Abstract
Gene regulation involves many different types of transcription control mechanisms, including mechanisms based on reiterative transcription in which nucleotides are repetitively added to the 3′ end of a nascent transcript due to upstream transcript slippage. In these mechanisms, reiterative transcription is typically modulated by interactions between RNA polymerase and its nucleoside triphosphate substrates without the involvement of regulatory proteins. This review describes the current state of knowledge of gene regulation involving reiterative transcription. It focuses on the methods by which reiterative transcription is controlled and emphasizes the different fates of transcripts produced by this reaction. The review also includes a discussion of possible new and fundamentally different mechanisms of gene regulation that rely on conditional reiterative transcription.
Introduction
Reiterative transcription (also known as RNA polymerase stuttering, transcript slippage, and pseudo-templated transcription) is a reaction catalyzed by bacterial, viral, and eukaryotic RNA polymerases in which nucleotides are repetitively added to the 3′ end of a nascent transcript due to slippage between the transcript and the DNA or viral RNA template. [1–4••]. Typically, slippage occurs between a homopolymeric sequence in the transcript and at least three complementary bases in the template [5,6]. The mechanism apparently involves one or more rounds of a one-base upstream shift of the transcript so that the same nucleotide in the template specifies multiple residues in the transcript [7,8]. It has also been shown that reiterative transcription can occur within dinucleotide [9••,10] and trinucleotide [11] repeat sequences in the template, apparently via two-base and three-base upstream shifts of the transcript, respectively. Reiterative transcription can occur during initiation, elongation, or termination and result in transcripts that are immediately released from the transcription complex [12,13••] or are extended by normal elongation after a switch to nonreiterative nucleotide addition [14,15••]. Although reiterative transcription can involve the addition of any nucleotide (at least under certain conditions), addition of U or A residues appears to occur most frequently. This preference presumably reflects a requirement in the reaction for disruption of the RNA-DNA hybrid within the transcription complex [16••,17••], which would be facilitated by relatively weak U:A or A:T base pairing. During transcription elongation, disruption of an 8- to 9-bp RNA-DNA hybrid is required [18,19]. Additionally, during elongation, transcript slippage can also occur in the downstream direction resulting in deletion of nucleotides [20,21•], a process that will not be discussed here.
Although reiterative transcription was first observed during the earliest biochemical characterizations of Escherichia coli RNA polymerase in the 1960s [22], it was regarded as an oddity and even an artifact for many years [1]. This perception changed when it was demonstrated that reiterative transcription plays a key role in gene expression, especially in viral systems. A classic example is programmed transcript slippage within the phosphoprotein gene of paramyxoviruses, which results in the addition of one to six extra (i.e., non-template encoded) G residues in the mRNA. These additions cause frameshifts that direct the synthesis in proper proportion of up to three different viral proteins with a common amino-terminal sequence [23]. Other interesting but distinct examples include the addition of a non-template encoded poly(A) tract at the 5′ end of vaccinia virus mRNAs, which appears to be required for translation of these transcripts [24], and polymerase slippage at vesicular stomatitis virus gene junctions to generate long 3′ poly(A) tails during transcript termination [12]. Related examples, particularly those in which reiterative transcription during elongation is used to express alternative open reading frames, have been observed in evolutionarily divergent bacteria, indicating the widespread nature of such mechanisms [7,14]. In some cases, these bacterial mechanisms are used to suppress expression of wild-type genes or to rescue the expression of genes containing frameshift mutations, including genes annotated as pseudogenes [7,25,26••]. Furthermore, reiterative transcription during elongation has been shown to affect expression of genes involved in human disease [2,27]. An excellent review describing a number of the examples listed above was recently published [16••].
In each of the preceding examples of reiterative transcription, the extent of repetitive nucleotide addition is effectively established or programmed by the sequence of the nucleic acid template. However, the extent of reiterative transcription can also be modulated over a wide range by metabolic factors, and the level of repetitive nucleotide addition can dramatically affect the expression of the resulting transcripts. This type of variable reiterative transcription can be used as a central element in gene regulation, and this review will focus on these regulatory mechanisms. At present, the known examples of this type of regulation occur in bacteria and employ metabolically sensitive reiterative transcription that occurs during transcription initiation.
Methods of controlling reiterative transcription and transcript fates
The known mechanisms of gene regulation involving reiterative transcription control repetitive nucleotide addition either directly or indirectly. In mechanisms involving direct control, the key regulatory element is competition between the repeating nucleotide(s) and the next normally templated nucleotide for addition to the 3′ end of the nascent transcript. In mechanisms involving indirect control, the extent of reiterative transcription is not affected by competition between the repetitively added and normally templated nucleotides. Instead, other regulatory factors, such as transcription start site selection, control the frequency at which RNA polymerase enters the reiterative or normally templated mode of transcription.
With either mechanism for controlling reiterative transcription, the transcripts containing extra nucleotides can have two distinct fates. They can be released from the transcription initiation complex after the addition of one or more extra nucleotides, which is referred to as nonproductive transcription, or they can be normally elongated following the addition of extra nucleotides, which is called productive transcription. The primary factor controlling these alternative fates is the identity of the repetitively added nucleotide. Generally, reiterative transcription with UTP produces transcripts that are released from the transcription initiation complex, while reiterative transcription with non-UTP substrates produces transcripts that are productively extended [17••].
Gene regulation involving direct control of reiterative transcription
Studies on the regulation of expression of the pyrBI operon of E. coli and of the pyrG operon of Bacillus subtilis have provided distinct examples of regulatory mechanisms in which reiterative transcription is controlled directly (Figure 1). Although reiterative transcription is controlled in a similar manner in each mechanism, the fates of the transcripts containing extra nucleotides are very different.
Figure 1.
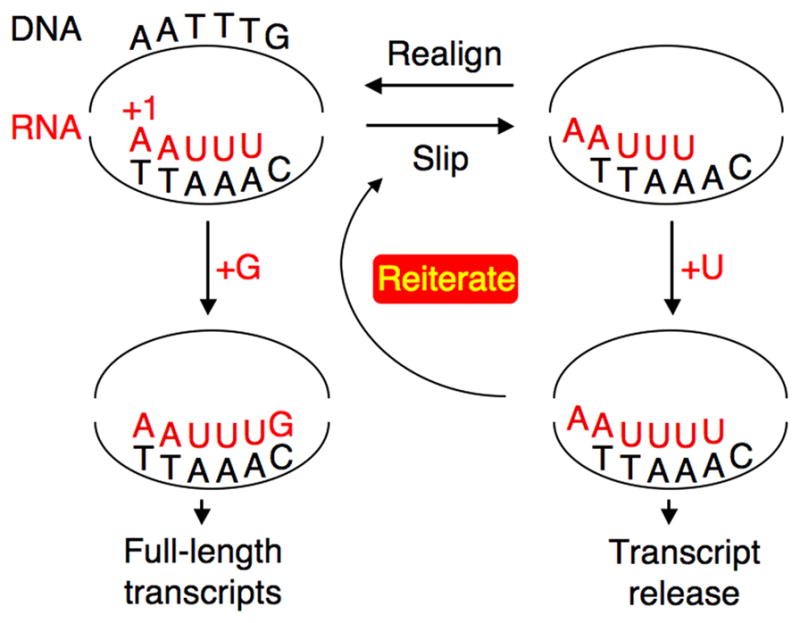
Direct control of reiterative transcription by competing NTP substrates. In this example, which uses the DNA sequence of the E. coli pyrBI initially transcribed region, the competing substrates are GTP and UTP. After synthesis of the AAUUU transcript, it can reversibly slip one base upstream due to a weak RNA-DNA hybrid. Addition of the template-encoded G residue at position +6 of the completely aligned AAUUU transcript results in an RNA-DNA hybrid that is stable enough to prevent further transcript slippage, allowing the AAUUUG transcript to be extended into full-length transcripts. Conversely, addition of a U residue at position +6 of the slipped transcript prevents addition of a G residue and entry into the productive mode of transcription through a mechanism that remains obscure. Subsequently, the AAUUUU transcript is either released from the transcription initiation complex or it slips upstream, allowing addition of another extra U residue. This process can be repeated many times, with each AAUUUUn transcript eventually released from the transcription complex.
pyrBI operon of E. coli
In E. coli, the pyrBI operon encodes the two subunits of the enzyme aspartate transcarbamylase, which catalyzes the first committed step in the de novo synthesis of pyrimidine nucleotides. Expression of this operon is negatively regulated over a sevenfold range by the intracellular concentration of UTP (which reflects pyrimidine availability) through a mechanism involving UTP-sensitive reiterative transcription during transcription initiation [13••,17••,28]. The pyrBI promoter region contains the sequence 5′-TATAATGCCGGACAATTTGCCG (nontemplate strand), with the −10 region and the physiologically relevant transcription start site underlined. The run of three T residues in the initially transcribed region is the site of reiterative transcription in the following regulatory model (Figure 1). After synthesis of the nascent transcript AAUUU, weak base pairing between the transcript and its DNA template allows a rapid and reversible one-base upstream shift (or slip) of the nascent transcript. When the intracellular level of UTP is high and the transcript is in the slipped position, the last (i.e., 5′) A in the AAA tract in the DNA template efficiently directs the addition of another U residue to the 3′ end of the transcript. This transcript can either be released from the transcription initiation complex or it can shift again. The cycle of slippage and U addition can occur repeatedly (up to at least 40 times), resulting in transcripts with progressively longer runs of U residues. However, all AAUUUUn (where n ≥ 1) transcripts are eventually released from the initiation complex, thereby preventing productive transcription of the pyrBI operon. On the other hand, when the intracellular level of UTP is low, slippage and correct repositioning of the nascent AAUUU transcript usually occur without extra U addition, which provides an opportunity for the addition of a template-encoded G residue to the 3′ end (i.e., at position +6) of the transcript. Once this addition occurs, more stable base pairing between the transcript and template prevents further slippage. The AAUUUG transcript is either released from the initiation complex as a simple aborted transcript or it is extended by the addition of a C residue, which apparently commits the transcription complex to the elongation mode. Therefore, high levels of full-length pyrBI transcripts are produced only when aspartate transcarbamylase is needed to synthesize more UTP. In this model, regulation of pyrBI expression can occur continuously over a range of intracellular UTP concentrations that modulate the frequency of nonproductive reiterative transcription.
pyrG operon of B. subtilis
The pyrG operon of B. subtilis encodes the pyrimidine biosynthetic enzyme CTP synthetase, which catalyzes the amination of UTP to form CTP. Expression of the pyrG operon is regulated over a greater than 20-fold range by a CTP-sensitive transcription attenuation control mechanism [29]. Attenuation occurs at an intrinsic transcription terminator (or attenuator) near the downstream end of the 189-bp pyrG leader region (i.e., the DNA between the pyrG promoter and the pyrG gene). The mechanism that causes conditional termination at the pyrG attenuator employs reiterative transcription involving the repetitive addition of G residues during transcription initiation (Figure 2). This reaction occurs during transcription of the pyrG initially transcribed region, which contains as its first five nucleotides the nontemplate strand sequence 5′-GGGCT (specifying the sequence 5′-GGGCU at the start of the pyrG transcript). Another key regulatory feature is an atypical sequence in the G+C-rich terminator stem-loop (or hairpin) specified by the pyrG attenuator. Nearly the entire upstream segment of the hairpin stem is comprised of a long pyrimidine tract (i.e., 5′-CUCCCUUUC). According to the well-established model for regulation [15••,30,31], when the intracellular level of CTP is high, nascent pyrG transcripts are faithful copies of the DNA template, and transcription elongation continues until termination at the attenuator. Therefore, when CTP is plentiful (reflecting ample pyrimidine availability), transcription of the pyrG gene is suppressed. On the other hand, when the intracellular level of CTP is low due to pyrimidine limitation, pyrG transcription pauses after the synthesis of the nascent transcript 5′-GGG (and before position +4C) because of insufficient substrate. Pausing provides time for the nascent transcript to slip upstream relative to the DNA template, which directs the addition of an extra G residue to the transcript. This process can be repeated up to at least nine times until eventually a C residue is inserted. The transcript is then elongated normally until RNA polymerase transcribes the attenuator sequence that specifies the upstream segment of the terminator hairpin, which includes the tract of nine pyrimidines. This tract will immediately base pair with the poly(G) tract at the 5′ end of the transcript, forming an antiterminator hairpin. Note that optimal antiterminator hairpin formation requires at least three extra G residues in the poly(G) tract [15••,30,31] and that G:U base pairing is permitted in RNA secondary structures [32]. As RNA polymerase continues to elongate the pyrG transcript, the antiterminator hairpin precludes formation of the terminator hairpin and full-length pyrG transcripts are formed. These transcripts are translated to make CTP synthetase, which is needed to overcome the CTP deficiency. Although the model describes pyrG expression at high and low intracellular concentrations of CTP, regulation can occur continuously over a wide range of CTP concentrations that control the extent of pausing at position +4. This incremental regulation is similar to that described for the pyrBI regulatory mechanism, in each case due to availability of NTP substrates. The key distinction between the pyrBI and pyrG regulatory mechanisms described above is that the repetitive addition of nucleotides precludes productive transcription elongation in the former case, but not in the latter.
Figure 2.
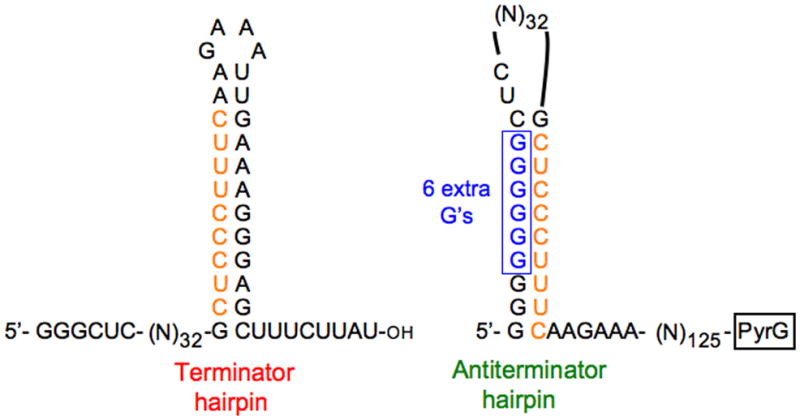
Alternative fates of transcripts synthesized from a promoter at which conditional reiterative transcription occurs. This example compares transcripts initiated at the pyrG promoter of B. subtilis. Under certain conditions, pyrG transcripts are subject to reiterative transcription that adds up to nine extra G residues following the run of three normally templated G residues at the 5′ end of the transcript. Transcripts containing extra G residues can be extended downstream following a switch from reiterative to normally templated transcription. On the left of the figure is the sequence of a transcript that does not contain extra G residues. This transcript contains a G+C-rich (terminator) hairpin immediately followed by a U-rich tract in the pyrG leader region. These RNA elements cause intrinsic transcription termination that precludes transcription of the downstream gene. On the right of the figure is the sequence of a transcript containing six extra G residues. This transcript forms an antiterminator hairpin that includes the run of nine G residues at the 5′ end of the transcript and the upstream segment of the terminator hairpin containing a run of nine C and U residues, both of which base pair with G in RNA. The antiterminator hairpin prevents terminator hairpin formation, allowing transcription of the downstream gene.
Gene regulation involving indirect control of reiterative transcription
A clear example of gene regulation involving the indirect control of reiterative transcription is provided by the mechanism controlling expression of the codBA operon of E. coli. In this example, the frequency at which RNA polymerase enters the reiterative mode of transcription is determined before transcription of the homopolymeric tract in the initially transcribed region (Figure 3).
Figure 3.
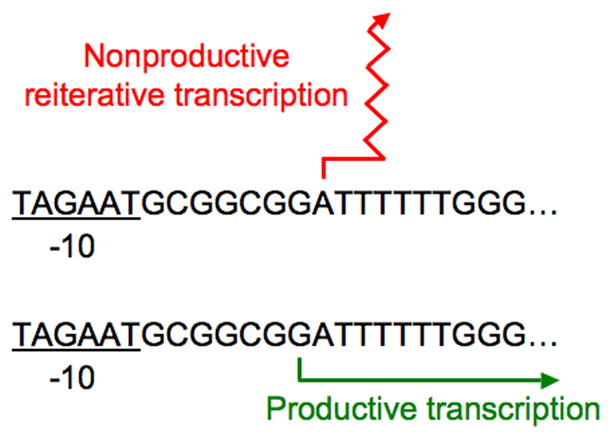
Indirect control of reiterative transcription. In this example, which uses the E. coli codBA promoter region, indirect control of reiterative transcription occurs through transcription start site switching. Under conditions of pyrimidine excess, codBA transcription initiation occurs predominantly at position A8 (counting downstream from the −10 region). Transcripts initiated at position A8 always engage in reiterative transcription, generating only nonproductive AUUUUn transcripts that are released from the transcription initiation complex. Under conditions of pyrimidine limitation, however, the major codBA transcription start site is position G7. For the most part, G7-initiated transcripts do not engage in reiterative transcription and are normally elongated to produce full-length codBA transcripts. G7-initiated transcripts avoid reiterative transcription because they form an RNA-DNA hybrid starting with a dC:rG base pair that is stable enough to prevent transcript slippage. Thus, in the case of the codBA operon, reiterative transcription is controlled by the selection of alternative transcription start sites that occurs prior to the possibility of repetitive nucleotide addition.
codBA operon of E. coli
The codBA operon of E. coli encodes cytosine permease (codB) and cytosine deaminase (codA), which are involved in cytosine uptake and cytosine utilization as a UTP precursor, respectively. Expression of the codBA operon is negatively regulated over an approximately 30-fold range by intracellular UTP levels [4••]. The nontemplate strand sequence of the codBA promoter region is 5′-TAGAATGCGGCGGATTTTTTGGG, with the −10 region and two alternative transcription start sites underlined. These start sites, which immediately precede a run of six T residues, are designated G7 and A8 (counting downstream from the −10 region). Position A8 is the inherently preferred transcription start site, provided the intracellular level of UTP is high [4••,33]. This dependence on UTP reflects the requirement for high concentrations of both the first and second nucleotide substrates for efficient formation of the first internucleotide bond of the transcript [17••,34,35]. Conversely, when the intracellular level of UTP is low, transcription initiation at position G7, which does not rely on UTP as the second nucleotide, is strongly favored [4••,33]. These observations together with experiments described elsewhere led to the following regulatory model (Figure 3) [4••,17••]. When the intracellular level of UTP is high, codBA transcription initiation occurs primarily at position A8. However, A8-initiated transcripts are not normally extended because, due to weak base pairing to the DNA template, they engage in nonproductive reiterative transcription within the run of six T residues in the initially transcribed region. Reiterative transcription apparently starts after the third residue in the T tract, and the final three residues in this tract guarantee that essentially all A8-initiated transcripts enter the reiterative mode of transcription before addition of a non-U residue to the 3′ end of the transcript is possible [5]. In contrast, when the intracellular level of UTP is low, transcription initiation occurs primarily at the G7 start site. For the most part, G7-initiated transcripts avoid reiterative transcription and can be normally elongated. The avoidance of reiterative transcription is apparently due to the fact that G7-initiated transcripts, from GAUUU through GAUUUUUU, form an RNA-DNA hybrid that is stable enough to preclude transcript slippage. Hybrid stability is due principally to the G:C base pair formed by the first nucleotide of the nascent transcript. Thus, pyrimidine mediated regulation of codBA expression occurs by UTP-sensitive selection of transcription start sites with different potentials for entering a nonproductive mode of reiterative transcription.
Conclusions
Other than the well-studied examples of gene regulation by reiterative transcription described in this review, not many examples of such mechanisms are known [17••]. Does this mean that few exist? The answer is almost certainly no. Bacterial promoters frequently contain a homopolymeric tract of at least three nucleotides at or near the start of the initially transcribed region. For example, approximately 10% of the several hundred well-characterized E. coli promoters contain three to eight T residues within two bases of the transcription start site [36]. These promoters, most of which are in operons unrelated to nucleotide metabolism, are all candidates for sites of regulated reiterative transcription. However, only a few of these promoters have been examined for this reaction. A complicating factor in the search for promoters regulated by reiterative transcript is that sequences other than the homopolymeric tract have profound effects on the reaction; in particular, sequences flanking this tract and core promoter sequences can have large effects on reiterative transcription [17••]. Many of the rules governing these effects remain to be established.
The few examples of gene regulation through reiterative transcription described above represent highly diverse mechanisms. It seems reasonable, therefore, that new mechanisms will be quite different from the known examples. In particular, these new mechanisms might respond to cellular signals other than nucleotides and regulate the expression of genes unrelated to nucleotide metabolism. The signals could be any small molecule or macromolecule in the cell that interacts with transcription complexes during any phase of transcription in a way that alters the extent of reiterative transcription. Candidate macromolecules include RNA polymerase binding factors. Interestingly, recent studies revealed that transcript slippage during initiation by human RNA polymerase II can be induced by the TATA-binding protein and transcription factor TFIIB [9••]. Other potential signals are factors that control transcription pausing, particularly during transcription elongation. Previous studies have provided ample evidence of the dependence of reiterative transcription on pausing, especially when the repetitively added nucleotide is not UTP [23,29]. It should be noted that the mechanisms of reiterative transcription during different phases of transcription are likely to be somewhat different and therefore possibly subject to different regulatory strategies.
A final prediction is that mechanisms of gene regulation involving reiterative transcription will be found in all kingdoms of life because the mechanisms of reiterative transcription catalyzed by bacterial, eukaryotic, and archeal RNA polymerases are likely to be highly conserved [37]. In fact, key features of the regulatory mechanisms described in this review have been uncovered recently in eukaryotes. For example, the mechanisms controlling expression of several genes involved in nucleotide metabolism in Saccharomyces cerevisiae employ transcription start site switching analogous to that described in the codBA regulatory mechanism of E. coli [38•,39•]. It is not difficult to imagine transcription start site switching in yeast or in any other eukaryote—including humans—that controls initiation at sites with different potentials for reiterative transcription. Perhaps the greatest obstacle to the discovery of new mechanisms of gene regulation requiring reiterative transcription is the lack of knowledge about existing mechanisms—an obstacle that should be diminished by this review.
Acknowledgments
Research on reiterative transcription performed in the author’s laboratory was supported by grants GM94466 and GM29466 from the National Institute of General Medical Sciences. I thank Bill McAllister, Misha Kashlev, Bob Switzer, Shawn Harmon, and Jeffery Vahrenkamp for excellent suggestions and comments and Evvie Allison for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacques JP, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- 2.Linton MF, Raabe M, Pierotti V, Young SG. Reading-frame restoration by transcriptional slippage at long stretches of adenine residues in mammalian cells. J Biol Chem. 1997;272:14127. doi: 10.1074/jbc.272.22.14127. [DOI] [PubMed] [Google Scholar]
- 3.Martin CT, Muller DK, Coleman JE. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 4••.Qi F, Turnbough CL., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. This paper is the original report of codBA regulation by reiterative transcription in E. coli, which is used in this review as the example of gene regulation involving indirect control of reiterative transcription. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y, Dylla SM, Turnbough CL., Jr A long T·A tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J Bacteriol. 2001;183:221–228. doi: 10.1128/JB.183.1.221-228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong XF, Reznikoff WS. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J Mol Biol. 1993;231:569–580. doi: 10.1006/jmbi.1993.1310. [DOI] [PubMed] [Google Scholar]
- 7.Baranov PV, Hammer AW, Zhou J, Gesteland RF, Atkins JF. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005;6:R25. doi: 10.1186/gb-2005-6-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo HC, Roberts JW. Heterogeneous initiation due to slippage at the bacteriophage 82 late gene promoter in vitro. Biochemistry. 1990;29:10702–10709. doi: 10.1021/bi00499a019. [DOI] [PubMed] [Google Scholar]
- 9••.Gilman B, Drullinger LF, Kugel JF, Goodrich JA. TATA-binding protein and transcription factor IIB induce transcript slipping during early transcription by RNA polymerase II. J Biol Chem. 2009;284:9093–9098. doi: 10.1074/jbc.M900019200. This study investigates the molecular determinants of transcript slipping (by two bases in this case) in early transcription by human RNA polymerase II. It establishes the contributions to transcript slipping of individual transcription factors, DNA elements, and transcript length. It was found that transcript slipping begins at position +5 in the DNA template and that slipping is induced by TATA-binding protein and transcription factor IIB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal M, Luse DS. Strong natural pausing by RNA polymerase II within 10 bases of transcription start may result in repeated slippage and reextension of the nascent RNA. Mol Cell Biol. 2002;22:30–40. doi: 10.1128/MCB.22.1.30-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabre E, Dujon B, Richard GF. Transcription and nuclear transport of CAG/CTG trinucleotide repeats in yeast. Nucleic Acids Res. 2002;30:3540–3547. doi: 10.1093/nar/gkf483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr JN, Wertz GW. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J Virol. 2001;75:6901–6913. doi: 10.1128/JVI.75.15.6901-6913.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Liu C, Heath LS, Turnbough CL., Jr Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 1994;8:2904–2912. doi: 10.1101/gad.8.23.2904. This paper reports the first example of gene regulation involving metabolite-sensitive reiterative transcription, which is used in this review as one of the examples of gene regulation involving direct control of reiterative transcription. [DOI] [PubMed] [Google Scholar]
- 14.Larsen B, Wills NM, Nelson C, Atkins JF, Gesteland RF. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc Natl Acad Sci USA. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Meng Q, Turnbough CL, Jr, Switzer RL. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc Natl Acad Sci USA. 2004;101:10943–10948. doi: 10.1073/pnas.0403755101. This paper describes the model for attenuation control of pyrG regulation in B. subtilis, which is used in this review as one of the examples of gene regulation involving direct control of reiterative transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Anikin M, Molodtsov V, Temiakov D, McAllister WT. Transcript slippage and recoding. In: Atkins JF, Gesteland RF, editors. Recoding: expansion of decoding rules enriches gene expression. Springer; 2010. pp. 409–432. [Gross HJ, Bujnicki JM (Series Editor): Nucleic Acids and Molecular Biology, vol 24.] This article provides a detailed review of transcript slippage in vivo and its implications for miscoding and recoding of genetic information. This paper also reviews experimental evidence for the mechanism of transcript slippage and discusses parameters that affect the frequency of this process. A highlight of this paper is a pictorial representation of base pairing between the RNA-DNA hybrid during reiterative transcription. [Google Scholar]
- 17••.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. This article provides a comprehensive description of bacterial gene regulation that does not involve regulatory DNA binding proteins, including regulation by conditional reiterative transcription. It provides extensive background and supporting information about the examples of gene regulation described in the current review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent T, Kashkina E, Anikin M, Temiakov D. Maintenance of RNA-DNA hybrid length in bacterial RNA polymerases. J Biol Chem. 2009;284:13497–13504. doi: 10.1074/jbc.M901898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 20.Maconald LE, Zhou Y, McAllister WT. Termination and slippage by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 21•.Ratinier M, Boulant S, Combet C, Targett-Adams P, McLauchlan J, Lavergne JP. Transcriptional slippage prompts recoding in alternate reading frames in the hepatitis C virus (HCV) core sequence from strain HCV-1. J Gen Virol. 2008;89:1569–1578. doi: 10.1099/vir.0.83614-0. This report describes the involvement of transcription slippage within an A10 tract in the RNA genome of HCV that accounts for frameshifts in the viral core protein open reading frame. It provides a clear example of transcript slippage leading to both nucleotide addition and deletion. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlin M, Berg P. Deoxyribonucleic acid-directed synthesis of ribonucleic acid by an enzyme from Escherichia coli. Proc Natl Acad Sci USA. 1962;48:81–94. doi: 10.1073/pnas.48.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hausmann S, Garcin D, Delenda C, Kolakofsky D. The versatility of paramyxovirus RNA polymerase stuttering. J Virol. 1999;73:5568–5576. doi: 10.1128/jvi.73.7.5568-5576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldick CJ, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penno C, Hachani A, Biskri L, Sansonetti P, Allaoui A, Parsot C. Transcriptional slippage controls production of type III secretion apparatus components in Shigella flexneri. Mol Microbiol. 2006;62:1460–1468. doi: 10.1111/j.1365-2958.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- 26••.Tamas I, Wernegreen JJ, Nystedt B, Kauppinen SN, Darby AC, Gomez-Valero L, Lundin D, Poole AM, Andersson SGE. Endosymbiont gene functions impaired and rescued by polymerase infidelity at poly(A) tracts. Proc Natl Acad Sci USA. 2008;105:14934–14939. doi: 10.1073/pnas.0806554105. This is an interesting study of reiterative transcription in host-dependent bacteria with extremely small genomes and very high A+T content. The genomes of these bacteria are riddled with poly(A) tracts containing 10 or more residues, which are sites of reiterative transcription. The addition of extra A residues rescues the functionality of genes with frameshift mutations and reduces the efficiency of expression of in-frame genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raabe M, Linton MF, Young SG. Long runs of adenines and human mutations. Am J Med Genet. 1998;76:101–102. doi: 10.1002/(sici)1096-8628(19980226)76:1<101::aid-ajmg19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Turnbough CL., Jr Multiple control mechanisms for pyrimidine-mediated regulation of pyrBI operon expression in Escherichia coli K-12. J Bacteriol. 1989;171:3337–3342. doi: 10.1128/jb.171.6.3337-3342.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Q, Switzer RL. Regulation of transcription of the Bacillus subtilis pyrG gene, encoding cytidine triphosphate synthetase. J Bacteriol. 2001;183:5513–5522. doi: 10.1128/JB.183.19.5513-5522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elsholz AKW, Jørgensen CM, Switzer RL. The number of G residues in the Bacillus subtilis pyrG initially transcribed region governs reiterative transcription-mediated regulation. J Bacteriol. 2007;189:2176–2180. doi: 10.1128/JB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen-MacAllister IE, Meng Q, Switzer RL. Regulation of pyrG expression in Bacillus subtilis: CTP-regulated antitermination and reiterative transcription with pyrG templates in vitro. Mol Microbiol. 2007;63:1440–1452. doi: 10.1111/j.1365-2958.2007.05595.x. [DOI] [PubMed] [Google Scholar]
- 32.Varani G, McClain WH. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Turnbough CL., Jr Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amiott EA, Jaehning JA. Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides. J Biol Chem. 2006;281:34982–34988. doi: 10.1074/jbc.M608638200. [DOI] [PubMed] [Google Scholar]
- 35.Sørensen KI, Baker KE, Kelln RA, Neuhard J. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J Bacteriol. 1993;175:4137–4144. doi: 10.1128/jb.175.13.4137-4144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hershberg R, Bejerano G, Santos-Zavaleta A, Margalit H. PromEC: An updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 2001;29:277. doi: 10.1093/nar/29.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. This is an interesting study showing the role of GTP-sensitive transcription start site switching in the regulation of a yeast gene encoding a guanine nucleotide biosynthetic enzyme. When GTP is sufficient, RNA polymerase II initiates transcription at TATA box-proximal G sites, producing attenuated transcripts. When GTP is limiting, transcription is initiated at an A downstream of the attenuation site, allowing synthesis of full-length transcripts. [DOI] [PubMed] [Google Scholar]
- 39•.Thiebaut M, Colin J, Neil H, Jacquier A, Séraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. This study describes regulation of pyrimidine nucleotide biosynthetic genes in yeast that relies on transcription start site switching. Transcription from TATA box-proximal start sites generates cryptic unstable URA2 transcripts. Uracil limitation activates selection of distal start sites that lead to the production of stable URA2 mRNAs. [DOI] [PubMed] [Google Scholar]


