Abstract
Previously, we demonstrated that skeletal mass, structure and biomechanical properties vary considerably among 11 different inbred rat strains. Subsequently, we performed quantitative trait loci (QTL) analysis in 4 inbred rat strains (F344, LEW, COP and DA) for different bone phenotypes and identified several candidate genes influencing various bone traits. The standard approach to narrowing QTL intervals down to a few candidate genes typically employs the generation of congenic lines, which is time consuming and often not successful. A potential alternative approach is to use a highly genetically informative animal model resource capable of delivering very high-resolution gene mapping such as Heterogeneous stock (HS) rat. HS rat was derived from eight inbred progenitors: ACI/N, BN/SsN, BUF/N, F344/N, M520/N, MR/N, WKY/N and WN/N. The genetic recombination pattern generated across 50 generations in these rats has been shown to deliver ultra-high even gene-level resolution for complex genetic studies. The purpose of this study is to investigate the usefulness of the HS rat model for fine mapping and identification of genes underlying bone fragility phenotypes. We compared bone geometry, density and strength phenotypes at multiple skeletal sites in HS rats with those obtained from 5 of the 8 progenitor inbred strains. In addition, we estimated the heritability for different bone phenotypes in these rats and employed principal component analysis to explore relationships among bone phenotypes in the HS rats. Our study demonstrates that significant variability exists for different skeletal phenotypes in HS rats compared with their inbred progenitors. In addition, we estimated high heritability for several bone phenotypes and biologically interpretable factors explaining significant overall variability, suggesting that the HS rat model could be a unique genetic resource for rapid and efficient discovery of the genetic determinants of bone fragility.
Keywords: Heterogeneous stock rat, Bone density, Bone strength, Osteoporosis, Genetics
Introduction
Osteoporosis is a common bone disease leading to increased susceptibility to fracture at multiple skeletal sites [1]. Bone mineral density, structure and strength are the major determinants for skeletal fracture [2-4]. Several studies demonstrated that these phenotypes are highly heritable [5-8]. Identification of genes underlying these phenotypes, particularly at the most common skeletal fracture sites, will provide valuable insights for understanding the genetics of osteoporosis and fracture risk.
Animal models have been widely used as a means to aid in the identification of genes contributing to complex human traits, such as osteoporosis and other bone-related phenotypes [7, 9-14]. Although the mouse is the most commonly used animal model for genetic studies, the rat offers several distinct advantages over mice due to their larger bone size, which enables the study of skeletal phenotypes at the hip and spine [18-22]. In addition, previous studies showed that the rat is a highly predictive model of skeletal fracture in human [15,16]. Furthermore, the existence of a large number of phenotypic databases along with the rapid growth of rat genomic resources enable researchers to use the rat as a crucial animal model for understanding the genetics of osteoporosis and other bone-related phenotypes.
Previously, we identified several inbred rat strains that are similar in body weight but vary considerably for different bone parameters, suggesting that these inbred rat models could provide valuable insights regarding the genetics of osteoporosis and fracture risk [17]. Subsequently, using four of these inbred strains we have discovered quantitative trait loci (QTLs) for different bone phenotypes [18-22] and identified candidate genes influencing these phenotypes [23-25]. However, these QTLs encompass broad chromosomal regions harboring hundreds of potential candidate genes. To narrow these critical QTL regions to a small chromosomal segment containing a few genes several alternative approaches such as the development of recombinant inbred and congenic lines have been attempted; however, these approaches have proven to be time-consuming and labor intensive and often they do not have enough resolution to detect the causal genes and variants.
The heterogeneous stock (HS) rat, a unique rat model, was developed by the National Institutes of Health (NIH) in 1984 [26]. These rats were originally derived from eight inbred founder strains: Agouti (ACI/N), Brown Norway (BN/SsN), Buffalo (BUF/N), Fischer 344 (F344/N), M520/N, Maudsley Reactive (MR/N), Wistar-Kyoto (WKY/N) and Wistar-Nettleship (WN/N) (Figure 1). Subsequently, these heterogeneous stock were bred for 50 generations using a rotational outbreeding regime to minimize the extent of inbreeding, drift and fixation [27]. Importantly, each of these HS rats represents a unique, genetically random mosaic of founding animal chromosomes due to recombinantions that have accumulated over many generations. It has been estimated that the average distance between recombination events in these rats is approximately 2 cM [28]. Thus, the HS rat is a unique genetic resource of animals for the fine mapping of QTLs to very small genomic regions. Recently, these rats have been successfully used to fine map QTLs for diabetes [29] and fear-related behavior phenotypes [30]. Whether HS rats could also provide the quality data for bone phenotypes for complex trait like osteoporosis remains to be determined.
Fig. 1.
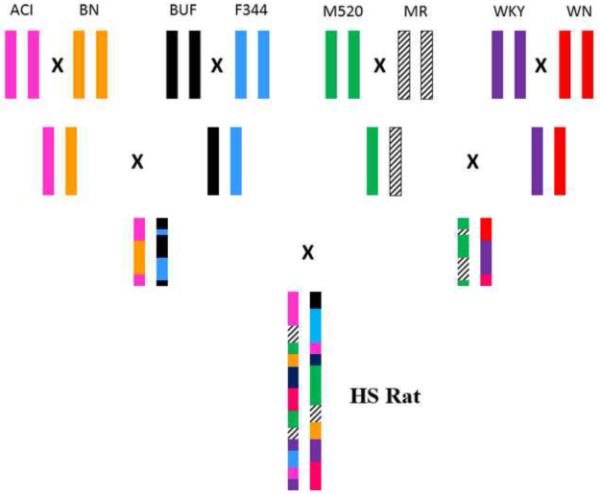
Schematic diagram of development of heterogeneous stock (HS) rats from 8 inbred strains of rats: Agouti (ACI/N), Brown Norway (BN/SsN), Buffalo (BUF/N), Fischer 344 (F344/N), M520/N, Maudsley Reactive (MR/N), Wistar-Kyoto (WKY/N) and Wistar-Nettleship (WN/N).
The purpose of this study is to investigate the usefulness of the HS rat model for potential fine mapping and identification of genes underlying bone fragility phenotypes. We hypothesize that HS rats possess considerable genetic variability and unique segregation pattern for bone geometry, density and strength phenotypes. We also hypothesize that several key bone phenotypes in these rats will have high heritability, making them an excellent model for genetic mapping studies for skeletal fracture.
Materials and Methods
Animals
We used a total of 667 HS rats (male n=319; female n=348) in this study. The HS rats were bred and grown at the Autonomous University of Barcelona. Microchips were implanted in these rats for proper identification and multiple phenotypes were obtained in the same animals at different time points to characterize primarily rat physiology and behavior. The rats were housed in cages in pairs (male) and trios (female) and maintained with food and water available ad libitum, under conditions of controlled temperature and a 12-h light-dark cycle. The HS rats were raised over 2.5 years in batches of approximately 250 animals per batch in accordance with the Spanish legislation on “Protection of Animals used for Experimental and Other Scientific Purposes” and the European Communities Council Directive (86/609/EEC).
Additionally, we used ACI, BN, BUF, F344 and WKY inbred female rats (n=6-7) in this study. The inbred rats were obtained from Harlan (Indianapolis, USA), kept at Indiana University and provided standard rat chow and water ad libitum under conditions of controlled temperature and a 12-h light-dark cycle as described previously [17]. The procedures performed throughout the experiment for these HS and inbred rats followed the guidelines of the Indiana University Animal Care and Use committee (IACUC).
Euthanasia and specimen collection
Inbred rats were euthanized at 20 weeks of age by cervical dislocation and HS rats were euthanized between 19 and 20 weeks of age by ether inhalation. The lower limbs and lumbar vertebrae (L1-6) were dissected out from these animals. The lower limbs on the right side were immediately stored at −20°C for subsequent biomechanical testing. The lower limbs on the left side were stripped of muscle, transferred to 70% ethyl alcohol and stored at 4°C for densitometry analyses.
Femur Geometry
Femoral length was measured from medial condyle to the femoral head. In addition, width of the femoral head, width of the femoral neck and axis length of the femoral neck measurements were made as described previously [17]. All of these femoral geometric measurements were performed using digital calipers accurate to 0.005 mm (Mitutoyo, Aurora, IL).
Dual energy X-ray absorptiometry (DXA)
The left femur and lumbar vertebrae 3-5 (L3-5) of inbred rats were scanned using DXA (Hologic QDR 1000/W; Hologic, Inc., MA, USA) with high-resolution mode (0.70-mm beam collimator and 0.25-mm step size). The same bones of HS rats were analyzed using DXA (PIXImus II mouse densitometer; Lunar Corp., Madison, WI, USA) with ultra-high resolution (0.18 × 0.18 mm/pixel). During scanning dissected bones were positioned on a platform supplied by manufacturer. After completion of scan of each bone mutually exclusive region of interest (ROI) boxes were drawn around the bone from which aBMD and BMC measurements were obtained.
Biomechanical testing
The frozen right femurs were brought to room temperature slowly in a saline bath. The femurs were tested in three-point bending by positioning them on the lower supports of a three-point bending fixture and applying load at the midpoint using a material testing machine (Alliance RT/5, MTS Systems Corp., Eden Prairie, MN, USA). The bones were held in place by small (1N) preload and then loaded in monotonic axial compression until fracture, at a crosshead speed of 20 mm/min. Load was applied midway between two supports that were 15 mm apart. After the long bones were fractured, cortical thickness was measured at the midshaft and 5 mm distal and proximal to the midshaft using digital calipers accurate to 0.01 mm, with a precision of + 0.005 mm (Mitutoyo, Aurora, IL). For femoral neck, the proximal half of each femur was mounted vertically in a special chuck that clamped the femoral shaft to the lower platen of the same materials testing machine. Load was applied downwards onto the femoral head at a crosshead speed of 20 mm/min until the femoral neck fractured. Force and displacement measurements were collected every 0.05 second. From the force vs. displacement curves, ultimate force (Fu; in N) and work to failure (W; in mJ) were calculated in TestWorks software, version 4.06. Fu reflects the strength of the bone or the maximum load that the bone can support before failing and W reflects the total energy the specimen can absorb before fracture.
Correlation analysis between different bone phenotypes
Pearson correlation coefficients was performed to obtain bone-specific (femur vs lumbar), phenotype-specific (geometry vs density; density vs biomechanics; geometry vs biomechanics), and site-specific (femur vs femoral neck; spine vs femoral neck) correlations from male and female HS rats. All statistical analyses were performed using the statistical package StatView (Abacus Concepts, Berkley, CA).
Statistics
Body weight was a significant predictor for all phenotypes; therefore, weight was used as a covariate for all statistical analysis. All results were expressed as mean ± standard deviation (SD). To detect significant differences for bone phenotypes among all rat strains, one-way analysis of variance (ANOVA) was performed, followed by Fischer‟s protected least-significance differences between strains. The level of significance was set at 0.05 or less for all phenotypes. Pearson correlation coefficients (r) were generated from linear regression analyses in HS rats to identify the underlying phenotypic relationships among all variables (bone geometry, mineral density and biomechanics).
Heritability estimation
Residuals of each skeletal phenotypic measure were computed with body weight as a covariate to obtain weight-adjusted values for the female rats. Several of the bone phenotypes (femur axis length, femur ultimate force and lumbar aBMD) demonstrated a mean difference across batches and the batch effect was adjusted as well for these phenotypes. The environmental variance component (Ve) was estimated as the error variance in the phenotypic residual among progenitors after covarying for weight and strain. The phenotypic variance component (Vp) was estimated as variance in female HS rats after covarying for weight (and batch as necessary, as indicated above). Heritability in the female rats was then calculated as 1-(Ve/Vp) [31].
Principal component analysis (PCA)
All phenotypic variables from the female HS rats were entered into a principal component analysis (PCA) to identify novel multivariate phenotypes for pleiotropic genetic effects using SAS version 9.1 (SAS Institute, Cary, NC, USA). Briefly, the first principal component (PC1) was derived from the correlation structure of the body weight and 12 bone phenotypes (Table 2) as a simple linear combination of the measured phenotypic values, explaining the maximum possible amount of common phenotypic variation among the phenotypes. Subsequent uncorrelated components (PC2, PC3 and PC4) were generated in a similar way, each explaining the maximum possible amount of the remaining variation among the phenotypes after consideration of previous components.
Table 2.
Heritability (H2) and principal component analysis (PCA) of body weight and bone phenotypes in female HS rats*
| Phenotype | H2 | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|---|
| Percent variation explained | 46.3 | 10.6 | 8.5 | 8 | |
| Body weight | 0.67 | 0.32 | −0.14 | −0.03 | −0.24 |
| Femur length | 0.82 | 0.31 | −0.28 | 0.17 | −0.02 |
| Head width | 0.90 | 0.24 | −0.10 | 0.59 | 0.16 |
| Neck width | 0.64 | 0.18 | 0.24 | 0.54 | 0.23 |
| Axis length | 0.90 | 0.15 | −0.16 | 0.36 | −0.25 |
| Femur aBMD | 0.71 | 0.34 | 0.04 | −0.20 | −0.10 |
| Femur BMC | 0.75 | 0.39 | −0.05 | −0.11 | −0.07 |
| Lumbar aBMD | 0.42 | 0.31 | −0.04 | −0.23 | −0.23 |
| Lumbar BMC | 0.34 | 0.36 | −0.10 | −0.13 | −0.18 |
| Femur UF | 0.69 | 0.26 | 0.07 | −0.21 | 0.53 |
| Femur W | 0.14 | 0.22 | 0.00 | −0.18 | 0.62 |
| Femur neck UF | 0.72 | 0.27 | 0.49 | −0.05 | −0.11 |
| Femur neck W | 0.67 | 0.06 | 0.74 | 0.07 | −0.17 |
H2 and PCA values > 0.3 are indicated in bold
Results
Body weight
Average body weight varied significantly among inbred progenitors and HS female rats. HS rats had significantly higher (p<0.05) mean body weight compared with BN, F344 rats and significantly higher (p<0.005) body weight compared with ACI rats (Figure 2). Among all inbred rats, body weight for BUF rats was significantly higher (p<0.05) compared with ACI rats.
Fig. 2.
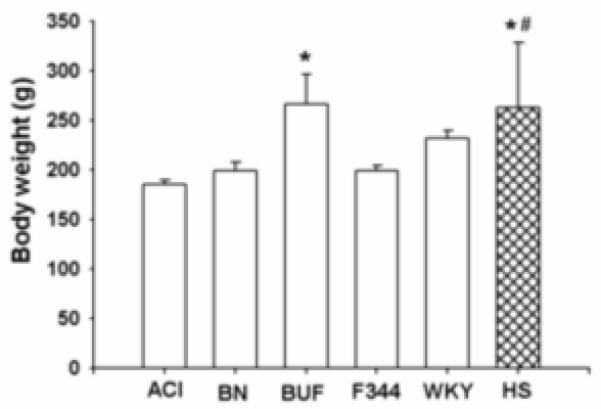
Body weight for female inbred progenitors (ACI, BN, BUF, F344 and WKY) and HS strains of rats. Data presented are mean ± SD (n=6-7 for inbred rats and n=348 for HS rats). *p<0.05 HS vs BN and F344; BUF vs ACI; #p<0.005 HS vs ACI
Femur geometry
Significant variability in femur length was detected among inbred progenitors and HS female rats. In general, longer femur length was observed in ACI, BUF and HS rats compared to BN, F344 and WKY rats (Figure 3). The shortest femur length was recorded for F344 rats.
Fig. 3.
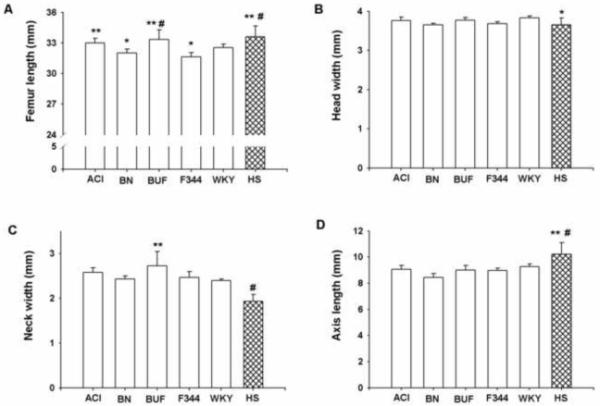
Femur length (A), head width (B), neck width (C) and axis length (D) for female inbred progenitors (ACI, BN, BUF, F344 and WKY) and HS strains of rats. Data presented are mean ± SD (n=6-7 for inbred rats and n=348 for HS rats). For femur length *p<0.05 BN vs ACI; F344 vs WKY; **p<0.005 ACI vs F344; BUF vs BN; HS vs WKY; #p<0.0001 BUF vs F344; HS vs BN and F344; head width *p<0.05 HS vs WKY; neck width *p<0.005 BUF vs BN, F344 and WKY; #p<0.0001 HS vs all others; axis length *p<0.005 HS vs all others except BN; #p<0.0001 HS vs BN
Femur length in HS rat was significantly higher (p<0.005) compared with WKY rats and significantly higher (p<0.0001) compared with F344 and BN rats. The average femur head width was not significantly different among inbred progenitors, but head width was significantly higher (p<0.05) in WKY rats compared with HS rats. HS rats also had significantly lower (p<0.001) femoral neck width compared with all progenitor inbred strains. Among all femur geometry measurements, the axis length of HS rats exceeded those from all progenitors: HS rat had significantly higher (p<0.005) axis length compared to ACI, BUF, F344 and WKY rats, and significantly higher (p<0.001) axis length compared to BN rats.
Femur and lumbar aBMD and BMC
Femur and lumbar DXA measurements varied greatly among inbred progenitors and HS female rats. Among all inbred progenitors, BUF rats had highest body weight adjusted aBMD and BMC in both femur and lumbar vertebrae compared with the other inbred progenitors (Figure 4). On the other hand, F344 rats had the lowest values for aBMD and BMC for both femur and lumbar vertebrae among all progenitors. The aBMD for both femur and lumbar was significantly lower (p<0.0001) in HS rats compared with their inbred progenitors. In contrast, femur BMC in HS rat was significantly higher (p<0.05) compared with BN and significantly higher (p<0.005) compared with WKY rats. HS rats also had significantly higher (p<0.05) values for lumbar BMC compared to WKY rats. The mean values for both femur and lumbar BMC in HS rats fell between those of the progenitor strains.
Fig. 4.
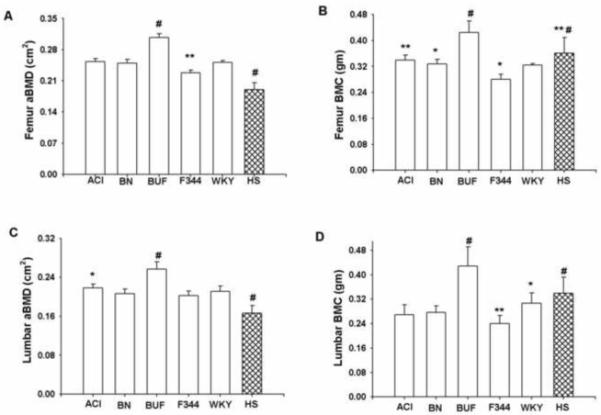
Femur aBMD (A), femur BMC (B), lumbar aBMD (C) and lumbar BMC (D) for female inbred progenitors (ACI, BN, BUF, F344 and WKY) and HS strains of rats. Data presented are mean ± SD (n=6-7 for inbred rats and n=348 for HS rats). For femur aBMD *p<0.005 F344 vs ACI; BN vs WKY; #p<0.0001 HS vs all others; BUF vs all others; femur BMC *p<0.05 BN vs F344 and HS; F344 vs WKY; **p<0.005 ACI vs F344; HS vs WKY; #p<0.0001 BUF vs all others; HS vs BUF and F344; lumbar aBMD **p<0.005 ACI vs F344; #p<0.0001 BUF vs all others; HS vs all others; lumbar BMC *p<0.05 HS vs WKY; **p<0.005 F344 vs WKY; #p<0.0001 BUF vs all others; HS vs all others except WKY
Femur and femoral neck biomechanics
The femur and femoral neck biomechanical properties showed significant variability among inbred progenitors and HS female rats. The values for body weight adjusted ultimate force (UF) and work to failure (W) at both femur and femoral neck were in the middle range in HS rats compared to the inbred progenitors (Figure 5). Among all inbred progenitors, F344 rats had the weakest femur biomechanical properties. On the other hand, BUF rats had the highest values for both femur and femoral neck UF.
Fig. 5.
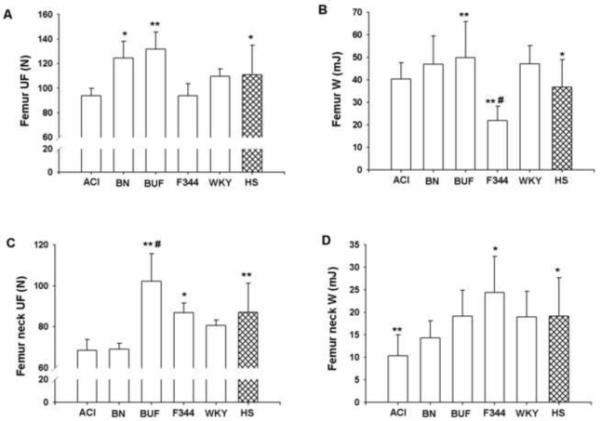
Femur UF (A), femur W (B), femur neck UF (C) and femur neck W (D) for female inbred progenitors (ACI, BN, BUF, F344 and WKY) and HS strains of rats. Data presented are mean ± SD (n=6-7 for inbred rats and n=348 for HS rats). For femur UF *p<0.05 BN vs ACI and F344; HS vs BUF; **p<0.005 BUF vs ACI and F344; femur W *p<0.05 HS vs BN and WKY; **p<0.005 F344 vs ACI and HS; BUF vs HS; #p<0.0001 F344 vs BN, BUF and WKY; femur neck UF *p<0.05 F344 vs ACI, BN and BUF; **p<0.005 BUF vs WKY and HS; HS vs ACI and BN; #p<0.0001 BUF vs ACI and BN; femur neck W *p<0.05 HS vs ACI; F344 vs BN; **p<0.005 ACI vs F344
Femur ultimate force and work to failure were significantly lower (p<0.05) in HS rats compared with BUF rats. In addition, femur work to failure was significantly lower (p<0.05) in HS rats compared with BN and WKY rats. In contrast, the value for the same phenotype was significantly higher (p<0.005) in HS rats compared with F344 rats. Femur neck UF was significantly lower (p<0.05) in ACI and BN compared to HS rats. Additionally, femoral neck W was significantly lower (p<0.05) in ACI compared to HS rats. In contrast, BUF rats had significantly higher (p<0.05) values for femur neck UF compared to HS rats.
Sex-specific differences of weight-adjusted bone phenotypes in HS rats
To identify gender-specific differences for bone phenotypes, we compared bone phenotypes between 319 male and 348 female HS rats between 19-20 weeks of age. All bone phenotypes (femur geometry, femur aBMD and BMC, lumbar aBMD and BMC, femur and femoral neck ultimate force and work to failure) were significantly lower (p<0.0001) in female rats compared to male rats after adjustment for body weight (Figure 6).
Fig. 6.
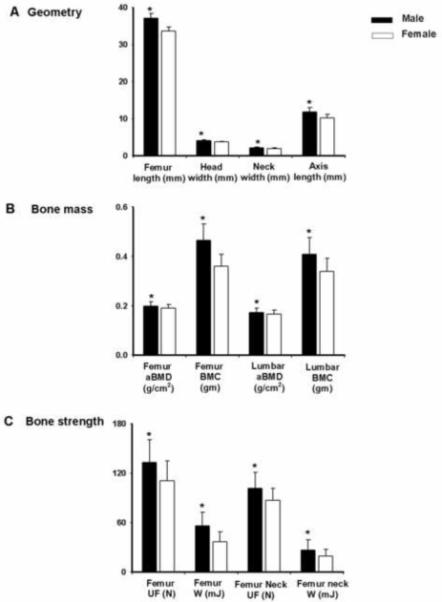
Geometry (A), bone mass (B) and bone strength (C) parameters for male and female HS strains of rats. Data presented are mean ± SD (n=319 for male and n=348 for female HS rats) *p<0.0001 between male and female
Correlation between different weight-adjusted bone phenotypes in HS rats
Correlation analysis in female HS rats showed that both aBMD and BMC were highly correlated (r=0.87 and 0.77, respectively) for femur and lumbar spine (Table 1). In addition, correlation coefficient values between femur and lumbar for aBMD and BMC were 0.68 and 0.86, respectively, suggesting high correlation for bone density and mineral content phenotypes between these two skeletal sites. Correlation analysis between femur densitometry and biomechanics also indicated high correlation between femur aBMD and ultimate force (r=0.50). The strength of correlation was good to high between femur BMC and ultimate force (r=0.56) and femur BMC and work to failure (0.48). Femur strength phenotypes, ultimate force and work to failure (UF and W), had high correlation (r=0.60) with each other. Similarly, femoral neck UF showed good correlation (r=0.49) with neck W. However, while femur aBMD and BMC had high correlation with femoral neck UF (r=0.58 and 0.60, respectively), none of these femur bone phenotypes had any significant correlation with femoral neck W (r=0.10 and 0.07, respectively).
Table 1.
Correlation (r-values) between different bone phenotypes in female and male HS rats*
| Female Phenotypes | Body weight | Femur length | Head width | Neck width | Axis length | Femur aBMD | Femur BMC | Lumbar aBMD |
Lumbar BMC | Femur UF | Femur W | Femur neck UF |
Femur neck W |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Body weight | 1.00 | ||||||||||||
| Femur length | 0.65 | 1.00 | |||||||||||
| Head width | 0.40 | 0.57 | 1.00 | ||||||||||
| Neck width | 0.21 | 0.27 | 0.46 | 1.00 | |||||||||
| Axis length | 0.29 | 0.34 | 0.27 | 0.07 | 1.00 | ||||||||
| Femur aBMD | 0.60 | 0.55 | 0.38 | 0.31 | 0.17 | 1.00 | |||||||
| Femur BMC | 0.73 | 0.73 | 0.47 | 0.34 | 0.30 | 0.87 | 1.00 | ||||||
| Lumbar aBMD | 0.56 | 0.48 | 0.30 | 0.25 | 0.22 | 0.68 | 0.73 | 1.00 | |||||
| Lumbar BMC | 0.72 | 0.68 | 0.42 | 0.29 | 0.28 | 0.77 | 0.86 | 0.77 | 1.00 | ||||
| Femur UF | 0.36 | 0.39 | 0.27 | 0.28 | 0.14 | 0.50 | 0.56 | 0.45 | 0.45 | 1.00 | |||
| Femur W | 0.31 | 0.39 | 0.29 | 0.14 | 0.15 | 0.37 | 0.48 | 0.26 | 0.41 | 0.60 | 1.00 | ||
| Femur neck UF | 0.43 | 0.33 | 0.30 | 0.32 | 0.16 | 0.58 | 0.60 | 0.45 | 0.51 | 0.36 | 0.34 | 1.00 | |
| Femur neck W | 0.06 | −0.07 | 0.01 | 0.17 | 0.06 | 0.10 | 0.07 | 0.06 | 0.04 | 0.09 | 0.04 | 0.49 | 1.00 |
| Male Phenotypes | Body weight | Femur length | Head width | Neck width | Axis length | Femur aBMD | Femur BMC | Lumbar aBMD |
Lumbar BMC | Femur UF | Femur W | Femur neck UF |
Femur neck W |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Body weight | 1.00 | ||||||||||||
| Femur length | 0.63 | 1.00 | |||||||||||
| Neck width | 0.28 | 0.50 | 1.00 | ||||||||||
| Neck width | 0.14 | 0.20 | 0.28 | 1.00 | |||||||||
| Axis length | 0.31 | 0.29 | 0.17 | 0.09 | 1.00 | ||||||||
| Femur aBMD | 0.65 | 0.56 | 0.31 | 0.17 | 0.18 | 1.00 | |||||||
| Femur BMC | 0.78 | 0.77 | 0.41 | 0.23 | 0.30 | 0.86 | 1.00 | ||||||
| Lumbar aBMD | 0.59 | 0.49 | 0.21 | 0.12 | 0.20 | 0.61 | 0.71 | 1.00 | |||||
| Lumbar BMC | 0.70 | 0.64 | 0.36 | 0.14 | 0.23 | 0.74 | 0.84 | 0.73 | 1.00 | ||||
| Femur UF | 0.44 | 0.44 | 0.23 | 0.15 | 0.14 | 0.50 | 0.58 | 0.44 | 0.48 | 1.00 | |||
| Femur W | 0.29 | 0.42 | 0.29 | 0.13 | 0.13 | 0.33 | 0.44 | 0.28 | 0.36 | 0.62 | 1.00 | ||
| Femur neck UF | 0.54 | 0.38 | 0.23 | 0.18 | 0.17 | 0.61 | 0.66 | 0.52 | 0.57 | 0.40 | 0.30 | 1.00 | |
| Femur neck W | 0.26 | 0.15 | 0.15 | 0.07 | 0.13 | 0.19 | 0.27 | 0.25 | 0.21 | 0.19 | 0.16 | 0.54 | 1.00 |
r-values equal or above 0.5 are indicated in bold
We also analyzed how femoral geometry and strength parameters are correlated to each other in female HS rats. The length of the femur was not strongly correlated to either femur UF (r= 0.39) or femur W (r=0.39), suggesting that this measure of femur macro-architecture is not a significant predictor for femur strength. Similarly, femoral neck geometry had low to no correlation with neck biomechanics (r<0.32 for all). Similar phenotypic correlation was also observed in male HS rats (Table 1).
Heritability
Heritability of various bone phenotypes in female rats revealed that bone geometry, density and strength had high heritability based on the comparison of HS variability with that of progenitor strains. Because high heritability of body weight as well as strong correlation of body weight and bone phenotypes in these rats, we adjusted the bone phenotypes for weight prior to heritability calculation to best capture weight-independent genetic effects. Heritability of adjusted bone geometry phenotypes ranged from 64% to 90% (Table 2). Femur density and ultimate force as well as femoral neck strength (ultimate force and work to failure) also had high heritability (67% to 75%). On the other hand, lumbar density and mineral content showed lower heritability, 42% and 34%, respectively.
Pleiotropy in HS rats
To identify novel phenotypes and possible pleiotropic genetic effects, principal component analysis (PCA) was performed in female HS rats. We included body weight in PCA with non-weight-corrected variables to identify how weight loaded with different bone phenotypes in the analysis. The first PC (PC1) explained about 46% of the common phenotypic variation and was most heavily weighted for body weight, femur and lumbar density and mineral content measures, suggesting that body weight and overall skeletal mass at both femur and spine have significant common influence (Table 2). PC2 explained a smaller proportion (11%) of the common phenotypic variation and was primarily weighted for femur neck biomechanical measures (femur neck ultimate force and work to failure). PC3 explained about 9% of the common phenotypic variation and was strongly weighted for femur geometry measures (head width, neck width and axis length), while PC4 accounted for 8% of overall variability and was influenced primarily by biomechanical measures at the mid-femur (femur ultimate force and work to failure) (Table 2).
Discussion
Our results clearly demonstrate that substantial variability for bone geometry, density and strength phenotypes at femur, hip and spine exist in HS rats, representing the bone phenotypic variability in their inbred progenitors. In addition, we observed strong heritability for several of these skeletal phenotypes, suggesting that the HS rat will be a unique genetic resource for dissecting the complex genetics underlying bone fragility.
In this study, we observed some interesting variability for bone phenotypes in HS rats compared with their inbred progenitors. While most of the bone phenotypes in HS rats were intermediate compared with the inbred progenitors, the mean of several bone phenotypes in HS rats exceeded those from the inbred rats. For example, the mean axis length was significantly higher whereas the mean femoral neck width, femur aBMD and lumbar aBMD were significantly lower in HS rats compared with all progenitor inbred strains, suggesting that these phenotypes in HS rats might have been influenced by the combined effect and interaction of different alleles from the progenitor rats.
Compared with HS rat, BN and WKY rats had shorter femur length and lower femur BMC but femur biomechanical parameters exhibit equal or superior properties, suggesting that BN and WKY rats might have high bone quality in the femur compared with HS rats. In the HS, femur neck width was shorter and axis length was longer compared with BUF and F344 rats, which represent as lower femoral neck bone strength in HS rats, compared with these progenitor strains. Interestingly, with similar femur neck geometry (shorter neck width and longer axis length) in HS rats compared with ACI and BN rats, femoral neck strength in HS rats was either equal or superior, suggesting better microarchitecture (trabecular orientation, cortical thickness) or intrinsic tissue properties (ultimate stress, toughness) at femoral neck in HS rats compared with these inbred progenitors rats.
Among all inbred progenitors BUF rats had the highest aBMD and BMC for both femur and lumbar vertebrae. These rats also had larger body size, femoral length and higher bone strength for femur and femoral neck. As body size might be related to these observed bone phenotypes, we analyzed these phenotypes using body size as a covariate. However, even after adjustment for body weight, the skeletal phenotypic differences persist, indicating that genetics might play a potential role for this robust bone mass in BUF rats. In contrast, F344 rats had the lowest mean values for aBMD and BMC both for femur and lumbar vertebrae as well as femur length among all progenitors, which is reflected by their lower femoral bone strength, indicating that F344 rats possess genes for overall increased fragility.
Correlation of different bone phenotypes in HS analysis showed that density and mineral content had strong correlation (r=0.81-0.86) within a bone (femur or lumbar spine), but this correlation becomes modest (r= 0.43-0.52) between different bones such as femur versus lumbar spine for both of these phenotypes. These data suggest that, besides common genes, there are distinct sets of genes affecting different traits in a site-specific manner. Also the correlation between density/mineral content and strength phenotypes within a bone is strong (r=0.45-0.64). This correlation becomes low to good (r=0.21-0.56) if density/mineral content from whole bone is correlated with a specific part of the bone (femur versus femoral neck), suggesting site-specific genetic regulation of bone phenotypes. In addition, low to no correlation (r=0.37-0.02) of both femur and femoral neck geometry with femur and femoral neck strength phenotypes, respectively, indicates that distinct sets of genes are influencing these phenotypes.
The results from principal component analysis showed that the bulk (73%) of the overall phenotypic variability could be explained by the first four principal components. The weighting of the first principal component on the phenotypes suggest that HS rats possess common genes underlying variation in overall body size and skeletal mass and strength. This principal component also indicates strong evidence of pleiotropy among major bone phenotypic groups (geometry, density and strength) in HS rats. Interestingly, factors uniquely influencing femur neck and femur strength phenotypes were discovered from the second and fourth components, respectively, while geometry measures were particularly strongly weighted on the third component.
In humans, bone geometry, density and strength at the site of high incidence of osteoporotic fracture such as hip and spine have shown to be highly heritable [7]. In our previous studies, we found high heritability for these bone phenotypes in the F2 rats derived from both F344 X LEW and COP X DA crosses [18-22]. In this study, HS rats showed significant heritability for femur and femoral neck geometry, femur and spine bone density and strength (Table 2), suggesting that genetic analysis for these skeletal phenotypes in HS and inbred rats will be very useful for identification of genes that confer susceptibility to osteoporotic fracture. In addition, the strength of these heritability values might be considered as important determinants to prioritize bone phenotypes for subsequent analyses in this rat model.
There are several limitations in the current study. We could not analyze skeletal phenotypes in MR/N, M520 and WN/N progenitor strains due to unavailability of these inbred rats from commercial sources. However, both MR/N and WN/N rats were derived from Wistar colony and since our analysis of 5 other inbred strains included WKY, which is also a Wistar-derived rat, we anticipate that the phenotypic distribution in MR/N and WN/N rats will be similar to WKY rats. In addition, we could not compare male bone phenotypes among inbred progenitors and HS rats because our previous inbred rat data were obtained from female only. As sex-specific QTL regulation has been demonstrated to be important in multiple studies [22,23,32,33], we need to extend our analyses to include male for further investigations in the future.
We did not compare cortical versus cancellous bone phenotypes between HS rats and their progenitors. However, we have obtained these phenotypes in our HS rats for genetic analysis in future. In addition, further analysis involving bone growth, modeling and bone turnover will be necessary as all of these phenotypes might have a genetic basis for bone fragility. Although many of the measures of BMD and bone structure are independent of bone growth or length, young growing animals in a rapid bone acquisition period will be valuable for the identification of the role of bone growth on peak bone mass gain in the adult animal. In addition, measurement of serum and urine biomarkers as well as histomorphometric measurements of bone will provide valuable insights into the process of normal and abnormal bone modeling, remodeling and bone turnover both at cortical and trabecular settings. We are currently genotyping the HS rats for single nucleotide polymorphisms (SNPs) distributed throughout the genome. Our goal is to identify QTLs contributing to different bone phenotypes in these rats within very narrow chromosomal regions, which will greatly facilitate the identification of causative genes for osteoporosis. Moreover, we plan to evaluate these genes in human populations in future.
In conclusion, we found significant variability for femur and lumbar density, femur geometry as well as femur and femoral neck strength phenotypes in HS rats compared with their inbred progenitors. In addition, we identified a strong genetic component for several of these bone phenotypes. Thus, the HS rat model will provide us a unique resource to dissect the complex genetic relationships for different skeletal phenotypes underlying bone fragility.
Research Highlights.
-
➢
We compared bone phenotypes between HS and inbred progenitor rats.
-
➢
We estimated heritability for bone phenotypes in HS rats and progenitor strains.
-
➢
Significant variability for multiple bone phenotypes exist in HS rats.
-
➢
HS rats showed high heritability for several bone phenotypes.
-
➢
HS rat model is a unique resource for the genetic determinants of bone fragility.
Acknowledgments
This work was supported by the US National Institutes of Health through the following grants: AR047822 and AG018397. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement N° HEALTH-F4-2010-241504 (EURATRANS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: All authors have no conflicts of interest.
References
- [1].Kanis JA, Melton LJ, Christiansen C, Johnston CC, Khaltaev N. The diagnosis of osteoporosis. J Bone Miner Res. 1994;9:1137–41. doi: 10.1002/jbmr.5650090802. [DOI] [PubMed] [Google Scholar]
- [2].Cummings SR, Black DM, Nevitt MC, Browner W, Cauley J, Ensrud K, Genant HK, Palermo L, Scott J, Vogt TM, The Study of Osteoporotic Fractures Research Group Bone density at various sites for prediction of hip fractures. Lancet. 1993;341:72–75. doi: 10.1016/0140-6736(93)92555-8. [DOI] [PubMed] [Google Scholar]
- [3].Faulkner KG, Cummings SR, Black D, Palermo L, Gluer CC, Genant HK. Simple measurement of femoral geometry predicts hip fracture: the study of osteoporotic fractures. J Bone Miner Res. 1993;8:1211–17. doi: 10.1002/jbmr.5650081008. [DOI] [PubMed] [Google Scholar]
- [4].Peacock M, Turner CH, Liu G, Manatunga AK, Timmerman L, Johnston CC., Jr. Better discrimination of hip fracture using bone density, geometry and architecture. Osteoporos Int. 1995;5:167–73. doi: 10.1007/BF02106096. [DOI] [PubMed] [Google Scholar]
- [5].Arden NK, Baker J, Hogg C, Bann K, Spector TD. The heritability of bone mineral density, ultrasound of the calcaneus and hip axis length: a study of postmenopausal twins. J Bone Miner Res. 1996;11:530–34. doi: 10.1002/jbmr.5650110414. [DOI] [PubMed] [Google Scholar]
- [6].Garnero P, Arden NK, Griffiths G, Delmas PD, Spector TD. Genetic influence on bone turnover in postmenopausal twins. J Clin Endocrinol Metab. 1996;81:140–46. doi: 10.1210/jcem.81.1.8550741. [DOI] [PubMed] [Google Scholar]
- [7].Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:378–83. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- [8].Ralston SH. Genetic determinants of osteoporosis. Curr Opin Rheumatol. 2005;17:475–79. doi: 10.1097/01.bor.0000166385.62851.92. [DOI] [PubMed] [Google Scholar]
- [9].Klein RF. Genetics of osteoporosis--utility of mouse models. J Musculoskelet Neuronal Interact. 2008;8:287–90. [PubMed] [Google Scholar]
- [10].Liu YJ, Shen H, Xiao P, Xiong DH, Li LH, Recker RR, Deng HW. Molecular genetic studies of gene identification for osteoporosis: a 2004 update. J Bone Miner Res. 2006;21:1511–35. doi: 10.1359/JBMR.051002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zmuda JM, Sheu YT, Moffett SP. The search for human osteoporosis genes. J Musculoskelet Neuronal Interact. 2006;6:3–15. [PubMed] [Google Scholar]
- [12].Lelovas PP, Xanthos TT, Thoma SE, Lyritis GP, Dontas IA. The laboratory rat as an animal model for osteoporosis research. Comp Med. 2008;58:424–30. [PMC free article] [PubMed] [Google Scholar]
- [13].Xiong Q, Han C, Beamer WG, Gu W. A close examination of genes within quantitative trait loci of bone mineral density in whole mouse genome. Crit Rev Eukaryot Gene Expr. 2008;18:323–43. doi: 10.1615/critreveukargeneexpr.v18.i4.20. [DOI] [PubMed] [Google Scholar]
- [14].Rosen CJ, Beamer WG, Donahue LR. Defining the genetics of osteoporosis: using the mouse to understand man. Osteoporos Int. 2001;12:803–10. doi: 10.1007/s001980170030. [DOI] [PubMed] [Google Scholar]
- [15].Wronski TJ, Walsh CC, Ignaszewski LA. Histologic evidence for osteopenia and increased bone turnover in ovariectomized rats. Bone. 1986;7:119–23. doi: 10.1016/8756-3282(86)90683-6. [DOI] [PubMed] [Google Scholar]
- [16].Bonjour JP, Ammann P, Rizzoli R. Importance of preclinical studies in the development of drugs for treatment of osteoporosis: a review related to the 1998 WHO guidelines. Osteoporos Int. 1999;9:379–93. doi: 10.1007/s001980050161. [DOI] [PubMed] [Google Scholar]
- [17].Turner CH, Roeder RK, Wieczorek A, Foroud T, Liu G, Peacock M. Variability in skeletal mass, structure, and biomechanical properties among inbred strains of rats. J Bone Miner Res. 2001;16:1532–39. doi: 10.1359/jbmr.2001.16.8.1532. [DOI] [PubMed] [Google Scholar]
- [18].Alam I, Sun Q, Liu L, Koller DL, Carr LG, Econs MJ, Foroud T, Turner CH. Sex-specific genetic loci for femoral neck bone mass and strength identified in inbred COP and DA rats. J Bone Miner Res. 2008;23:850–59. doi: 10.1359/JBMR.080221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sun Q, Alam I, Liu L, Koller DL, Carr LG, Econs MJ, Foroud T, Turner CH. Genetic loci affecting bone structure and strength in inbred COP and DA rats. Bone. 2008;42:547–53. doi: 10.1016/j.bone.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alam I, Sun Q, Liu L, Koller DL, Fishburn T, Carr LG, Econs MJ, Foroud T, Turner CH. Identification of a quantitative trait locus on rat chromosome 4 that is strongly linked to femoral neck structure and strength. Bone. 2006;39:93–99. doi: 10.1016/j.bone.2005.12.009. [DOI] [PubMed] [Google Scholar]
- [21].Alam I, Sun Q, Liu L, Koller DL, Fishburn T, Carr LG, Econs MJ, Foroud T, Turner CH. Whole-genome scan for linkage to bone strength and structure in inbred Fischer 344 and Lewis rats. J Bone Miner Res. 2005;20:1589–96. doi: 10.1359/JBMR.050512. [DOI] [PubMed] [Google Scholar]
- [22].Koller DL, Alam I, Sun Q, Liu L, Fishburn T, Carr LG, Econs MJ, Foroud T, Turner CH. Genome screen for bone mineral density phenotypes in Fischer 344 and Lewis rats. Mammalian Genome. 2005;16:578–86. doi: 10.1007/s00335-004-2459-0. [DOI] [PubMed] [Google Scholar]
- [23].Alam I, Sun Q, Koller DL, Liu L, Liu Y, Edenberg HJ, Foroud T, Turner CH. Genes influencing spinal bone mineral density in inbred F344, LEW, COP and DA rats. Functional and Integrative Genomics. 2009;10:63–72. doi: 10.1007/s10142-009-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alam I, Sun Q, Koller DL, Liu L, Liu Y, Edenberg HJ, Li J, Foroud T, Turner CH. Differentially expressed genes strongly correlated with femur strength in rats. Genomics. 2009;94:257–62. doi: 10.1016/j.ygeno.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alam I, Sun Q, Liu L, Koller DL, Liu Y, Edenberg HJ, Econs MJ, Foroud T, Turner CH. Genomic expression analysis of rat chromosome 4 for skeletal traits at femoral neck. Physiological Genomics. 2008;35:191–96. doi: 10.1152/physiolgenomics.90237.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res. 1984;8:477–9. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- [27].Boucher W, Cotterman CW. On the classification of regular systems of inbreeding. J Math Biol. 1990;28:293–305. doi: 10.1007/BF00178778. [DOI] [PubMed] [Google Scholar]
- [28].Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97:12649–54. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Solberg Woods LC, Holl K, Tschannen M, Valdar W. Fine-mapping a locus for glucose tolerance using heterogeneous stock rats. Physiol Genomics. 2010;41:102–8. doi: 10.1152/physiolgenomics.00178.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Johannesson M, Lopez-Aumatell R, Stridh P, Diez M, Tuncel J, Blázquez G, Martinez-Membrives E, Cañete T, Vicens-Costa E, Graham D, Copley RR, Hernandez-Pliego P, Beyeen AD, Ockinger J, Fernández-Santamaría C, Gulko PS, Brenner M, Tobeña A, Guitart-Masip M, Giménez-Llort L, Dominiczak A, Holmdahl R, Gauguier D, Olsson T, Mott R, Valdar W, Redei EE, Fernández-Teruel A, Flint J. A resource for the simultaneous high-resolution mapping of multiple quantitative trait loci in rats: the NIH heterogeneous stock. Genome Res. 2009;19:150–8. doi: 10.1101/gr.081497.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Falconer DS, Mackay TFC. Introduction to Quantita tive Genetics. Ed 4 Longmans Green, Harlow; Essex, UK: 1996. [Google Scholar]
- [32].Peacock M, Koller DL, Lai D, Hui S, Foroud T, Econs MJ. Bone mineral density variation in men is influenced by sex-specific and non sex-specific quantitative trait loci. Bone. 2009;45:443–8. doi: 10.1016/j.bone.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peacock M, Buckwalter KA, Persohn S, Hangartner TN, Econs MJ, Hui S. Race and sex differences in bone mineral density and geometry at the femur. Bone. 2009;45:218–25. doi: 10.1016/j.bone.2009.04.236. [DOI] [PMC free article] [PubMed] [Google Scholar]


