Abstract
Purpose
To determine the differential gene expression between oral squamous cell carcinoma (OSCC) with and without metastasis to cervical lymph nodes and to assess prediction of nodal metastasis using molecular features.
Experimental Design
We used Affymetrix U133 2.0 plus arrays to compare the tumor genome-wide gene expression of 73 node-positive OSCC with 40 node-negative (≥18 months) OSCC. Multivariate linear regression was used to estimate the association between gene expression and nodal metastasis. Stepwise logistic regression and Receiver Operating Characteristics (ROC) analysis were used to generate predictive models and to compare these with models using tumor size alone.
Results
We identified five genes differentially expressed between node-positive and node-negative OSCC after adjusting for tumor size and Human Papillomavirus status: REEP1, RNF145, CTONG2002744, MYO5A and FBXO32. Stepwise regression identified a four-gene model (MYO5A, RFN145, FBXO32 and CTONG2002744) as the most predictive of nodal metastasis. A leave-one-out ROC analysis revealed that our model had a higher Area Under the Curve (AUC) for identifying occult nodal metastasis compared to that of a model using tumor size alone (respective AUC: 0.85 and 0.61; p = 0.011). A model combining tumor size and gene expression did not further improve prediction of occult metastasis. Independent validation using 31 metastatic and 13 non-metastatic cases revealed a significant under-expression of CTONG2002744 (p = 0.0004).
Conclusions
These results suggest that our gene expression markers of OSCC metastasis hold promise for improving current clinical practice. Confirmation by others and functional studies of CTONG2002744 are warranted.
Keywords: oral squamous cell carcinoma, oral cancer, genetic expression profiles, gene expression profiling, metastasis, microarrays
Introduction
In oral cancer, as with other head and neck squamous cell carcinomas, metastasis occurs first in the cervical lymph nodes and is an important predictor of poor prognosis (1,2). One of the impediments to the effective management of OSCC is that for patients with stage I/II disease (non-metastatic), our only means for predicting risk for cervical lymph node spread is tumor size and location. Overall, the metastatic potential is 30% or above for oral tumors of T2 to T4 staging (3,4). The rate of metastatic potential can vary according to site of a particular tumor within the oral cavity (5,6). Thus, the management of the patient without clinically apparent neck nodal metastasis (the so-called N0 neck) can range from close observation with clinical and radiographic exams (patients with T1 tumors) to surgery or radiation to the neck (T2-4 tumors). For this reason, accurate risk stratification is of paramount importance to either avoid the potential morbidity of over-treatment or prevent further progression of disease.
Recent evidence has suggested that the process of metastasis may be associated with specific pattern of genetic changes within a tumor that enables certain cells to invade surrounding tissue and/or travel to and seed in regional or distant locations (7,8). Further, our previous work has shown that gene expression changes seen in tumor cells from metastatic lymph nodes could be detected in the primary site as well (9). In an effort to identify genes whose expression change is associated with nodal metastasis, we assessed the genome-wide differential expression of metastatic vs. non-metastatic OSCC. We then evaluated our top candidate genes for their ability to detect occult nodal metastasis among clinically node-negative OSCC. We also explored whether gene expression profiles found to distinguish OSCC with or without nodal metastasis were associated with a difference in survival.
Materials and Methods
Study population
We identified English-speaking patients 18 year of age or older with a first, primary OSCC or dysplasia between December 16th, 2003 and April 17th, 2007 at any of three University of Washington-affiliated hospitals as previously described (10). The RNA was isolated and hybridized onto Affymetrix HG-U133 Plus 2.0 arrays as previously described (10). Out of 187 recruited subjects, we had quality-controlled array results for 167 subjects. The DNA was typed for Human Papillomavirus (HPV) as previously described (11). Normalization and filtering of gene expression data were done as previously described (10), after which ~25,000 probe sets remained for differential expression analyses.
Determining differential gene expression associated with nodal metastasis
Among the 167 OSCC cases with gene expression data, nodal metastasis cases were defined as those OSCC patients with pathologically-confirmed nodal metastasis (n=73). Non-metastatic cases were OSCC patients who either 1) presented without clinical evidence of nodal metastasis (as determined by physical examination and computer tomography (CT) scans ± positron emission tomography (PET)), or 2) if pathologic nodal status was unknown (only for T1 tumors), patients had to be free of nodal metastasis for at least 18 months from diagnosis and not have received radiation or chemotherapy to the neck nodes (n=40). To determine the association between OSCC nodal status and gene expression, we utilized a regression-based analysis implemented by GenePlus software adjusting also for tumor size (T1-4) and HPV status (negative/low risk vs. high risk) (12). For this comparison, we used the number of false discovery (NFD) ≤1 as the type I error selection criterion (13).
Prediction modeling
To build a gene expression prediction model for metastatic OSCC, we performed stepwise logistic regression based on the probe sets differentially expressed between OSCC cases with nodal metastases vs. those without using SAS version 9.2. For the stepwise regression, the significance level for entrance and exit were set at 0.01 and 0.05, respectively. We performed a jackknife leave-one-out analysis to quantify the ability of the model to predict nodal metastasis, which attempts to reduce the over-optimism of ROC and AUC estimates due to using the same data both to estimate and assess the predictive ability of risk scores (14). Coefficient estimates for the risk model were obtained after excluding one subject, and the resulted risk model was used to generate a risk score using the excluded subject’s gene expression and/or stage characteristics. This process was repeated until risk scores were assigned to each subject. ROC and AUC estimates were calculated for these jackknife risk scores.
Comparing prediction of occult metastasis by tumor size vs. gene expression models
We identified a subset of cases with occult nodal metastasis (as determined by physical examination and imaging studies) to determine how gene expression compared with tumor size as the main criteria by which to predict nodal metastasis (n=20). Occult metastatic disease was determined by the presence of pathologic lymph nodes following a prophylactic lymphadenectomy in patients deemed to be clinically node-negative at presentation by physical examination and CT scan. Non-metastatic cases were used as the comparison group (n=40). Risk scores were generated using a jackknife leave-one-out analysis for the gene expression model, and two other models, one based on tumor size alone and another combining gene expression with tumor size. For each model, we constructed ROC curves and calculated AUC to quantify the ability of each model to predict occult metastasis. One thousand bootstrap samples were generated to estimate standard errors and 95% confidence intervals for AUC estimates, and to obtain p-values for testing the null hypothesis that specific gene expression values do not add to ability of tumor size to predict occult metastasis.
Survival analysis
To determine if the probe sets found to distinguish OSCC cases with and without nodal metastases were also associated with survival, we analyzed expression array results for the subset of the 167 OSCC patients for which (n=150) we had at least four months of follow-up time. The requirement for our study participants to have at least four months follow-up refers to the starting point at which we began to capture events. This was done because we did not want to include any events until participants had completed treatment to avoid capturing deaths from patients who died due to co-morbidities rather than tumor biology. Follow-up time for analyses of survival for the 150 OSCC cases was calculated from the date of surgery to the date of death, loss-to-follow-up, or April 30, 2007, whichever came first, according to the Kaplan-Meier method. Cox-proportional hazards regression model was used to estimate overall and OSCC-specific survival associations with gene expression, age, sex (male vs. female) and tumor size (T1-4). These statistical analyses were conducted using STATA software version 10.
Validation of expression array results by qRT-PCR
We performed validation testing by RT-PCR for CTONG2002744 (EST uc001pfj.1, UCSC build Human Mar. 2006 (hg18)), FBX032, MYO5A, and RNF145. To determine whether the correlation between Affymetrix expression data and qRT-PCR, we assessed the correlation between these two platforms for all four genes. We performed a scatterplot, a simple correlation and a linear regression adjusted for nodal metastasis status in a random set of 30 samples (from the training set) from which we chose to re-test the expression of the four probe sets via qRT-PCR. Using this adjusted linear regression, we obtained a p-value for the association between Affymetrix expression values and qRT-PCR (the scatterplot results are shown Supplementary Information Figure S1). To obtain a testing set for independent validation, we then applied the same eligibility criteria above to identify eligible cases that had been recruited from the beginning of the study until March 31st, 2009. We used the RT-PCR data from this subset of subjects to estimate the statistical power to detect a difference in mean expression between this independent cohort of metastasis (n=31) and non-metastatic (n=13) cases that would provide a full validation of the Affymetrix platform. Total RNA from these samples was processed in the same fashion as those in the training set. For CTONG2002744, primers were designed using online primer design software (http://www.ncbi.nlm.nih.gov/tools/primer-blast). To confirm target specificity of the primers, three samples were sequenced and confirmed against sequences identified in the UCSC Genome Browser, NCBI, and Affymetrix NetAffx databases. The primer sequences 5’- ATGCCCTCCACATGAATCAG-3’ and 5’-TTCCTTCTGCATCTGTCCAA-3’ amplified a 215-bp amplicon. For FBXO32, MYO5A, RNF145, and the reference gene ACTB, bioinformatically validated QuantiTect primers (Qiagen, Valencia, CA) were used. The FBOX32 primers amplified a 106-bp amplicon spanning exons 8 and 9. The MYO5A primers amplified a 74-bp amplicon spanning exons 9 and 10. The RNF145 primers amplified a 148-bp amplicon spanning exons 3,4, and 5. The BACT primers amplified a 146-bp amplicon spanning exons 3 and 4. Each sample was assayed in triplicate in 10 µl reactions using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA), and the QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, CA). All samples were run on a 7900HT Sequence Detection System (ABI, Foster City, CA). The cycling conditions were as follows: 1) RT reaction with a 2 minute incubation at 42° C, 2 minute on ice, 30 minute incubation at 42° C, and 3 minute at 95° C; and 2) cDNA amplification – 95° C for 15 minutes, followed by 40 cycles of 15 seconds at 94° C, 30 seconds at 55° C, and 30 seconds at 72° C. Ten point standard curves were generated using Total RNA – Human Normal Tissue – Tongue (Biochain Institute, Inc., Hayward, CA) for the four test genes and Universal Human Reference RNA (Stratagene, La Jolla, CA) for ACTB. The linear correlation coefficient (R2) was 0.99 or greater for all runs. The mean Ct values were calculated from the triplicate Ct values. Samples with standard deviation of the Ct values greater than 0.3 in the triplicate test were repeated. Mean Ct values were further adjusted in relation to the mean Ct value of the ACTB gene. For each probe set the, we calculated the difference between the mean Ct value of the probe set and the mean Ct value of the ACTB reference gene. This represented the delta Ct value for each probe set. Given that lower Ct values pertain to higher expression for any given cDNA, we multiplied the delta Ct value by (-1) so that a higher "transformed" value correlated with higher expression values. Differences in “transformed” Ct values between metastatic vs. non-metastatic cases were calculated using a two-tailed t-test of unequal variance in Microsoft Office Excel 2007.
Results
Associations with nodal metastasis
The selected characteristics of OSCC cases with and without nodal metastases are described in Table 1. Cases with nodal metastasis had a higher proportion of T2, whereas non-metastatic cases were more likely to have T1 tumors. About one third of the cases presented with clinically node-negative disease, but later were found to have occult metastasis upon prophylactic lymphadenectomy. Cases with and without nodal metastasis had a similar age, sex, and HPV DNA distribution (Table 1). Cases without nodal metastases had a higher proportion of oral cavity vs. oropharyngeal tumors than cases with nodal metastases. Since the tissue processed represented a biopsy sample from the patients’ tumors with a mixture of tumor cells and stroma, we also examined percent of tumor content. We had detailed tumor content data for 101 out of 113 cases. The proportion of tumor content for metastatic and non-metastatic cases is presented in Table S1, Supplementary Information. Overall, although non-metastatic cases appear to have a higher invasive percentage (33% have 75%+ invasive, compared to 15.4% of metastatic cases), non-metastatic cases also have more that have low invasive percent (19.4% vs. 9.2% have <10% invasive). The mean for metastatic cases is 46.2% vs. 51.2% for non-metastatic cases. A t-test comparing invasive percentage did not reveal a statistically significant difference between these two groups (p-value for the difference is 0.4038).
Table 1.
Characteristics of the 113 OSCC Cases With and Without Nodal Metastasis
| OSCC without nodal metastasis |
OSCC with nodal metastasis |
|||
|---|---|---|---|---|
| (n=40) | (n=73) | |||
| n | (%) | n | (%) | |
| Age at reference (years) | ||||
| 23–49 | 7 | (17.5) | 17 | (23.3) |
| 50–59 | 13 | (32.5) | 26 | (35.6) |
| 60–69 | 12 | (30.0) | 17 | (23.3) |
| 70–88 | 8 | (20.0) | 13 | (17.8) |
| Sex | ||||
| M | 26 | (65.0) | 50 | (68.5) |
| F | 14 | (35.0) | 23 | (31.5) |
| Tumor Stage | ||||
| 1 (<2cm) | 19 | (47.5) | 14 | (19.2) |
| 2 (≥2cm,<4cm) | 8 | (20.0) | 32 | (43.8) |
| 3 (≥4cm) | 3 | (7.5) | 10 | (13.7) |
| 4 (invading adjacent structures) | 10 | (25.0) | 17 | (23.3) |
| Nodal Stage (Pathologic) | ||||
| Not determined | 6 | (15.0) | 0 | 0.0 |
| 0 | 34 | (85.0) | 0 | 0.0 |
| 1 | 0 | 0.0 | 19 | (26.0) |
| 2 | 0 | 0.0 | 52 | (71.2) |
| 3 | 0 | 0.0 | 2 | (2.7) |
| HPV 16 DNA | ||||
| Negative | 30 | (75.0) | 46 | (63.0) |
| Positive | 10 | (25.0) | 27 | (37.0) |
| Tumor Site | ||||
| Oral | 35 | (87.5) | 50 | (68.5) |
| Oropharyngeal | 5 | (12.5) | 23 | (31.5) |
Probe sets associated with nodal metastasis
Results from our regression analysis adjusting for tumor size and HPV status identified five genes (six probe sets) that were differentially expressed between nodal metastasis cases and non-metastatic cases (NFD ≤ 1): 1) REEP1 probe set id 204364_s_at and 204365_s_at; 2) RNF145; 3) clone CTONG2002744 probe set id 225123_at; 4) MYO5A probe set id 227761_at and 5) FBXO32 probe set id 232573_at. Stepwise regression identified a model with four of these probe sets (MYO5A, RFN145, FBXO32 and clone CTONG2002744 as the most predictive of nodal metastasis with an AUC of 0.87 (see Table S2 in Supplemental Information for normalized, log-transformed gene expression values for these four probe sets).
We evaluated how using expression data for these probe sets compares with the use of tumor size (< 2 cm (T1) vs. ≥ 2 cm (T2-4)), a general clinical guideline, to decide prophylactive treatment to cervical lymph nodes for OSCC patients with clinically node-negative disease. Among the 113 OSCC cases used to determine the differential expression in nodal metastasis, 61 patients presented with clinically node-negative OSCC. Two of these subjects did not have imaging data. One of 61 patients had unknown clinical T staging. Out of the remaining 58 patients, 49 had tumors greater than 2 cm as measured by clinical exam, and all had a prophylactive neck dissection on this basis. Of these, 18 had occult metastatic nodes as determined pathologically. The sensitivity and specificity of clinical tumor stage (T1 vs. T2-T4) to predict occult nodal metastasis was 94.7% (18 out of 19) and 20.5% (i.e., only 8 out of 39 would be correctly diagnosed and 31 would have been misdiagnosed), respectively. Only 37% (18 out of 49) of all cervical lymphadenectomies performed showed positive lymph nodes. To compare clinical criteria with gene expression in diagnosing occult metastasis, we compared the scores from the 4-gene signature model with those of a model using tumor size alone in our cohort of patients with clinically node-negative OSCC. A significantly higher AUC was observed for the 4-gene signature model than for a model using tumor size alone (AUC= 0.85 vs. 0.61, respectively (p<0.011)) (Figure 1). From this analysis, we determined that setting a threshold cutoff point so that the sensitivity of the 4-gene signature model is no worse than a model using tumor size alone (i.e. 95%), then the specificity of the 4-gene signature model would be 76.9%. Thus, if applied to our cohort, the 4-gene signature model would have missed 1 out of 20 occult metastatic cases, and in turn, only 9 out of 39 node-negative cases would have been incorrectly diagnosed as node-positive cases.
Figure 1.
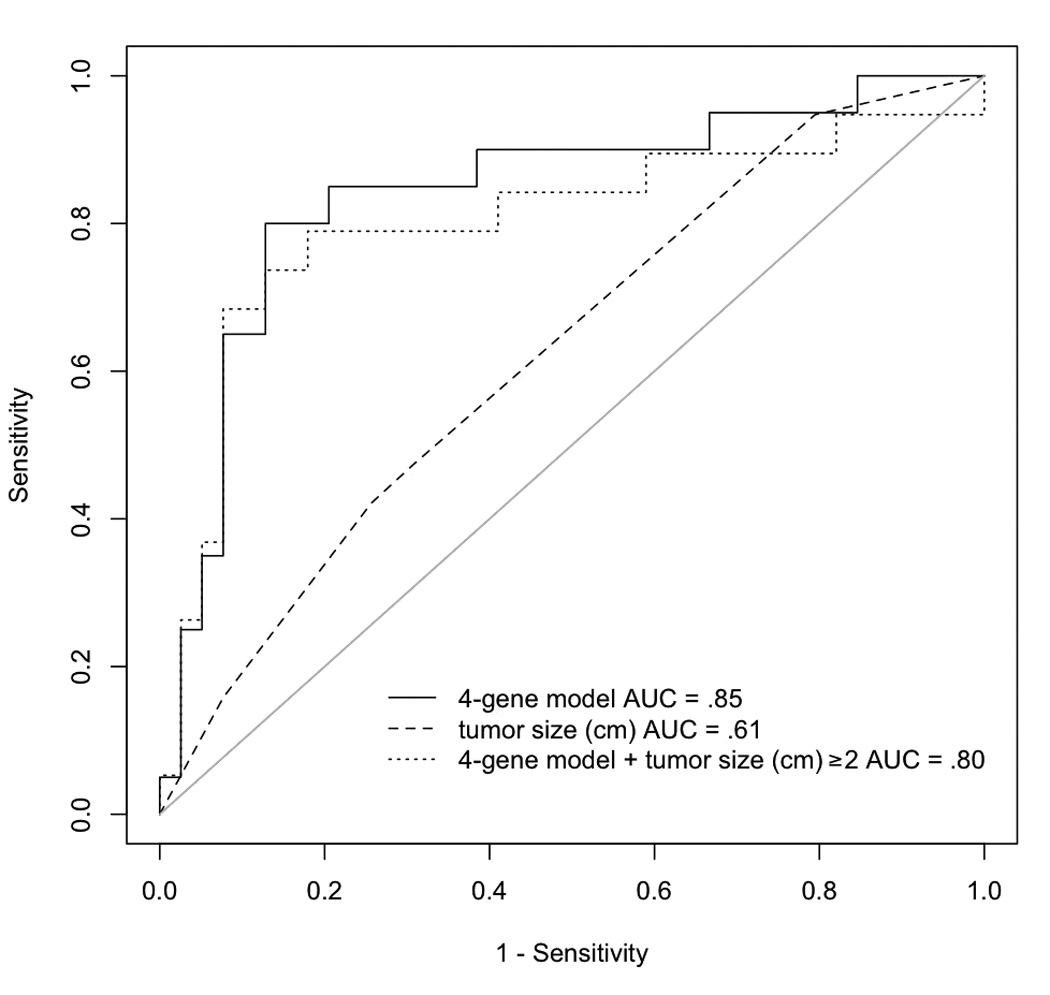
Receiver Operating Characteristic analysis comparing the prediction of occult metastasis for Tumor Stage and gene expression data.
Survival analysis
The baseline characteristics of the 150 OSCC patients included in the survival analysis were presented previously in Mendez et al. (Table S1, Supplementary Information) (15). The range of follow-up time for patients known to be alive at the end of the study was 10.7 to 38.7 months, with a median of 22.2 months. Forty-one patients had died by the end of the follow-up period; there were 27 OSCC-specific deaths, 10 non-OSCC-specific deaths and four deaths of unknown causes. To test the hypothesis that these four probe sets associated with nodal status were associated with survival, we estimated hazard ratios (HR) for both overall and OSCC-specific mortality adjusting for age, sex and tumor size. Results from these analyses are shown in Table 2. The 4-gene signature model was independently associated with overall and OSCC-specific survival (HR (95% CI) = 1.21 (1.07–1.36) and 1.19 (1.02–1.38), respectively). In addition, consistent with the direction of gene expression change associated with metastasis, down-regulation of the CTONG2002744 clone and up-regulation of the FBXO32 gene were associated with worse overall and OSCC-specific survival.
Table 2.
Multivariate Analysis for Overall and OSCC-specific Survival
| Overall | OSCC-specific | |||
|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Age | 1.01 (0.98–1.04) | 0.37 | 1.01 (0.97–1.05) | 0.66 |
| Sex (Female vs. Male) | 0.74 (0.36–1.52) | 0.42 | 0.67 (0.27–1.65) | 0.38 |
| Tumor size* | 1.29 (0.99–1.69) | 0.061 | 1.47 (1.06–2.05) | 0.02 |
|
Gene expression |
||||
| Probe sets |
||||
| CTONG2002744 | 0.63 (0.45–0.88) | 0.007 | 0.64 (0.42–0.97) | 0.04 |
| RNF145 | 1.57 (0.93–2.67) | 0.09 | 1.77 (0.93–3.37) | 0.08 |
| FBOX32 | 1.83 (1.28–2.63) | 0.001 | 1.74 (1.10–2.74) | 0.02 |
| MYO5A | 1.31 (0.88–1.96) | 0.19 | 1.41 (0.86–2.33) | 0.18 |
| 4-gene signature | 1.21 (1.07–1.36) | 0.002 | 1.19 (1.02–1.38) | 0.02 |
T1/2 vs. T3/4
HR: Hazard Ratio
P -value derived from a Wald test for each variable in the model
CI: Confidence Interval
Validation of expression array results by qRT-PCR
The correlation coefficients for the CTONG2002744 clone, MYO5A, RNF145 and FBXO32 between microarray and qRT-PCR expression data for the 30 samples assayed from the training set were 0.87 (p <0.000; 95% CI: 0.6766082, 1.116981), 0.29 (p<0.475; 95% CI: - 0.1958988, 0.409744), 0.38 (p<0.005; 95% CI: 0.1227414, 0.6184207) and 0.66 (p<0.000; 95% CI: 0.1546326, 0.4521232) (see Figure S1, Supplemental Information). Thus, microarray and qRT-PCR correlation was greater for CTONG2002744 clone and FBXO32. To validate the finding of differential expression for these markers on an independent cohort of patients, we identified an additional 44 cases, 31 of which were metastatic vs. 13 non-metastatic OSCC, using the same criteria to identify cases in the training set. We then estimated the power to detect a difference in mean expression between metastatic vs. non-metastatic cases that would provide a full validation of the Affymetrix platform. Eighty percent power (at alpha = 0.05) was possible only for CTONG2002744, and thus only this transcript was assayed by RT-PCR on this 44 sample set (see Table S3, Supplemental Information). The mean 1/Ct value for this transcript in the metastatic vs. the non-metastatic cases was 2.08 vs. 3.79, respectively (p = 0.0004) (see Table S4, Supplemental Information).
Discussion
To date, options to determine which patients with Stage I/II OSCC (i.e., non-metastatic) harbor occult metastatic lymph nodes and would thus, benefit from a cervical lymphadenectomy are limited. Tumor depth and sentinel lymph node biopsies have been both proposed to address this issue and are under investigation, but there are no genetic biomarkers currently used to help inform clinicians on this clinical scenario. In this study, our goal was to identify biomarkers of metastasis from biopsy samples containing a mixture of both tumor and stroma since 1) there is ample evidence showing the contribution of tumor stroma in the process of carcinogenesis and thus, both cell sources could potentially contain informative biomarkers; and 2) laser capture microdissection or other methods to isolate tumor cells from biopsy samples are not likely to be adopted in clinical practice soon and therefore we wanted our methods to reflect sampling more likely to be performed in a clinic setting. We identified six probe sets which were associated with OSCC nodal status in a multivariate analysis adjusting for tumor size and HPV status. We also noted that our prediction model based on the expression of MYO5A, RFN145, FBXO32 and CDNA FLJ33813 fis, clone CTONG2002744 had a significantly higher AUC for predicting occult metastasis than tumor size, the current clinical criterion by which to offer a prophylactic lymphadenectomy in node-negative OSCC patients. Moreover, a model combining tumor size and gene expression did not predict occult metastasis better than a model using gene expression alone. Lastly, this 4-gene signature model was also predictive of overall and OSCC-specific survival, a finding consistent with the notion that lymph node spread in OSCC is associated with shorter survival. Validation of the mRNA signal with qRT-PCR showed that this finding was not platform dependent for at least CTONG2002744 clone and FBXO32. In addition, differential down-regulation of CTONG2002744 was validated in an independent cohort of 44 OSCC patients. These findings suggest that further studies to examine the functional role of CTONG2002744 in OSCC metastasis are warranted.
Although not much has been published on these probe sets, evidence thus far points to biologic relevance for CTONG2002744 clone and FBXO32. The source of the CTONG2002744 clone is a tongue tumor tissue (http://www.ncbi.nlm.nih.gov/nuccore/21749430), but there are no published articles on this gene’s function. FBXO32 encodes a member of the F-box protein family which is highly expressed during muscle atrophy. Two studies thus far have linked this gene with cancer. In one study, FBXO32 was found to have a potential tumor suppressor function (16). Another study showed that FBXO32 (MAFbx) was up-regulated in gastrointestinal stromal tumor after treatment with Gleevec and that its expression was regulated by ERK 1/2-dependent pathway (17). A functional role in cancer for the other probe sets that we found to predict nodal metastases has not yet been found. However, it is interesting that the two probe sets for which there is a potential functional role in cancer are also the two probe sets with significant associations not only with nodal metastasis but with OSCC survival (Table 2).
Comparing gene expression models with tumor size criteria showed that gene expression was a better predictor of nodal metastasis even in metastatic cases which were clinically labeled as node-negative. To depict a more realistic clinical scenario for which this 4-gene signature would be used, we decided to test the model’s performance in correctly diagnosing occult nodal metastasis in a subset of patients which presented with clinically node-negative OSCC. In this case, the 4-gene signature model would have missed 1 out of 20 occult metastatic cases (i.e., no worse than tumor size criteria), and in turn, only 9, out of 39, as opposed to 31 out of 39 node-negative cases, would have been incorrectly diagnosed as node-positive cases by the current guideline based on tumor size ≥ 2cm (T2-4). Thus, assuming that a 4-gene signature score cutoff that does not miss any more occult metastatic cases than tumor size alone, and that these results validate in independent datasets of bigger samples sizes, the most notable improvement of using gene expression data is realized in the higher specificity it affords over current clinical guidelines.
We acknowledge the potential for over-optimistic AUC values since the same data set was used to obtain probe sets associated with nodal metastasis and to evaluate the ability of the probe sets to predict nodal metastasis. An attempt to correct this over-optimism was done by performing a jack-knife leave-one-out validation, but these findings will need to be further validated with independent data sets as they become publicly available. In addition, we validated the differential expression for the CTONG2002744 clone in an independent dataset of 44 OSCC patients.
Other gene expression studies have developed profiles associated with head and neck cancer metastasis (18,19). There was little or no overlap between these signatures, nor with our six probe sets. A recent study by Rickman et al. found a 4-gene model of distant metastasis for OSCC (20). As with the previous studies, there is no overlap between our genes and theirs, but this is most likely due to the fact that the endpoint was different. In fact, “controls” (non-metastatic cases) in that study included patients who, despite being free of distant metastasis, had late stage disease due to loco-regional tumor burden (including nodal metastasis). The reason for the lack of overlap between all these studies is likely multifactorial, including differences in how metastatic cases were defined, tissue collection protocols, array platforms used, and statistical analyses.
The probe sets presented in this study were selected based on a stringent NFD threshold and after adjustment for potential confounders such as tumor size and HPV status. As more patients are recruited, we will conduct subgroup analysis by site (oral cavity vs. oropharynx) and HPV status to investigate the generalizability of our gene signature and to determine if the molecular pathways by which tumor cells gain a lymphotropic phenotype are site- or HPV-specific. Nonetheless, to our knowledge, this is the largest prospective study comparing N+ vs. N0 tumors. We only included as nodal metastases those N+ cases that were pathologically confirmed, and compared them to N0 cases whom had baseline CT scans, pathologic confirmation available for all tumors ≥ 2 cm (T2-4) and at least 18 months follow-up to ensure no occult nodal disease. That only a few genes were found to be differentially expressed is consistent with our previous finding of only one differentially expressed gene (ribosomal protein S13) between N+ vs. N0 OSCCs in our previous, smaller study (21). Thus, given the multiple comparison penalty imposed by high-throughput genome-wide analyses, it may be that the current samples sizes used in these studies do not allow unmasking of more pronounced differential gene expression when comparing phenotypes like nodal metastasis.
We expected that any probe sets that are truly associated with nodal metastasis might also be associated with disease-specific survival. Testing of this hypothesis on the 150 patient cohort, which included 37 patients not used to determine the probe sets associated with nodal metastasis, showed that indeed, the 4-gene signature model was predictive of OSCC-specific survival. We note that the probe sets reported in this study did not overlap with our previously reported survival expression signature for OSCC (15). This despite the fact that the patients who were classified by gene expression profiling to have a high risk of poor survival in that report had more advanced disease (including nodal tumor burden) compared to those who were not so classified. However, the probe sets in our survival signature were not significantly associated with nodal metastasis (unpublished data). The main reason for these observations may be that the survival signature was derived from probe sets initially shown to discriminate between OSCC and normal oral epithelium while the 4-gene metastasis signature is not. Since all the patients with nodal metastasis receive treatment for it, it is possible that the prognostic significance of our survival signature is not reflective of the extent of nodal involvement, but of some other feature in those patients (such as treatment responsiveness, local recurrence, etc.) associated with a more aggressive phenotype.
It is possible that the prediction of occult metastatic disease using clinical tumor size criterion might have been improved by using both pathologic tumor size and tumor depth. In particular, tumor depth greater than 4 mm has been shown to be an independent predictor of nodal metastasis (22). However, these two measurements are done post-operatively, at which point the decision of whether to offer a prophylactive cervical lymphadenectomy would have already been made. Moreover, comprehensive tumor depth assessment is not routinely done at most centers. Thus, a depth assessment of < 4 mm at one or a few loci does not exclude the possibility that other foci of disease reached this threshold.
In conclusion, we found a 4-gene model predictive of occult metastasis; that this prediction is superior to that afforded by clinical tumor size alone; and that this model is also predictive of disease-specific survival. If validated in larger studies, this 4-gene signature model could be used to improve decision-making regarding prophylactic cervical lymph node treatment in patients with node-negative OSCC.
Translational Relevance
Oral squamous cell carcinoma (OSCC) can be a devastating disease which typically spreads first to cervical lymph nodes. This sentinel event has been shown to be associated with survival. However, to date, little is known about the molecular mechanisms that may play a role in this process and there are no biomarkers to help inform on those patients either harboring occult metastasis or at risk of developing it. In this study, we have used a genome-wide expression profiling approach to determine the differential gene expression between metastatic vs. non-metastatic OSCC. We only included as nodal metastases those N+ cases that were pathologically confirmed, and compared them to N0 cases who had baseline CT scans, pathologic confirmation available for all tumors ≥ 2 cm (T2-4) and had at least 18 months of follow-up to ensure the absence of occult nodal disease. To our knowledge, this is the largest study of this kind for oral cancer. The findings presented herein describe a 4-gene signature model that may help inform upon those OSCC patients with occult metastatic disease – a key and novel contribution to the field of oral cancer.
Supplementary Material
Acknowledgements
We thank the study participants and the clinical and research staff.
Grant Support: This study was supported by grant R01CA095419 from the National Cancer Institute, National Institutes of Health, Bethesda, MD; grant 1K1L2RR025015-01 from the National Center for Research Resources (NCRR); the Harold Amos Medical Faculty Development Program from the Robert Wood Johnson Foundation; by funds from the Fred Hutchinson Cancer Research Center; and by resources from and use of facilities at the VA Puget Sound Health Care System, University of Washington Medical Center and Harborview Medical Center, Seattle, Washington.
References
- 1.Johnson JT, Barnes EL, Myers EN, et al. The extracapsular spread of tumors in cervical node metastasis. Arch Otolaryngol. 1981;107:725–729. doi: 10.1001/archotol.1981.00790480001001. [DOI] [PubMed] [Google Scholar]
- 2.Pinsolle J, Pinsolle V, Majoufre C, et al. Prognostic value of histologic findings in neck dissections for squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 1997;123:145–148. doi: 10.1001/archotol.1997.01900020023003. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham MJ, Johnson JT, Myers EN, Schramm VL, Jr., Thearle PB. Cervical lymph node metastasis after local excision of early squamous cell carcinoma of the oral cavity. Am J Surg. 1986;152:361–366. doi: 10.1016/0002-9610(86)90305-3. [DOI] [PubMed] [Google Scholar]
- 4.Ho CM, Lam KH, Wei WI, Lau SK, Lam LK. Occult lymph node metastasis in small oral tongue cancers. Head Neck. 1992;14:359–363. doi: 10.1002/hed.2880140504. [DOI] [PubMed] [Google Scholar]
- 5.Shah JP, Candela FC, Poddar AK. The patterns of cervical lymph node metastases from squamous carcinoma of the oral cavity. Cancer. 1990;66:109–113. doi: 10.1002/1097-0142(19900701)66:1<109::aid-cncr2820660120>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 6.Lindberg R. Distribution of cervical lymph node metastases from squamous cell carcinoma of the upper respiratory and digestive tracts. Cancer. 1972;29:1446–1449. doi: 10.1002/1097-0142(197206)29:6<1446::aid-cncr2820290604>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Hynes RO. Metastatic potential: generic predisposition of the primary tumor or rare, metastatic variants-or both? Cell. 2003;113:821–823. doi: 10.1016/s0092-8674(03)00468-9. [DOI] [PubMed] [Google Scholar]
- 9.Mendez E, Fan W, Choi P, et al. Tumor-specific genetic expression profiles of metastatic oral squamous cell carcinoma. Head Neck. 2007;29:803–814. doi: 10.1002/hed.20598. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Mendez E, Houck J, et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohavanichbutr P, Houck J, Fan W, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–188. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas JG, Olson JM, Tapscott SJ, Zhao LP. An efficient and robust statistical modeling approach to discover differentially expressed genes using genomic expression profiles. Genome Res. 2001;11:1227–1236. doi: 10.1101/gr.165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu XL, Olson JM, Zhao LP. A regression-based method to identify differentially expressed genes in microarray time course studies and its application in an inducible Huntington's disease transgenic model. Hum Mol Genet. 2002;11:1977–1985. doi: 10.1093/hmg/11.17.1977. [DOI] [PubMed] [Google Scholar]
- 14.Wasson JH, Sox HC, Neff RK, Goldman L. Clinical prediction rules. Applications and methodological standards. N Engl J Med. 1985;313:793–799. doi: 10.1056/NEJM198509263131306. [DOI] [PubMed] [Google Scholar]
- 15.Mendez E, Houck JR, Doody DR, et al. A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res. 2009;15:1353–1361. doi: 10.1158/1078-0432.CCR-08-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolov A, Chahwan S, Ochs M, et al. Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol Cancer Ther. 2003;2:699–709. [PubMed] [Google Scholar]
- 18.Roepman P, Wessels LF, Kettelarij N, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37:182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Temam S, Oh M, et al. Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia. 2006;8:925–932. doi: 10.1593/neo.06430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickman DS, Millon R, De Reynies A, et al. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27:6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 21.Mendez E, Cheng C, Farwell DG, et al. Transcriptional expression profiles of oral squamous cell carcinomas. Cancer. 2002;95:1482–1494. doi: 10.1002/cncr.10875. [DOI] [PubMed] [Google Scholar]
- 22.Sparano A, Weinstein G, Chalian A, Yodul M, Weber R. Multivariate predictors of occult neck metastasis in early oral tongue cancer. Otolaryngol Head Neck Surg. 2004;131:472–476. doi: 10.1016/j.otohns.2004.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


