA1 VIGS in all cultivars we tested. Ultimately, these approaches could be used to systematically characterize gene functions at genome-wide level and look for genes that make cotton unique and important.
The genetic requirement for resistance to Verticillium wilt in cotton is largely unknown. By using the VIGS approach, we demonstrated that cotton NDR1 and MKK2 orthologs are required for cotton with partial resistance to V. dahliae. We also showed that silencing a RLP, GhVe1, partially compromised resistance to V. dahliae. An interesting test will be to determine whether GhNDR1 and GhMKK2 function downstream of GhVe1, which might perceive an unknown PAMP from V. dahliae. The current knowledge of the molecular mechanisms of disease resistance has been mainly derived from the study of a few reference plants, such as Arabidopsis. Many disease resistance components and immune signaling pathways have been well studied in Arabidopsis by genetic and biochemical approaches. Given the conservation of Arabidopsis and cotton at the genome organization level (Rong et al., 2005), it is tempting to speculate that the defense signaling and gene functions are sufficiently conserved between these two species. Systemic characterization of the cotton orthologs by loss-of-function and gain-of-function will provide a wealth of information on the molecular mechanisms of cotton resistance to fungi, bacteria, nematodes and viruses.
Plants respond to multiple PAMPs and activate the convergent immune signaling. By using a protoplast transient transfection in combination with immunocomplex kinase assay, we demonstrated that cotton cells could respond to different PAMPs, including flg22, PGN and chitin, leading to the activation of the evolutionarily conserved MAPK cascade. Another potent bacterial elicitor, elf18, did not activate MAPK in cotton, consistent with the report that elf18 seems to be recognized only by members of the Brassicaceae (Zipfel et al., 2006). It is interesting to note that flg22 activated MPK6, but not MPK4 in cotton. Blast analysis against the Gossypium unigenes database suggested the existence of a cotton MPK4 homolog with 89% identities and 94% similarities with AtMPK4 at the amino acid level. In Arabidopsis, MEKK1 (MAPKKK) and MKK1/MKK2 function upstream of MPK4 (Gao et al., 2008, Qiu et al., 2008, Suarez-Rodriguez et al., 2007). In the mekk1 mutants, MPK4 activation by different stimuli was absent (Ichimura et al., 2006). The mekk1 mutants display abnormal plant growth independent of its kinase activity (Ichimura et al., 2006). It will be particularly interesting to learn whether cotton possesses an MEKK1 ortholog and whether it exhibits kinase activity.
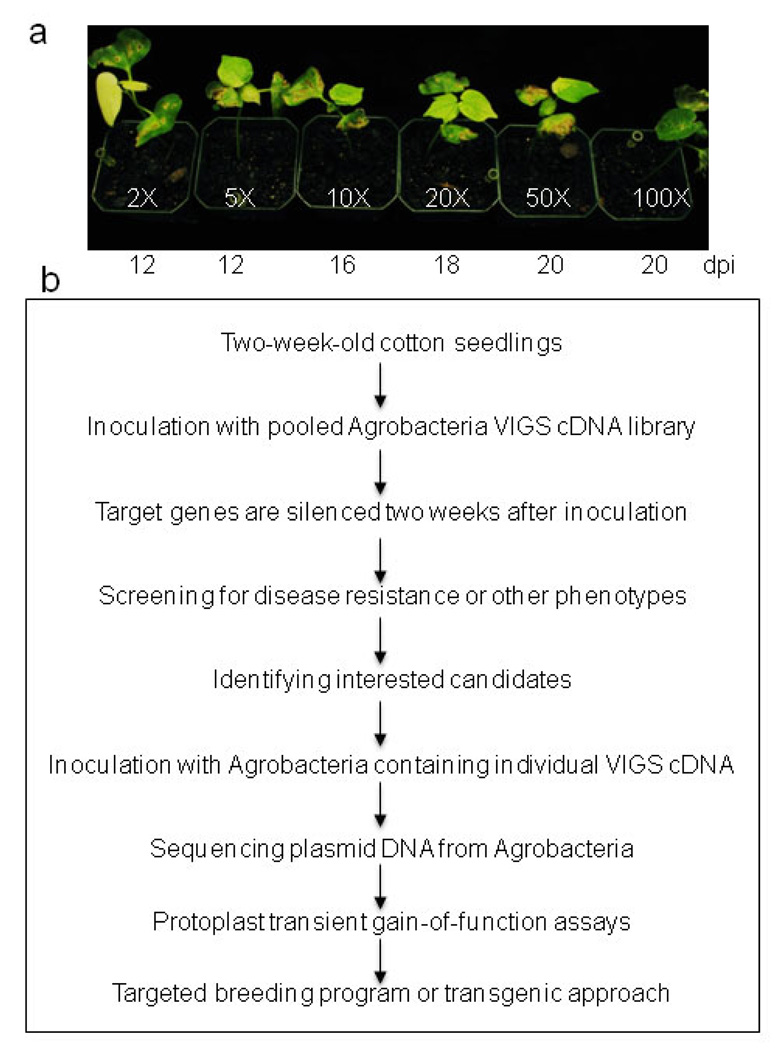
The rapidly innovative sequencing technologies are vastly expanding the sequence database for important plant species; the majority of which are difficult to manipulate for functional genomics studies. VIGS provides a rapid and powerful tool to dissect gene functions in species not amenable to stable genetic transformation. The large-scale VIGS experiments were easily adopted as a fast-forward genetics approach to screen for interesting phenotypes (Burch-Smith et al., 2004, Baulcombe, 1999). This method has been successfully used in VIGS-efficient plants, such as Nicotiana benthamiana. For example, HSP90 was identified as an important component in plant disease resistance by screening about 5000 cDNA clones, and BECLIN 1 was identified in autophagy-regulated programmed cell death through screening 1500 cDNA clones in N. benthamiana (Liu et al., 2005, Lu et al., 2003a). Since the high efficiency of Agrobacterium-mediated VIGS could be readily achieved in different cotton genotypes, this fast-forward genetics approach could be further adopted for high-throughput screening of important genes involved in abiotic and biotic stresses in cotton. A random cDNA library will be constructed into pTRV2 vector for screening of interesting phenotypes (Fig. 8). Since it takes considerable space and effort to grow a large number of cotton seedlings for VIGS, we sought to determine whether one plant could be inoculated with multiple clones containing different cDNA. We determined that VIGS still occurred effectively even if the pTRV2-CLA1 Agrobacterium culture was mixed with pTRV2 control vector Agrobacterium culture at 1:49 ration (50 fold dilution) for infiltration (Fig. 8a). This suggests that we could pool at least 50 individual clones in a mixture for the initial screening to minimize the amount of time and effort for VIGS assays (Fig. 8b). After identifying the pool that attributes interesting phenotypes, we will further test individual VIGS cDNA clones from the pool. The nature of genes linked to the phenotype will be revealed by sequencing the plasmid DNA recovered from Agrobacteria. Protoplast transient assay will further facilitate the study of the gene/protein functions through protein localization, protein-protein interaction, protein degradation, and gene transcription and regulation. The interested genes will provide valuable resources for targeted breeding programs and genetic engineering of cotton cultivars with desired traits (Fig. 8b).
Figure 8. Fast forward genetics and functional genomics to study cotton gene functions at a large-scale.
a. Agrobacterium culture containing 1/50 of pTRV2-GhCLA1 is sufficient to trigger gene silencing. Agrobacterium culture containing different dilutions of pTRV2-GhCLA1 was inoculated into cotton cotyledon. pTRV2 was substituted to make pTRV1 and pTRV2 at 1:1 ratio in the Agrobacterium culture. The number below photograph indicates the days after infiltration when 100% of plants showed photobleaching phenotype. b. The scheme of high throughput study of cotton gene functions by VIGS and protoplast transient assays.
EXPERIMENTAL PROCEDURES
Plant materials and growth
Cotton (Gossypium hirsutum) cultivars Deltapine 90, R135, Phytogen 480WR, Phytogen 425RF, FM 832, PSC 355 and FM 9160B2F were grown in pots containing Metro Mix 700 (SunGR, Beavile, WA) at 23°C under 12 hr/12 hr light conditions.
Construction of VIGS vectors and Agrobacterium-mediated VIGS
GhCLA1, GhNDR1, GhVe1, GhMKK2, and GhNPR1 cDNA was amplified by PCR from the cDNA library of G. raimondii leaf tissues, and inserted into pYL156 (pTRV-RNA2) vector with restriction enzymes EcoRI and KpnI digestion. GhCLA1-F, 5’-GGAATTCCACAACATCGATGATTTAG-3’, GhCLA1-R, 5’-GGGGTACCATGATGAGTAGATTGCAC-3’; GhNDR1-F,5’-CGGAATTCGGATACATACTTCAAACCCC-3’, GhNDR1-R,5’-GGGGTACCGCTCCAAGCAGCACCACAC-3’; GhVe1-F, 5’-CGGAATTCCTGACACATTTCCAGAGAAC-3’, GhVe1-R, 5’-GGGGTACCGTCAATGGAGGTAAACACCG-3’; GhMKK2-F, 5’-CGGAATTCGCCATCGGAAGCTGACAATGACG-3’; GhMKK2-R, 5’-GGGGTACCGCTAGCCCTGAGGTGCTTGTC-3’; GhNPR1-F, 5’-GGAATTCATGGATATAGCTCAAGTGGA-3’, GhNPR1-R, 5’-GGGGTACCTTACCTTTAGGCCCGGTCAC-3’.
The plasmids containing binary TRV vectors pTRV-RNA1, pTRV-RNA2 (pYL156), and pYL156 derivatives were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Agrobacterium culture was grown for overnight at 28°C in LB medium containing antibiotics Kanamycin and Gentamicin, as well as10 mM MES and 20 uM acetosyringone. The cells were pelleted by centrifugation and re-suspended in infiltration culture containing 10 mM MgCl2, 10 mM MES and 200 uM acetosyringone. Cell suspensions were incubated at room temperature for at least 3 hr. The Agrobacterium culture containing pTRV-RNA1 and pTRV-RNA2 or its derivatives was mixed at 1:1 ratio and infiltrated into two fully expanded cotyledons of 2-week-old plants with a needleless syringe. To facilitate the infiltration, small holes were punched with a needle on the underside of cotyledon. VIGS experiments were repeated at least three times with more than six plants for each construct per repeat.
Verticillium dahliae infection of cotton plants
Verticillium dahliae (isolate King) was grown on potato dextrose (PDA) agar at room temperature (23°C) for 3–4 days. The spore suspensions were prepared at 1×106 conidia/ml in a solution containing 0.001% Tween-20, and stem inoculated into both control and VIGS silenced plants using a syringe needle (22G11/2 gauge), at a position approximately 1 cm below cotyledons (Bolek et al., 2005).
Cotton protoplast isolation and transient gene expression assays
Cotton protoplasts were isolated from green cotyledons, etiolated cotyledons and young leaves according to He et al. (2007) with modifications. Cotyledons and true leaves were excised from 1-, 2- or 3-week-old plants, sliced with a razor blade and digested in an enzyme solution supplemented with 2% sucrose for 0.5 hr under vacuum. Subsequently, the enzyme solution was incubated without vacuum at room temperature for 3 hr or 12 hr. The protoplasts were released by filtering through a 30-µm nylon mesh, washed with W5 solution and diluted in MMG solution to a density of 2 × 105 cells/ml.
Reporter gene assay was conducted as described by He et al. (2007). The protoplasts were transfected with reporter genes, incubated at room temperature for 1 hr, and then treated with different phytohormones at the indicated concentrations for another 5 hr. For protein localization, cotton protoplasts were transfected with C-terminal GFP fusion of AvrPto, AXR2, or WRKY28 under the control of 35S promoter. 35S-GFP was used as a control. Protein localization was observed 12 hr later after transfection using a fluorescence microscopy. To confirm the nuclear localization, the transfected cells were stained with DNA stain dye Hoechst 33342 (Sigma) at the concentration of 1 mg/ml for 5 min.
Immunocomplex kinases assay and Western blot analysis
GhMPK6 was PCR amplified from G. hirsutum with primers 5’-CG GGATCCATGGAAGGCGGAGGACCACC-3’ and 5’-GAAGGCCTCTGCTGCAGATATTCTGGGTTG-3’, and cloned into a plant expression vector. 200 µl of cotton protoplasts were transfected with cotton or Arabidopsis MAPKs, and incubated for 6 hr. The protoplasts were treated with different PAMPs at the indicated concentrations for 15 min. Immunocomplex kinase and Western blot assays were performed as reported (Lu et al., 2010). The expression of cotton and Arabidopsis MAPK proteins was detected by an anti-HA antibody.
Cotton RNA isolation and RT-PCR analysis
Cotton total RNA was extracted from leaves or stems of VIGS silenced plants at approximately two weeks post VIGS using Spectrum™ Plant Total RNA Kit (Sigma) according to the manufacturer’s protocol. RNA was then treated with DNase (Invitrogen) to remove genomic DNA. 1 µg of purified RNA was subsequently reverse transcribed with First-Strand cDNA Synthesis kit (Promega) using oligo-dT as a primer. The primers of genes used for semi-quantitative RT-PCR are as following: GhCLA1-F, 5’-GCCCTTTGTGCATCTTC-3’, GhCLA1-R, 5’-CTCTAGGGGCATTGAAG-3’; GhNDR1-F, 5’-CACTTTTCATGTGGCTATG-3’, GhNDR1-R, 5’-GCAAGAAATTGAACAAAG-3’; GhVe1-F, 5’-GAAGGGCCAATACCAGAAG-3’, GhVe1-R, 5’-GAGGAAGCCCACACAATCC-3’; GhMKK2-F, 5’-GCCTGAAGCTCCTCCGCC-3’; GhMKK2-R, 5’-CCCAAGCTCCAAATGTCAC-3’; GhNPR1-F, 5’-GGAATCGGATGACGTCGAG-3’, GhNPR1-R, 5’-CATCACCAGCGATGGCAAG-3’. PCR amplification was run for 35 cycles (each cycle: 45 sec at 95 °C, 30 sec at 52 °C, and 1 min at 72 °C). GhActin9 was used as an internal standard control (Djonovic et al., 2006).
Measurement of ROS production
ROS burst was determined by a luminol-based assay as described by Felix et al. (1999) with modifications. The first true leaf of three-week-old cotton plants was excised into leaf discs of 0.25cm2, following an overnight incubation in 96-well plate with 100 µl of H2O to eliminate the wounding effect. H2O was replaced by 100 µl of reaction solution containing 20 µM of luminol and 10 µg/ml of horseradish peroxidase (Sigma) supplemented with 100 nM of flg22. The measurement was conducted immediately after adding the solution with a luminometer (Perkin Elmer, 2030 Multilable Reader, Victor X3), with a 1 min interval reading time for a period of 15 min. The measurement values for ROS production from 20 leaf discs per treatment were indicated as means of RLU (Relative Light Units). The experiments were repeated twice with similar results.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Jim Starr for encouragement and support for the project, and the critical reading of the manuscript. We thank Dr. S.P. Dinish-Kumar for pTRV-VIGS vectors, Dr. J. Sheen for pTRV2-AtCLA1 and various reporter constructs, Dr. X. Chen for VdNEP construct, Dr. S. Wu for AXR2-GFP construct, Drs. M. Cabbage and M. Dickman for sharing their greenhouse space and Dr. H. Koiwa for the use of microscope. We also thank Dr. Jim Starr, Dr. Richard Percy and Bayer CropScience for providing cotton seeds and Dr. T. Isakeit for a V. dahliae isolate for initial setting up the bioassay. L.S. and P.H. were supported by Texas A&M Start-up funds and NSF (IOS-1030250) to L.S. and NIH (R01GM092893) to P.H.
REFERENCES
- Baulcombe DC. Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- Becker A, Lange M. VIGS--genomics goes functional. Trends Plant Sci. 2010;15:1–4. doi: 10.1016/j.tplants.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Bhojwani SS, Power JB, Cocking EC. Isolation, culture and division of cotton callus protoplasts. Plant Sci Lett. 1977;8:85–89. [Google Scholar]
- Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, et al. Mapping of verticillium wilt resistance genes in cotton. Plant Sci. 2005;168:1581–1590. [Google Scholar]
- Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Ecker JR. Plant genomics: the third wave. Annu Rev Genomics Hum Genet. 2004;5:443–477. doi: 10.1146/annurev.genom.5.061903.180017. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci U S A. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007;145:1303–1310. doi: 10.1104/pp.107.107672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Ecker JR, Briggs S, Caboche M, Coruzzi GM, Cook D, et al. National Science Foundation-Sponsored Workshop Report: "The 2010 Project" functional genomics and the virtual plant. A blueprint for understanding how plants are built and how to improve them. Plant Physiol. 2000;123:423–426. doi: 10.1104/pp.123.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y. Virus-induced gene silencing. Methods Mol Biol. 2003;236:287–294. doi: 10.1385/1-59259-413-1:287. [DOI] [PubMed] [Google Scholar]
- Djonovic S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM. Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant Microbe Interact. 2006;19:838–853. doi: 10.1094/MPMI-19-0838. [DOI] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 2003;36:905–917. doi: 10.1046/j.1365-313x.2003.01944.x. [DOI] [PubMed] [Google Scholar]
- Ellendorff U, Fradin EF, de Jonge R, Thomma BP. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot. 2009;60:591–602. doi: 10.1093/jxb/ern306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez JM, Cantero A, Romero C, Kawaide H, Jimenez LF, Kuzuyama T, et al. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000;124:95–104. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Fradin EF, Thomma BP. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol. 2006;7:71–86. doi: 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, Nazar RN, Robb J, et al. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150:320–332. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, et al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- Grover CE, Kim H, Wing RA, Paterson AH, Wendel JF. Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 2004;14:1474–1482. doi: 10.1101/gr.2673204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Sheen J. The use of protoplasts to study innate immune responses. Methods Mol Biol. 2007;354:1–9. doi: 10.1385/1-59259-966-4:1. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem. 2006;281:36969–36976. doi: 10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, et al. NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell. 2002;3:291–297. doi: 10.1016/s1534-5807(02)00205-8. [DOI] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, et al. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci U S A. 2001;98:6511–6515. doi: 10.1073/pnas.091114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001;127:1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Lin L, Pierce GJ, Bowers JE, Estill JC, Compton RO, Rainville LK, et al. A draft physical map of a D-genome cotton species (Gossypium raimondii) BMC Genomics. 2010;11:395. doi: 10.1186/1471-2164-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002a;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002b;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, et al. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003a;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. Virus-induced gene silencing in plants. Methods. 2003b;30:296–303. doi: 10.1016/s1046-2023(03)00037-9. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Martienssen RA. Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci U S A. 1998;95:2021–2026. doi: 10.1073/pnas.95.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard L, Yanofsky MF. Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J. 2004;39:682–696. doi: 10.1111/j.1365-313X.2004.02149.x. [DOI] [PubMed] [Google Scholar]
- Parkhi V, Kumar V, Campbell LM, Bell AA, Shah J, Rathore KS. Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 2010 doi: 10.1007/s11248-010-9374-9. [DOI] [PubMed] [Google Scholar]
- Paterson AH. Cotton Genomics. In: Zehr UB, editor. Cotton, Biotechnology in Agriculture and Forestry 65. Berlin Heidelberg: Springer-Verlag; 2010. pp. 45–63. [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, et al. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008;148:212–222. doi: 10.1104/pp.108.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilis J, Penas G, Messeguer J, Brugidou C, San Segundo B. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant Microbe Interact. 2008;21:1215–1231. doi: 10.1094/MPMI-21-9-1215. [DOI] [PubMed] [Google Scholar]
- Robinson SJ, Parkin IA. Bridging the gene-to-function knowledge gap through functional genomics. Methods Mol Biol. 2009;513:153–173. doi: 10.1007/978-1-59745-427-8_9. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Rong J, Bowers JE, Schulze SR, Waghmare VN, Rogers CJ, Pierce GJ, et al. Comparative genomics of Gossypium and Arabidopsis: unraveling the consequences of both ancient and recent polyploidy. Genome Res. 2005;15:1198–1210. doi: 10.1101/gr.3907305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Anand A, Kang L, Mysore KS. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 2004;40:322–331. doi: 10.1111/j.1365-313X.2004.02211.x. [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Hema R, Anand A, Kang L, Udayakumar M, Mysore KS. A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol. 2007;176:782–791. doi: 10.1111/j.1469-8137.2007.02225.x. [DOI] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001;127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci U S A. 2006;103:18054–18059. doi: 10.1073/pnas.0605389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolinder NL. Genetic Engineering of Cotton. In: Paterson AH, editor. Genetics and Genomics of Cotton, Plant Genetics and Genomics: Crops and Models 3. Springer Science+Business Media, LLC; 2009. pp. 187–207. [Google Scholar]
- Tuttle JR, Idris AM, Brown JK, Haigler CH, Robertson D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008;148:41–50. doi: 10.1104/pp.108.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Narasimhan ML, Stevenson RA, Zhu JK, Weller SC, Subbarao KV, et al. Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J. 2003;35:574–587. doi: 10.1046/j.1365-313x.2003.01830.x. [DOI] [PubMed] [Google Scholar]
- Wang JY, Cai Y, Gou JY, Mao YB, Xu YH, Jiang WH, et al. VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl Environ Microbiol. 2004;70:4989–4995. doi: 10.1128/AEM.70.8.4989-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Wheeler TA, Woodward JE. The effects of Verticillium wilt on cotton cultivars. Beltwide Cotton Conferences; Jan. 4–7; New Orleans, Louisiana. 2010. pp. 314–324. [Google Scholar]
- Yang C, Guo WZ, Li GY, Gao F, Lin SS, Zhang TZ. QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 2008;174:290–298. [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Zhang HB, Li Y, Wang B, Chee PW. Recent advances in cotton genomics. Int J Plant Genomics. 2008a;2008 doi: 10.1155/2008/742304. 742304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Cheng C, Gao Q, Liu J, Guo X. Molecular cloning and characterization of GhNPR1, a gene implicated in pathogen responses from cotton (Gossypium hirsutum L.) Biosci Rep. 2008b;28:7–14. doi: 10.1042/BSR20070028. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.