Abstract
A recombinant protein termed CLS, which corresponds to the C-terminal portion of human L-selectin and contains its entire transmembrane and cytoplasmic domains (residues Ser473-Arg542), has been produced and its oligomeric state in detergents characterized. CLS migrates in the SDS polyacrylamide gel at a pace that is typically expected from a complex twice of its molecular weight. Additional studies revealed however that this is due to residues in the cytoplasmic domain, as mutations in this region or its deletion significantly increased the electrophoretic rate of CLS. Analytical ultracentrifugation and fluorescence resonance energy transfer studies indicated that CLS reconstituted in dodecylphosphocholine detergent micelles is monomeric. When the transmembrane domain of L-selectin is inserted into the inner membrane of Escherichia coli as a part of a chimeric protein in the TOXCAT assay, little oligomerization of the chimeric protein is observed. Overall, these results suggest that transmembrane and cytoplasmic domains of L-selectin lack the propensity to self-associate in membranes, in contrast to the previously documented dimerization of the transmembrane domain of closely related P-selectin. This study will provide constraints for future investigations on the interaction of L-selectin and its associating proteins.
Keywords: L-selectin, transmembrane domain, membrane protein, TOXCAT, fluorescence resonance energy transfer, SDS-PAGE
Introduction
Cell adhesion receptors not only mediate cell-matrix interactions and cell-cell contacts via their extracellular domains, but also transmit signals across the cell membrane. Cell adhesion receptors typically consist of a large extracellular domain, a single transmembrane (TM) domain and a relatively short cytoplasmic domain devoid of any enzymatic activities. Whereas the extracellular domain often binds to extracellular matrix proteins or co-receptors, the cytoplasmic domain can interact with a multitude of intracellular proteins with signaling implications. In addition to providing an anchor in the plasma membrane, the TM helix in many adhesion receptors is capable of lateral association with other TM helices, contributing to receptor function and regulation [1].
Selectins are a family of cell adhesion receptors that are characterized by the presence of a C-type lectin domain at their N-terminal extracellular end. They are responsible for recruiting leukocytes into inflammatory tissues and returning them from circulation to lymphoid tissues [2–4]. Through the lectin domain, selectins recognize and bind to their counter-receptors containing the sialyl Lewisx tetrasaccharide [5, 6]. In addition to the interaction between lectin domains and polysaccharides from opposite cells, lateral association or clustering of selectins on the surface of host cell also contributes to the interaction between leukocytes and their interacting cells and thus helps to mediate leukocyte capture and rolling. For instance, P-selectin, which becomes expressed on activated endothelial cells or platelets, is a non-covalent homodimer in the cell membrane [7]. Dimerization of P-selectin is mediated by its TM domain, because full-length P-selectin purified from platelets forms dimers and oligomers in detergent solutions whereas the recombinant P-selectin ectodomain does not [8]. Moreover, the TM domain of P-selectin contains the GxxxG sequence, a common motif that mediates association of TM helices [9, 10]. Many GxxxG-containing TM proteins can form high-molecular weight oligomers in the presence of SDS [9, 11], which explains the ability of full-length P-selectin to form dimer species in SDS-PAGE [8]. Coincidentally, P-selectin glycoprotein ligand-1 (PSGL-1), the counter-receptor for selectins, is a disulfide-linked homodimer on the cell surface [12, 13]. Mutating the membrane-proximal residue Cys320 to either Ser or Ala abolishes formation of the disulfide bond between PSGL-1, but the mutant receptor remains as a non-covalent dimer [14]. When the TM domain of PSGL-1 is replaced with that of CD43, PSGL-1 cannot dimerize in the membrane, indicating that the TM domain of PSGL-1 is important for receptor dimerization [14]. Comparison of the rolling behavior of cells expressing dimeric or monomeric PSGL-1 on dimeric or monomeric P-selectin surface indicates that the receptor dimerization does not influence significantly the initial tethering but can stabilize subsequent cell rolling by facilitating rebinding [15].
Similar to studies on P-selectin, cross-linking L-selectin on the cell surface through a dimerization domain appended to the cytoplasmic domain of L-selectin greatly increases the number of lymphocytes rolling on the endothelial walls [16]. In another study, a monoclonal antibody induces L-selectin dimerization and enhanced its tethering to purified L-selectin ligands [17]. These studies suggest the possible existence of dimeric or oligomeric L-selectin on the cell surface. Here, we tested whether the TM domain of L-selectin, like that of P-selectin, is capable of self-association in the membrane and may thus contribute to the clustering of host receptor.
Materials and Methods
Materials
Human L-selectin cDNA was purchased from ATCC (Manassas, VA). Escherichia coli BL21 and DH5α competent cells were purchased from Stratagene (La Jolla, CA). Thrombin was from GE Healthcare (Piscataway, NJ). 99.9% pure D2O was from Cambridge Isotope Laboratories (Andover, MA). Tetramethylrhodamine(TMR)-5-maleimide was from AnaSpec (Fremont, CA). Fluorescein-5-maleimide was purchased from Invitrogen (Carlsbad, CA). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP·HCl) was purchased from Pierce (Rockford, IL). All lipids and phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL).
Expression and purification of CLS and variants
To produce the His-tagged glutathione S transferase-CLS fusion protein (His-GST-CLS), the gene fragment encoding residues S473-R542 of human L-selectin was amplified from its cDNA. The fragment was inserted into the pHex vector as a BamHI-XhoI fragment as described before [18]. Standard PCR reactions were performed to obtain desired mutations in CLS. Truncation mutants were obtained by inserting a stop codon at the desired position. All DNA sequences were confirmed by sequencing (Lone Star Labs, Houston, TX). Overexpression of His-GST-CLS fusion protein from E. coli BL21 cells and its purification by Ni-affinity chromatography followed previously published protocols [18, 19]. After the fusion protein was cleaved by thrombin (10–20 units/mg of fusion protein) at room temperature overnight, the generated CLS protein was separated from the His-GST fragment by preparative reverse-phase HPLC and stored at −80 °C as lyophilized powder. The purity and identity of each CLS protein was assessed and confirmed by SDS-PAGE, analytical HPLC and mass spectrometry. The molar extinction coefficient for CLS is calculated to be 8747.5 cm−1·M−1 at 280 nm [20].
SDS-PAGE
Purified CLS was dissolved in hexafluoroisopropanol (HFIP) and its concentration determined using its extinction coefficient. Approximately 2–5 µg of CLS in HFIP was transferred to a cone-bottomed glass vial, dried gently under nitrogen gas, and dissolved in the SDS-containing sample buffer. The sample was either loaded directly onto a 15% Tris-glycine SDS polyacrylamide gel or boiled for 5 min before being loaded. After electrophoresis at 150 V for 90 min, the gel was stained with colloidal Coomassie G-250 (SimplyBlue SafeStain, Invitrogen).
TOXCAT
Gene fragments encoding the L-selectin TM domain in various lengths were amplified by PCR from the pHex-CLS vector described above, and inserted respectively into the pccKAN plasmid [21]. The resulting pccKAN-based plasmids were transformed to E.coli MM39 cells. The topology of the ToxR-TM-MBP fusion protein expressed in MM39 cells was checked with the maltose complementation test [21, 22]. Expression of the fusion protein was measured by Western blot using an anti-MBP monoclonal antibody (Sigma). The activity of CAT expressed in MM39 cells was measured as described [23].
Circular dichroism (CD) spectroscopy
Approximately 33 µg of purified CLS dissolved in HFIP was mixed with 100 µl of 30 mM desired detergent dissolved in ethanol in a glass vial, dried gently with nitrogen gas, and placed under vacuum overnight. The dried CLS/detergent film, with a molar ratio of 1/600, was dissolved in 500 µl of 1 mM MOPS buffer, pH 7.4 or a buffer otherwise specified. The final concentration of CLS in the aqueous buffer was confirmed by the absorbance at 280 nm. Far-UV CD spectra (190–260 nm) were collected on a JASCO J815 spectrometer using a 0.1-cm quartz cuvette at 20 °C. The step-wise wavelength was set to 0.2 nm/step. Each spectrum was scanned 10 times and corrected for background signal from the MOPS buffer containing 6 mM detergent.
Analytical ultracentrifugation
Various amounts of CLS dissolved in HFIP was individually mixed in a glass vial with DPC dissolved in ethanol to achieve protein/detergent ratios of 1/300 and 1/600. Nitrogen gas was gently applied to the mixture in glass vials to evaporate organic solvents before the glass vials were placed under vacuum overnight. Each dried CLS/DPC mixture (or DPC without CLS for background correction) was then rehydrated with 1 mL buffer (50 mM Tris·HCl, 100 mM NaCl, pH 7.4) containing 53.4% D2O to achieve the final DPC concentration of 15 mM. The density of the buffer matched that of the detergent (data not shown), eliminating contribution of the buoyant molecular weight of the protein/detergent complex. Equilibrium analytical ultracentrifugation experiments were performed at 25 °C using the Beckman XL-A ultracentrifuge located at Rice University. After the samples were centrifuged at desired rotor speeds (30000, 40000 and 45000 rpm) for 16 hours and have reached the equilibrium, its absorption profile at 280 nm was collected as a function of radius from the rotor axis and subtracted against the readings of 15 mM DPC without CLS in density-matched buffer. To analyze the data, the molecular mass and the partial specific volume of CLS in density matched buffer, calculated using SEDENTERP [24], are 6510.0 Da and 0.7578, respectively. The radial distribution profiles were globally fit as previously described [25].
Thiol-specific conjugation of fluorophore to CLS_S329C
The S329C mutant CLS was expressed and purified as the wild type. Approximately 1–2 mg of purified CLS_S329C was dissolved in 5.5 ml conjugation buffer (50% isopropanol, 25% acetonitrile, 25% water, 40 mM HEPES, 0.4 mM TCEP·HCl, pH 7.6) at room temperature. Maleimide-containing fluorophore, at 10× excess, was dissolved in 50 µl N,N’-dimethylformamide and immediately added to the peptide solution to start conjugation. To follow the extent of conjugation, a 90-µl aliquot was taken at various time points, quenched immediately with 10 µl of 1 M dithiothreitol, and analyzed by HPLC. When the conjugation was complete, the fluorophore-conjugated CLS peptide was purified by preparative HPLC and stored as lyophilized powder at −80 °C. The mass spectrometric analysis was carried out on an Applied Biosystems MALDI-TOF instrument located at the proteomics core facility at the University of Texas M. D. Anderson Cancer Center, using sinapinic acid as the matrix. The extinction coefficients of fluorescein- and TMR-CLS are 68,000 M−1·cm−1 at 492 nm and 62,000 M−1·cm−1 at 542 nm, respectively.
Fluorescent resonance energy transfer (FRET) measurement
Purified TMR- and fluorescein-CLS, as well as unlabeled CLS, were individually dissolved in ethanol and mixed with dodecylphosphocholine (DPC) in a glass vial to desired protein/detergent molar ratios. Each mixture was dried under gentle nitrogen stream before being placed under vacuum overnight. The dried mixture was dissolved in and, if necessary, later diluted by 10 mM MOPS, pH 7.4 to a desired final concentration.
FRET experiments were performed using a 1-mL cuvette on a PTI QuantaMaster fluorimeter (Photon Technology International, Birmingham, NJ) at 20°C. In the titration experiment, the initial measurement was carried out for a sample containing 2 µM fluorescein-CLS, 8 µM unlabeled CLS and 5 mM DPC. Subsequently for each titration point, a small aliquot was removed from this sample and replaced with the same volume of a solution containing 2 µM fluorescein-CLS, 8 µM TMR-CLS and 5 mM DPC. Thus, the donor fluorophore concentration, the overall protein concentration and the overall protein/detergent ratio were kept constant throughout the titration. The solution was mixed thoroughly and incubated for at least 5 min before multiple emission scans were taken to confirm equilibrium had been reached. The emission scan of 505–595 nm was obtained with excitation at 492 nm, with slid widths set to 0.2 nm, and corrected for buffer background. FRET efficiency was calculated as (F0-F)/F0, where F is the fluorescein emission intensity at 522 nm in a given condition and F0 is that in the absence of TMR-CLS. To measure the effect of increasing protein concentration on FRET, the initial measurement was carried out for a sample containing 10 µM fluorescein-CLS, 20 µM TMR-CLS and 15 mM DPC. Subsequently, after each aliquot of 10 mM MOPS buffer was added to the sample to lower the overall protein and detergent concentration but keep constant the protein/detergent ratio, it was mixed thoroughly and an emission scan obtained as described above. The extent of FRET was tracked by the FRET ratio, the ratio of fluorescence emission at 522 nm (fluorescein emission) versus that at 575 nm (TMR emission).
Results
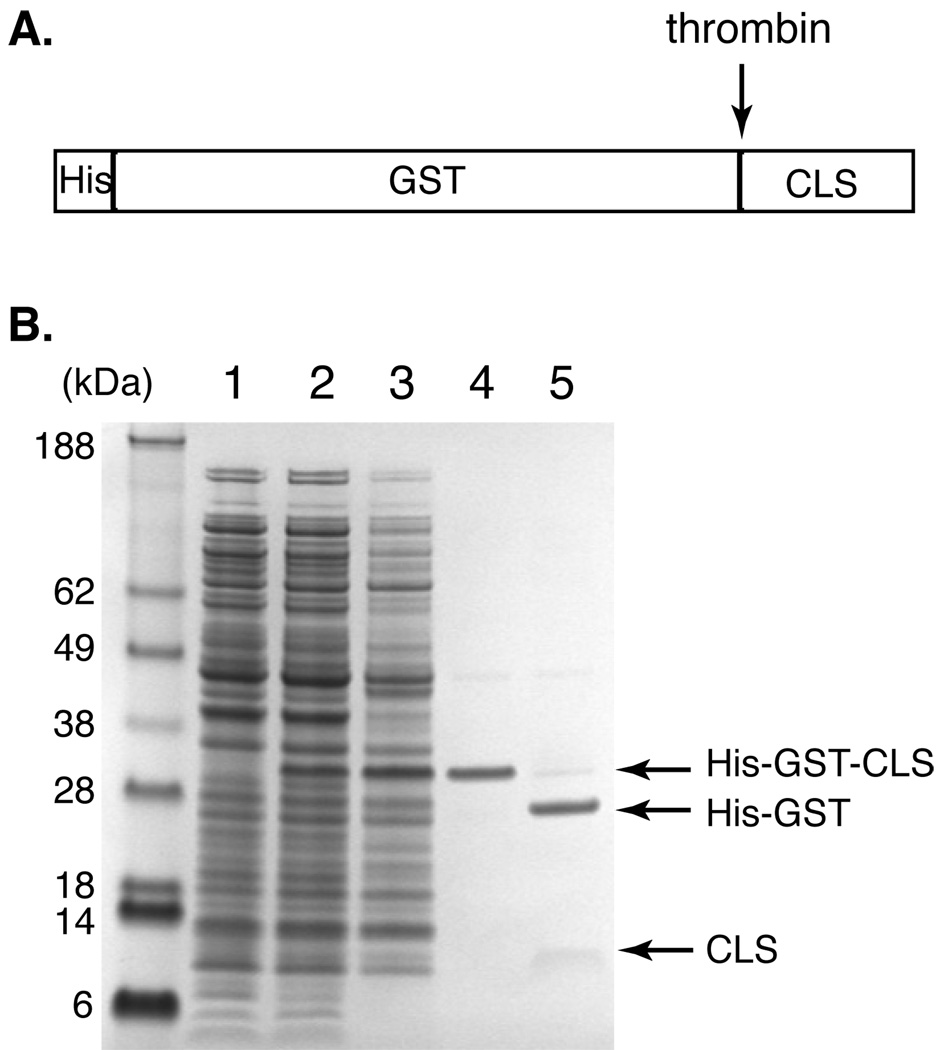
This study started with characterization of a recombinant protein we termed CLS (Table 1). Containing the entire TM and cytoplasmic domains of human L-selectin, CLS had been produced for studies of the interaction of L-selectin with its intracellular binding partners. Expressed as a part of the His-tagged GST-fusion protein, CLS was separated from the His-GST portion by thrombin cleavage and further purified by reverse-phase HPLC (Fig. 1). In the SDS gel, CLS migrated with an apparent molecular weight of 12 kDa, which is approximately twice its molecular weight (Fig. 1B). For many membrane peptides or proteins, a slower migration in SDS gel than what is expected from their molecular weight is often attributed to the formation of protein oligomers mediated by their TM domains [11, 26]. Since CLS contains the TM domain of L-selectin, the apparently slow migration of CLS suggested that the L-selectin TM domain may be capable of dimerization.
Table 1.
Sequences of L-selectin-derived TM peptides used in this study.
| Peptide name | amino acid sequencea |
|---|---|
| CLS | GSKLDKSFSMIKEGDYNPLFIPVAVMVTAFSGLAFIIWLARRLKKAKKSKRSMNDPY |
| CLS_S329C | GSKLDKSFSMIKEGDYNPLFIPVAVMVTAFSGLAFIIWLARRLKKAKKSKRCMNDPY |
| CLS_326Δ | GSKLDKSFSMIKEGDYNPLFIPVAVMVTAFSGLAFIIWLARRLKKAKKS |
| CLS_317Δ | GSKLDKSFSMIKEGDYNPLFIPVAVMVTAFSGLAFIIWLA |
| CLS_L320E | GSKLDKSFSMIKEGDYNPLFIPVAVMVTAFSGLAFIIWLARREKKAKKSKRSMNDPY |
The TM sequence is indicated in bold and mutated residues underlined.
Figure 1. Anomalous migration of CLS (C-terminal fragment of L-selectin) in SDS-PAGE revealed during CLS purification.
(A) Diagram showing the domain structure of His-GST-CLS fusion protein. A thrombin-cleavage site is placed between the His-tagged GST domain and CLS. (B) A SDS polyacrylamide gel depicting expression and purification of CLS, revealing its slow electrophoretic rate. Molecular weight markers are shown on the left labeled with corresponding sizes in kDa. Lane 1: E. coli whole cell lysate before IPTG induction; 2: cell lysate after IPTG induction; 3: solubilized inclusion body from the cell lysate; 4: His-GST-CLS fusion protein eluted from the Ni-NTA column; 5: thrombin cleavage of the fusion protein. Note that CLS is more diffuse in the gel and does not stain very well by colloidal Coomassie blue.
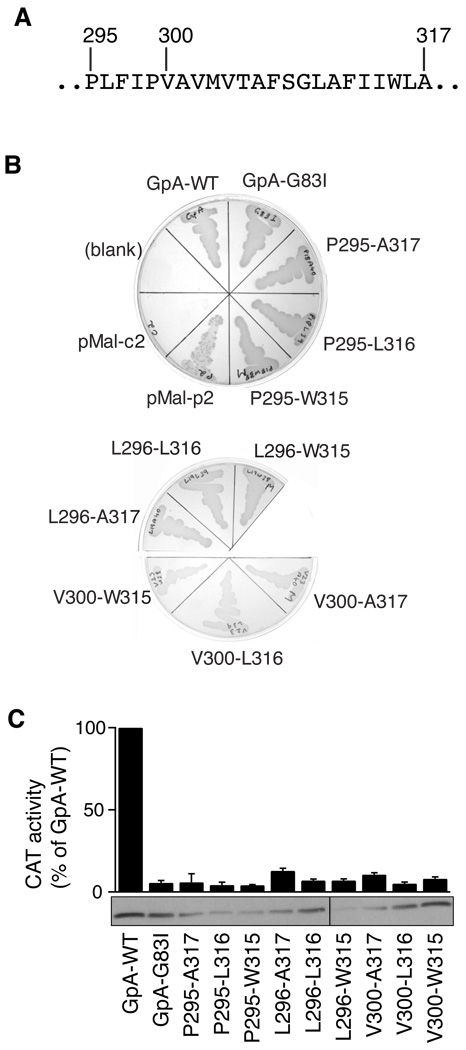
To assess its propensity to dimerize in a cell membrane, the TM domain of L-selectin was inserted into the ToxR´-TM-MBP chimeric protein in the TOXCAT assay. In this assay [21], if the TM domain in the chimeric protein self-associates in the inner membrane of E. coli, the connected ToxR´ domain, a DNA-binding domain, will be brought into proximity with another ToxR´ domain, thereby leading to transcriptional activation of a gene encoding chloramphenicol acetyl transferase (CAT). Thus, the expression level of CAT in the cell, as indicated by its enzymatic activity, can be used as an indicator for the extent of TM helix-mediated dimerization in the membrane [21]. Since the dimerization of a TM helix can be influenced by its length [22, 27], the L-selectin TM domain was tested in the TOXCAT assay at various lengths, each of which was identified by the starting and ending residues of L-selectin (Fig. 2). The MalE complementation test showed that all the chimeric constructs were inserted correctly into the inner membrane of E. coli and had the expected membrane topology (Fig. 2B). Furthermore, Western blotting of E. coli cell lysates showed that the expression level of each L-selectin chimeric protein was comparable to those of GpA-WT and GpA-G83I, the well-documented control constructs for highly dimeric and monomeric TM helices, respectively [21]. However, none of the L-selectin TM constructs induced significant expression of CAT as GpA-WT did, indicating that the L-selectin TM domain does not dimerize in the membrane (Fig. 2C).
Figure 2. The L-selectin TM domain lacks the ability to self-associate in the cell membrane.
(A) Sequence of the L-selectin TM domain. Residue numbers marking the sequences analyzed by the TOXCAT assay are labeled on the top. (B) MalE complementation test showing correct topology of chimeric ToxR’-TM-MBP proteins. Each L-selectin TM sequence tested in the assay is identified by its starting and ending residue numbers as in the mature protein. On M9 minimum media, where maltose is the only carbon source, only cells with MBP expression in the periplasm survive. MalE-deficient MM39 E. coli cells transformed with pMal-c2 and pMal-p2 vectors (New England Biolabs), which express MBP in the cytoplasm and the periplasm, respectively, were included as controls. (C) The enzymatic activity of CAT induced by self-association of the tested L-selectin TM sequences and expressed as the percentage of that induced by GpA-WT. The GpA-WT and GpA-G83I constructs were used as positive and negative controls, respectively. The data are shown as mean ± s.d. of 3 independent measurements. The lower panel shows the expression levels of the chimeric protein probed by Western blot against MBP.
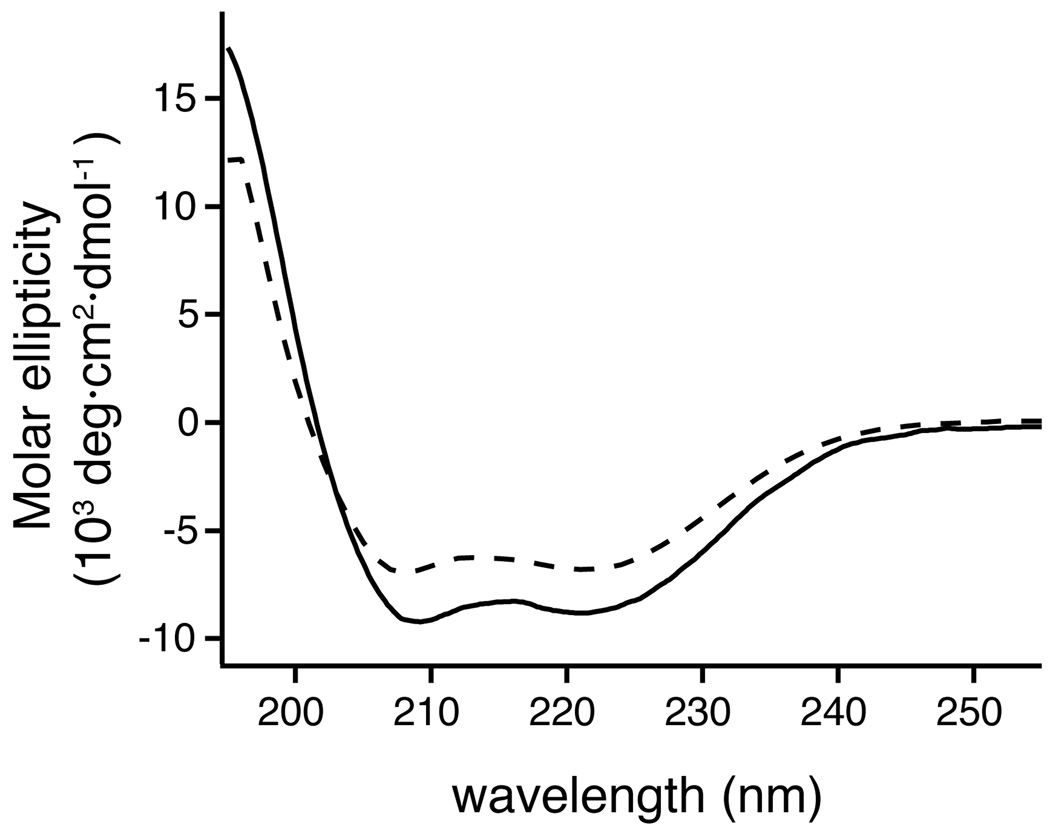
To resolve the contradicting implications from SDS-PAGE and TOXCAT studies, CLS was reconstituted into dodecylphosphocholine (DPC) micelles for further biophysical characterization. The CD spectra of CLS, both in SDS and DPC micelles, had minima at 208 and 222 nm, indicating that CLS is properly dispersed in the detergent micelle with substantial α-helical conformation (Fig. 3). Moreover, the CD spectrum in SDS micelles exhibits more molar ellipticity than that in DPC, suggesting that the anionic head group of SDS can stabilize the α-helical structure of CLS or induce additional ones.
Figure 3. Far-UV CD spectra of CLS in detergent micelles.
Solid line: CLS in 6 mM SDS/1 mM MOPS, pH 7.4. Dashed line: CLS in 6 mM DPC/10 mM MOPS, pH 7.4. The CLS concentration was approximately 9.5 µM.
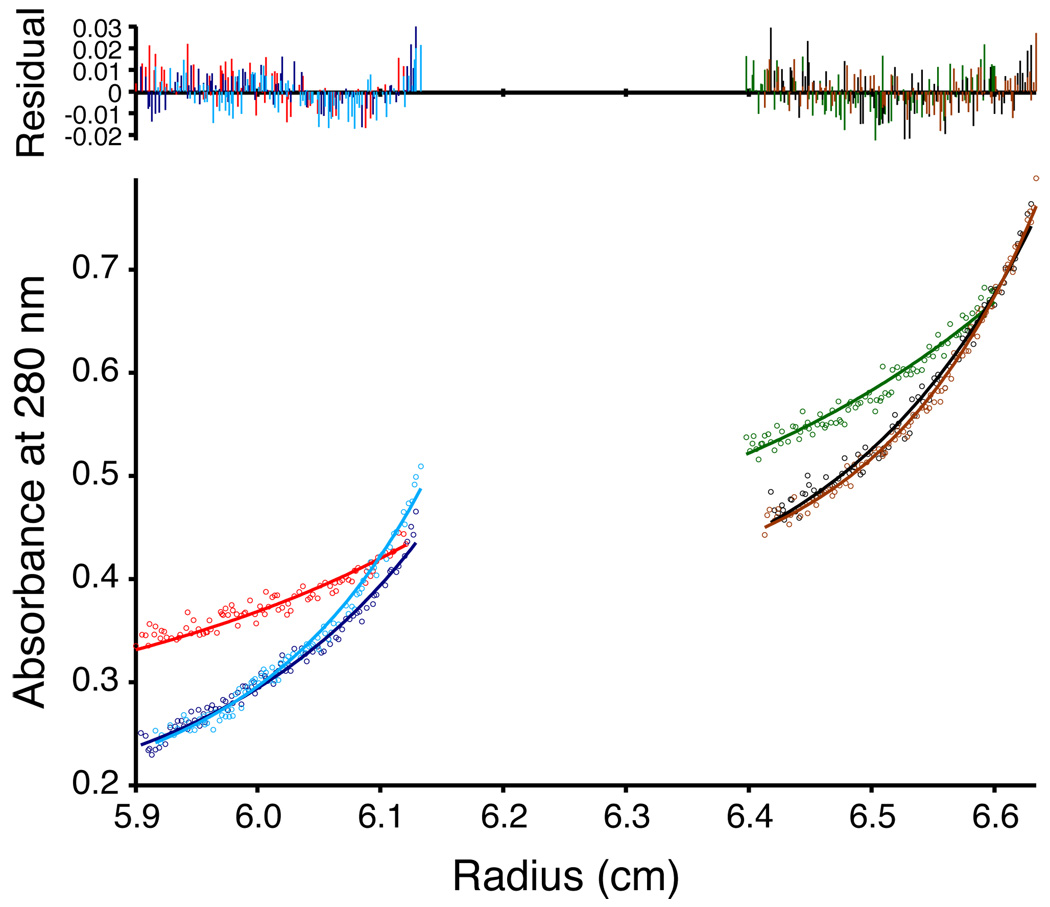
Analytical ultracentrifugation was used to analyze possible CLS oligomerization. The equilibrium sedimentation experiment was carried out in 15 mM DPC at protein/detergent ratios of 1/300 and 1/600 (Fig. 4). The buffer was mixed with heavy water to match its density to that of DPC. The equilibrium radial concentration profile was obtained for CLS and fit with various monomer-n-mer reversible equilibrium models. The best fit was a monomer-dimer equilibrium, but with a rather large dissociation constant (pKd was 1.49 in molar fraction units), indicating that for the range of protein/detergent ratios in the study, CLS was mostly a monomer.
Figure 4. Sedimentation equilibrium of CLS in 15 mM DPC at pH 7.4.
Bottom panel: equilibrium absorbance at 280 nm versus radius profiles for two cell compartments with different peptide/detergent ratios at 30, 40 and 45 krpm. The lines represent the best fit. Top: residual plots showing the difference between the calculated fit and experimental data.
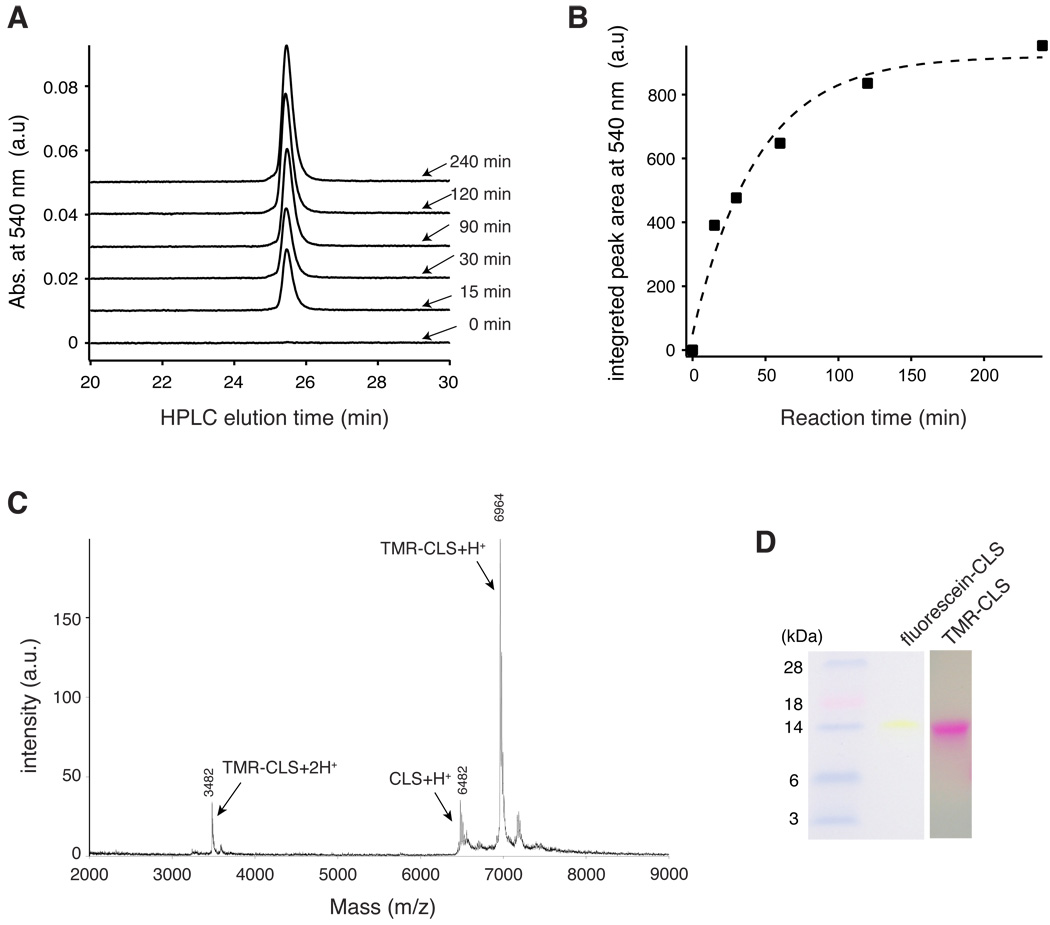
Fluorescence resonance energy transfer (FRET) is an established technique to detect and characterize association of membrane proteins [28–32]. In order to maximize the chances to detect CLS oligomerization by FRET, the pair of fluorescein and tetramethylrhodamine (TMR), with its relatively large Förster radius of 50 Å [33], was chosen for this study. Since CLS does not contain an endogenous cysteine residue, residue Ser329 in the cytoplasmic domain of CLS was mutated to cysteine to allow for a specific thiol-dependent conjugation with fluorescein or TMR (Table 1). Figure 5 depicts the conjugation process. Although unlabeled CLS_S329C coelutes with both fluorescein-conjugated CLS_S329C (termed fluorescein-CLS) and TMR-conjugated CLS_S329C (TMR-CLS) in analytical HPLC, the extent of conjugation could be monitored by the increase of absorbance at the visible wavelength unique to the fluorophore (Fig. 5A,B). After the reaction was complete, fluorophore-conjugated CLS was purified by HPLC. Characterization by mass spectrometry confirmed the conjugation and also showed that the labeling efficiency was approximately 90% (Fig. 5C). Finally, both fluorescein-CLS and TMR-CLS exhibited a slow migrational rate in SDS gels similar to that of unlabeled CLS, indicating that conjugation of these fluorescent groups to CLS does not significantly alter its electrophoretic property (Fig. 5D).
Figure 5. Thiol-specific conjugation of a fluorophore to CLS_S329C.
(A) Overlaid HPLC traces of aliquots of a TMR-conjugating reaction that were taken at indicated time points. Only the TMR-conjugated peptide had the absorbance at 540 nm. For 0-min time point, an aliquot was taken prior to the addition of TMR-5-maleimide. (B) Pseudo-first-order kinetics of the conjugating reaction. The area under the HPLC peak at 25.6 min is plotted against the time at which the aliquot was taken from the reaction and quenched. The plot was fit to a single exponential curve (dashed line) with a time constant of 30.5 min. (C) MALDI-TOF mass spectrum of TMR-CLS. Major peaks are labeled with the mass and the assigned peptide. The ratio of the TMR-CLS peak intensity versus the unlabeled CLS peak is approximately 90%. (D) SDS polyacrylamide gels showing fluorescence conjugation of CLS does not alter the slow electrophoretic rate of CLS. Both fluorescein-CLS and TMR-CLS were not stained by Commassie blue, but directly visualized. Pre-stained molecular weight markers were shown on the left.
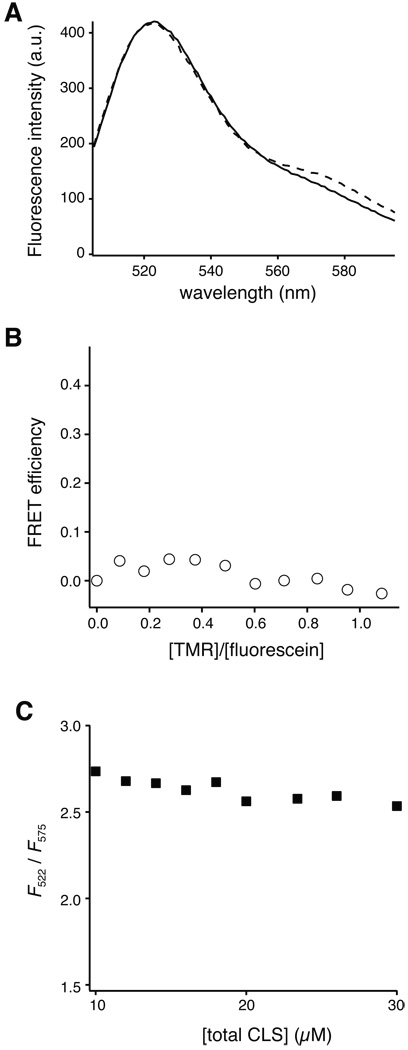
To determine the oligomeric state of CLS in DPC micelles by FRET, the fluorescence intensity of fluorescein in fluorescein-CLS was monitored with titration of TMR-CLS as the acceptor fluorophore. When the total protein concentration and the protein/detergent ratio are kept constant during the titration, the FRET efficiency will increase proportionally with the acceptor/donor molar ratio if the protein is dimeric [30, 32, 34]. However, little or no FRET was observed between fluorescein-CLS and TMR-CLS that were reconstituted in DPC micelles at a 1/500 protein/detergent molar ratio (Fig. 6A). It did not increase significantly even when the TMR-CLS concentration became higher than fluorescein-CLS (Fig. 6B). Moreover, when the total protein and detergent concentrations were increased but with the protein/detergent molar ratio kept constant, only a small degree of non-specific FRET was observed (Fig. 6C). Thus, these results indicate that fluorescein-CLS and TMR-CLS do not associate with each other in DPC micelles. In other words, CLS is a monomer in DPC micelles.
Figure 6. Lack of fluorescence resonance energy transfer between fluorophore-conjugated CLS.
(A) Fluorescence emission spectra of fluorescently labeled CLS in 5 mM DPC and 10 mM MOPS, pH 7.4 at 20 °C. Emission spectra were collected with excitation at 492 nm. Solid line, 2 µM fluorescein-CLS and 8 µM unlabeled CLS; dashed line, 2 µM fluorescein-CLS, 1.7 µM TMR-CLS and 6.3 µM unlabeled CLS. (B) Lack of FRET between fluorescein-CLS and TMR-CLS, even with increasing ratios of TMR over fluorescein. TMR-CLS as the acceptor fluorophore was titrated to fluorescein-CLS in 5 mM DPC with fluorescein-CLS and overall CLS concentrations and the CLS/DPC molar ratio being kept constant (see Materials and Methods for details). FRET efficiency was calculated as (F0-F)/F0, where F is the fluorescein emission intensity at 522 nm in a given condition and F0 is that in the absence of TMR-CLS. (C) The extent of FRET as a function of total CLS concentration, with the CLS/DPC molar ratio kept constant at 1/500. The ratio of fluorescein-CLS and TMR-CLS was 1:2. The extent of FRET is indicated by F522/F575, the ratio of emission intensity at 522 nm (fluorescein emission) versus that at 575 nm (TMR emission).
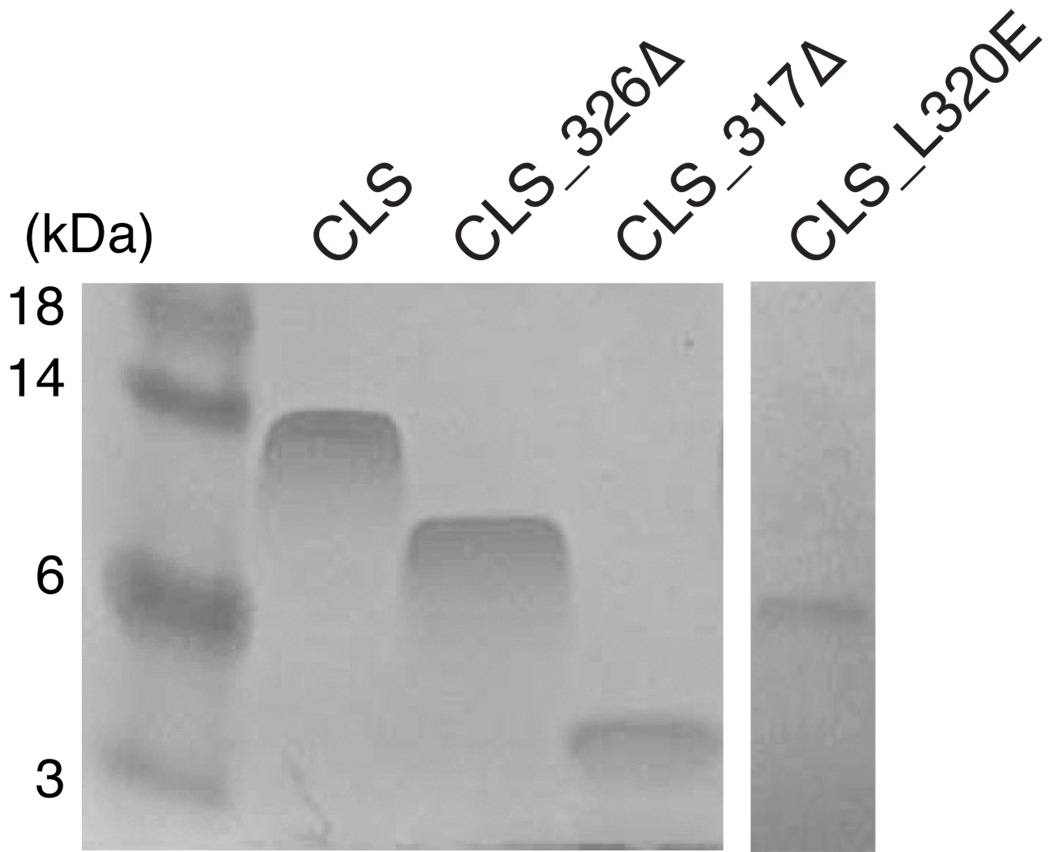
Finally, mutagenesis was used to identify the residues that confer CLS its anomalous electrophoretic rate in SDS-PAGE. As shown in Figure 7, although the mutant protein CLS_326Δ lacked residues in the membrane-distal region of the cytoplasmic domain and electrophoresed at a faster rate than the wild-type CLS, its migration in the SDS gel (at the approximately 8–9 kDa) was still significantly slower than what would be expected from its molecular weight of 5,475 Da. In contrast, the CLS_317Δ protein, which lacks the entire cytoplasmic domain, electrophoresed at a rate that was much faster than CLS and CLS_326Δ but consistent with its molecular weight of 4,378 Da. Moreover, changing residue Leu320 in CLS to Glu abolished the ability of CLS to migrate slowly in the SDS gel (Fig. 7). These results suggest that residues located in the membrane-proximal region of the cytoplasmic domain of CLS are responsible for its slow electrophoretic rate in SDS-PAGE.
Figure 7. Truncations or mutation of the juxtamembrane region of CLS abolished its slow electrophoretic rate in SDS-PAGE.
Equal amount (5 µg) of purified CLS proteins were loaded onto 15% Tris-glycine SDS polyacrylamide gel. After electrophoresis, the protein bands were stained by colloidal Coomassie blue. Molecular weight markers are shown on the left labeled with corresponding sizes in kDa.
Discussion
Here we have reported evidence to support the conclusion that L-selectin TM and cytoplasmic domains lack the ability to dimerize in cell membranes. TOXCAT studies indicated little dimerization activity of the L-selectin TM domain in a cell membrane. Equilibrium sedimentation and FRET analysis of CLS also showed it to be a monomer in zwitterionic detergent micelles. Although CLS migrates in SDS polyacrylamide gels at an abnormally slow speed, follow-up studies showed that such an anomalous electrophoretic rate is not due to protein dimerization in SDS because adding 6 M urea to the gel, boiling the CLS sample before loading, or varying the amount of CLS loaded did not alter significantly the electrophoretic rate of CLS (data not shown). It is instead due to the residues in the membrane-proximal region of the cytoplasmic domain in CLS because their removal or mutation led CLS to migrate in SDS-PAGE at a rate consistent with its molecular weight (Fig. 7).
For most proteins, SDS, a strong anionic detergent, is an effective denaturant. These proteins are in the denatured form during SDS-PAGE, which provides the foundation for the well-documented correlation between a protein’s electrophoretic mobility and its molecular weight. In contrast, a number of small membrane proteins are able to maintain their helical conformations in SDS presumably because to them SDS is more a detergent to provide a membrane-mimicking environment rather than a denaturant. In many cases, these membrane proteins can interact with one another through lateral helical interactions in SDS [35]. As a consequence, SDS-PAGE has been used extensively to detect and monitor the presence of membrane protein oligomers [e.g. 11, 22, 23, 36]. However, recent reports also revealed factors other than oligomerization that can influence electrophoretic mobility of a membrane protein in SDS-PAGE. Deber and colleagues showed that the electrophoretic rate of a helix-loop-helix sequence derived from human cystic fibrosis transmembrane conductance regulator protein in SDS-PAGE correlates largely with the amount of bound SDS [37]. Wimley and colleagues also showed that polar residues positioned in a transmembrane helix can decrease the electrophoretic mobility without affecting helix dimerization [38]. In CLS, the membrane-proximal region of the cytoplasmic domain is enriched with positively charged residues, which may influence the binding of SDS to the protein. This is consistent with our observation that the anionic head group of SDS can influence the conformation of CLS as judged by CD spectroscopy (Fig. 3). Thus, removal of the membrane-proximal region from CLS (i.e. CLS_317Δ) or adding a negatively charged residue to this region (i.e. CLS_L320E) could conceivably change SDS binding to CLS and as a consequence altered its electrophoretic rate in SDS gels.
Clustering of selectins on the cell surface helps to mediate leukocyte tethering and rolling under flow by facilitating the interaction between leukocytes and their interacting cells [15, 17]. L-selectin is found to cluster in the microvilli of leukocytes [39]. Recently the TM domain of L-selectin was shown to be important for targeting L-selectin to the microvilli [40]. The interaction of the L-selectin cytoplasmic domain with intracellular cytoskeletal proteins may not be essential to its microvillar positioning [41, 42]. Similarly, removal of the cytoplasmic domain of PSGL-1, L-selectin’s counter-receptor, does not disrupt localization of PSGL-1 in the microvilli [43]. However, unlike PSGL-1 and its close homolog, P-selectin, we showed in this study that TM domain of L-selectin are not capable of self-association in a membrane environment. Thus, it appears that TM dimerization may not be a prerequisite for microvillar positioning. Features in the TM domain of L-selectin that are responsible for targeting and clustering of L-selectin in the microvilli remain to be discovered. One possibility is that the TM domain of L-selectin associates directly with another membrane protein that is specifically localized and clustered in the microvilli. It is also possible that the TM domain influences or facilitates the interaction of its adjacent cytoplasmic residues with certain intracellular proteins that are involved in targeting proteins to the microvilli [42]. Nonetheless, our in vitro characterization of TM and cytoplasmic domains of L-selectin has provided constraints for future investigations and may help to shed insights on the mechanism of L-selectin interaction with its associating proteins.
Acknowledgement
We thank Dr. James Lear for sharing the analysis program for analytical ultracentrifugation. This work was supported by a National Institutes of Health grant GM084175.
Abbreviations
- CAT
chloramphenicol acetyl transferase
- CD
circular dichroism
- DPC
dodecylphosphocholine
- FRET
fluorescence resonance energy transfer
- GST
glutathione S-transferase
- HFIP
hexafluoroisopropanol
- HPLC
high performance liquid chromatography
- PSGL-1
P-selectin glycoprotein ligand-1
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCEP·HCl
Tris(2-carboxyethyl) phosphine hydrochloride
- TM
transmembrane
- TMR
tetramethylrhodamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moore DT, Berger BW, DeGrado WF. Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 3.Kansas GS. Selectins and their ligands: current concepts and controversies. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- 4.Patel KD, Cuvelier SL, Wiehler S. Selectins: critical mediators of leukocyte recruitment. Semin Immunol. 2002;14:73–81. doi: 10.1006/smim.2001.0344. [DOI] [PubMed] [Google Scholar]
- 5.Foxall C, Watson SR, Dowbenko D, Fennie C, Lasky LA, Kiso M, Hasegawa A, Asa D, Brandley BK. The three members of the selectin receptor family recognize a common carbohydrate epitope, the sialyl Lewis(x) oligosaccharide. J. Cell Biol. 1992;117:895–902. doi: 10.1083/jcb.117.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 7.Barkalow FJ, Barkalow KL, Mayadas TN. Dimerization of P-selectin in platelets and endothelial cells. Blood. 2000;96:3070–3077. [PubMed] [Google Scholar]
- 8.Ushiyama S, Laue TM, Moore KL, Erickson HP, McEver RP. Structural and functional characterization of monomeric soluble P-selectin and comparison with membrane P-selectin. J. Biol. Chem. 1993;268:15229–15237. [PubMed] [Google Scholar]
- 9.Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane α-helices. Nat. Struct. Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 10.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 11.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane α-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 12.Moore KL, Stults NL, Diaz S, Smith DF, Cummings RD, Varki A, McEver RP. Identification of a specific glycoprotein ligand for P-selectin (CD62) on myeloid cells. J. Cell Biol. 1992;118:445–456. doi: 10.1083/jcb.118.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez M, Joffraud M, Giraud S, Baisse B, Bernimoulin MP, Schapira M, Spertini O. Regulation of PSGL-1 interactions with L-selectin, P-selectin, and E-selectin: role of human fucosyltransferase-IV and -VII. J. Biol. Chem. 2005;280:5378–5390. doi: 10.1074/jbc.M410899200. [DOI] [PubMed] [Google Scholar]
- 14.Epperson TK, Patel KD, McEver RP, Cummings RD. Noncovalent association of P-selectin glycoprotein ligand-1 and minimal determinants for binding to P-selectin. J. Biol. Chem. 2000;275:7839–7853. doi: 10.1074/jbc.275.11.7839. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran V, Yago T, Epperson TK, Kobzdej MM, Nollert MU, Cummings RD, Zhu C, McEver RP. Dimerization of a selectin and its ligand stabilizes cell rolling and enhances tether strength in shear flow. Proc. Natl. Acad. Sci. USA. 2001;98:10166–10171. doi: 10.1073/pnas.171248098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Steeber DA, Tang ML, Farrar MA, Perlmutter RM, Tedder TF. Regulation of L-selectin-mediated rolling through receptor dimerization. J. Exp. Med. 1998;188:1385–1390. doi: 10.1084/jem.188.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dwir O, Steeber DA, Schwarz US, Camphausen RT, Kansas GS, Tedder TF, Alon R. L-selectin dimerization enhances tether formation to properly spaced ligand. J. Biol. Chem. 2002;277:21130–21139. doi: 10.1074/jbc.M201999200. [DOI] [PubMed] [Google Scholar]
- 18.Luo S-Z, Mo X, Afshar-Kharghan V, Srinivasan S, Lopez JA, Li R. Glycoprotein Ibα forms disulfide bonds with 2 glycoprotein Ibβ subunits in the resting platelet. Blood. 2007;109:603–609. doi: 10.1182/blood-2006-05-024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo SZ, Li R. Specific heteromeric association of four transmembrane peptides derived from platelet glycoprotein Ib-IX complex. J. Mol. Biol. 2008;382:448–457. doi: 10.1016/j.jmb.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc. Natl. Acad. Sci. USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS. Dimerization of the transmembrane domain of integrin αIIb subunit in cell membranes. J. Biol. Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 23.Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the "BH3-only" protein BNIP3 in membranes and detergent. J. Biol. Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 24.Laue T, Shaw BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, U. K.: The Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 25.Kochendoerfer GG, Salom D, Lear JD, Wilk-Orescan R, Kent SBH, DeGrado WF. Total chemical synthesis of the integral membrane protein influenza A virus M2: role of its C-terminal domain in tetramer assembly. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 26.Mackenzie KR. Folding and stability of α-helical integral membrane proteins. Chem. Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 27.Langosch D, Brosig B, Kolmar H, Fritz HJ. Dimerisation of the glycophorin A transmembrane segment in membranes probed with the ToxR transcription activator. J. Mol. Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 28.Rapaport D, Shai Y. Aggregation and organization of pardaxin in phospholipid membranes. A fluorescence energy transfer study. J. Biol. Chem. 1992;267:6502–6509. [PubMed] [Google Scholar]
- 29.Adair BD, Engelman DM. Glycophorin A helical transmembrane domains dimerize in phospholipid bilayers: a resonance energy transfer study. Biochemistry. 1994;33:5539–5544. doi: 10.1021/bi00184a024. [DOI] [PubMed] [Google Scholar]
- 30.Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 31.You M, Li E, Wimley WC, Hristova K. Forster resonance energy transfer in liposomes: measurements of transmembrane helix dimerization in the native bilayer environment. Anal. Biochem. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Reddy LG, Bennett R, Silva ND, Jr, Jones LR, Thomas DD. A fluorescence energy transfer method for analyzing protein oligomeric structure: application to phospholamban. Biophys. J. 1999;76:2587–2599. doi: 10.1016/S0006-3495(99)77411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P, Brand L. Resonance energy transfer: methods and applications. Anal. Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 34.Fisher LE, Engelman DM, Sturgis JN. Detergents modulate dimerization, but not helicity, of the glycophorin A transmembrane domain. J. Mol. Biol. 1999;293:639–651. doi: 10.1006/jmbi.1999.3126. [DOI] [PubMed] [Google Scholar]
- 35.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. The affinity of GxxxG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J. Biol. Chem. 2004 doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 36.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 37.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walkenhorst WF, Merzlyakov M, Hristova K, Wimley WC. Polar residues in transmembrane helices can decrease electrophoretic mobility in polyacrylamide gels without causing helix dimerization. Biochim Biophys Acta. 2009;1788:1321–1331. doi: 10.1016/j.bbamem.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Andrian UH, Hasslen SR, Nelson RD, Erlandsen SL, Butcher EC. A central role for microvillous receptor presentation in leukocyte adhesion under flow. Cell. 1995;82:989–999. doi: 10.1016/0092-8674(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 40.Buscher K, Riese SB, Shakibaei M, Reich C, Dernedde J, Tauber R, Ley K. The transmembrane domains of L-selectin and CD44 regulate receptor cell surface positioning and leukocyte adhesion under flow. J. Biol. Chem. 2010;285:13490–13497. doi: 10.1074/jbc.M110.102640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavalko FM, Walker DM, Graham L, Goheen M, Doerschuk CM, Kansas GS. The cytoplasmic domain of L-selectin interacts with cytoskeletal proteins via alpha-actinin: receptor positioning in microvilli does not require interaction with alpha-actinin. J. Cell Biol. 1995;129:1155–1164. doi: 10.1083/jcb.129.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivetic A, Florey O, Deka J, Haskard DO, Ager A, Ridley AJ. Mutagenesis of the ezrinradixin-moesin binding domain of L-selectin tail affects shedding, microvillar positioning, and leukocyte tethering. J. Biol. Chem. 2004;279:33263–33272. doi: 10.1074/jbc.M312212200. [DOI] [PubMed] [Google Scholar]
- 43.Miner JJ, Xia L, Yago T, Kappelmayer J, Liu Z, Klopocki AG, Shao B, McDaniel JM, Setiadi H, Schmidtke DW, McEver RP. Separable requirements for cytoplasmic domain of PSGL-1 in leukocyte rolling and signaling under flow. Blood. 2008;112:2035–2045. doi: 10.1182/blood-2008-04-149468. [DOI] [PMC free article] [PubMed] [Google Scholar]