Abstract
HLA DQB1*0602 is found in most individuals with hypocretin-deficient narcolepsy, a disorder characterized by a severe disruption of sleep and wake. Population studies indicate that DQB1*0602 may also be associated with normal phenotypic variation of rapid eye movement (REM) sleep. Disruption of REM sleep has been linked to specific symptoms of insomnia. We here examine the relationship of sleep and DQB1*0602 in older individuals (n=46) with primary insomnia, using objective (polysomnography, wrist actigraphy) and subjective (logs, scales) measures. DQB1*0602 positivity was similarly distributed in the older individuals with insomnia (24%) as in the general population (25%). Most sleep variables were statistically indistinguishable between DQB1*0602 positive and negative subjects except that those with the allele reported that they were significantly less well rested than those without it. When sleep efficiencies were lower than 70%, DQB1*0602 positive subjects reported being less well rested at the same sleep efficiency than those without the allele. Examination of EEG during REM sleep also revealed that DQB1*0602 positive subjects had EEG shifted towards faster frequencies compared with negative subjects. Thus, DQB1*0602 positivity is associated with both a shift in EEG power spectrum to faster frequencies during REM sleep and a diminution of restedness given the same sleep quantity.
Keywords: Human Leukocyte Antigens, insomnia, aging, electroencephalography
1. Introduction
More than 95% of individuals with hypocretin (orexin)-deficient narcolepsy, a disease characterized by significant disruption of sleep/wake and REM sleep regulation, carry the Human Leukocyte Antigen (HLA) DQB1*0602 allele, as compared to 25% of the general population (Bourgin et al., 2008). It has been suggested that DQB1*0602 is an important factor in the autoimmune attack hypothesized to be responsible for the destruction of hypocretin-expressing neurons in the lateral hypothalamus (Bourgin et al., 2008). In a population-based study of individuals without narcolepsy, DQB1*0602 positivity was associated with a shortened REM sleep latency (Mignot et al., 1998) and more frequent direct transitions between wake and REM sleep during naps in men (Mignot et al., 2006). It is possible that subtle phenotypic variations in REM sleep may result from changes in hypocretin function that are mediated through DQB1*0602.
Assessment of insomnia is frequently characterized by a dichotomy between subjective and objective measures of sleep quantity and quality. Differences in REM sleep have been hypothesized to be possible causes of this dichotomy, positing that greater amounts of REM sleep lead to a misperception of a greater amount of nocturnal wakefulness in both individuals with insomnia and good sleeping controls (Feige et al., 2008). More beta activity in the electroencephalogram (EEG) also has been correlated with lesser concordance between subjective and objective estimates of sleep time (Perlis et al., 2001).
Given the link between DQB1*0602 and REM sleep, as well as between characteristics of REM sleep and insomnia, we tested the hypothesis that DQB1*0602 positivity changes the dynamics of REM sleep and leads to worse insomnia symptoms. To examine this, we studied relevant data from a group of older individuals with primary insomnia.
2. Methods
2.1 Subjects
We analyzed data for individuals 55 years or older who had been diagnosed as having primary insomnia. Subjects were recruited specifically for participation in this study from the general community in the Bay Area. Other types of insomnia, sleep apnea (Respiratory Distress Index >10), use of sleep-active medication (except occasional use of anti-histamines), periodic limb movement disorder (periodic limb movements with arousals >10), dementia (Mini-Mental State Exam, MMSE, <25; (Folstein et al., 1975), poor executive function (Executive Interview, EXIT, >15; (Royall et al., 1992), and depression (Geriatric Depression Scale, GDS, ≥7 triggered an in-depth evaluation to exclude major depression; (Yesavage et al., 1982) were all excluded. Subjects did not report a history of hypagogic hallucinations or sleep paralysis. Non-exclusionary information collected during screening included the Penn State Worry Questionnaire (PSWQ; (Meyer et al., 1990), State-Trait Anxiety Inventory (STAI; (Spielberger et al., 1983), Epworth Sleepiness Scale (ESS; (Johns, 1991), Morningness-Eveningess Questionnaire (MEQ; (Horne and Östberg, 1976), and 36-Item Short Form Survey Instrument (SF-36; (Ware, Jr. and Sherbourne, 1992) scored for both Physical (PCS) and Mental Composite Scores (MCS). A blood sample was taken at baseline and DNA was extracted from the sample using the QIAamp DNA Blood Kit (QIAGEN, Germantown MD). The presence of the HLA DQB1*0602 allele was determined using standard single allele specific primer-polymerase chain reaction procedures (Mignot et al., 2006).
2.2 Protocol
During the first week of study, subjects slept ad libitum at home and completed daily sleep logs and a single-item restedness scale (“not at all rested” to “very rested” with a 7-point range, higher scores indicating feeling more rested) in the morning following each night's sleep episode. The following week, subjects continued to sleep ad libitum at home and wore a wrist actigraph (Actiwatch-L, MiniMitter, Bend OR) in addition to completing sleep logs. A wrist actigraph collects three-dimensional arm movement data; such data are a useful proxy for determining sleep and wake (Ancoli-Israel et al., 2003). Actigraphy data collected in 30-s epochs were analyzed with Actiware-Sleep (v.3.1, Mini-Mitter, Bend OR) to derive sleep efficiency (total sleep time divided by time in bed), using the high sensitivity scoring option (found to be most appropriate for the disturbed sleep of insomnia patients) (Kushida et al., 2001).
Subjects also had a two-night stay in a sleep research laboratory during which sleep state was determined using electromyography (EMG, chin), electro-oculography (EOG, above the right outer canthus and below the left outer canthus), and electroencephalography (EEG, scalp sites C3, C4, O1, O2, A1 and A2, according to the International 10-20 System of Electrode Placement). This sleep recording typically (74%) took place within a month of the actigraphy data collection and all occurred within three months. Data from leg EMG, breathing bands, pulse oximetry, and nasal cannulation were also used for the determination of Periodic Limb Movement Index and Respiratory Disturbance Index. EMG, EOG, and EEG data were collected using an Alice4 polysomnograph (Respironics, Murrysville PA). These data were scored into sleep states (wake, stage 1 non-REM, stage 2 non-REM, stage 3 non-REM, stage 4 non-REM, and REM) using a 30-s window by a single expert polysomnograph technician applying standard criteria (Rechtschaffen and Kales, 1968). Spectral analysis of EEG data was done using PRANA software (PhiTools, Strasbourg France). Data in each 30-s bin were Fourier transformed (2-s Hanning window) with Welch averaging and automatic artifact removal so as to derive the power spectra in 0.5 Hz resolution. Power spectra from C3-A2, C4-A1, O1-A2, and O2-A1 derivations were calculated. Data were further binned into spectral frequencies of delta (0.5-4.5 Hz), theta (4.5-8.5 Hz), alpha (8.5-12.5 Hz), sigma (12.5-15.5 Hz), beta (15.5-22.5 Hz), and gamma (22.5-35 Hz). The power in each frequency for each 30-s data bin was normalized to total power (generating so-called, “relative power”). Data were then grouped according to sleep state (non-REM, REM, or wake) and averaged over fifths of the night. Data were then averaged between the two recording nights within subjects. For visualization, data were then averaged across subjects.
2.3 Statistical Methods
Between group comparisons were made using Student's t-tests (Excel v.11.8231.8221, Microsoft, Redmond WA) or Fisher exact tests (java script located at graphpad.com/quickcalcs/contingency1.cfm). Analysis of variance (ANOVA, SAS v.9.1, Cary NC) was used for multilayered group comparisons. The threshold for significance in these comparisons involving six frequency bands was 0.0083 (Bonferroni correction of α/n where the overall test α was set at 0.05). Linear regression analyses were performed using Origin (v.6.1, OriginLab, Northampton MA). Data are presented as mean ± SD. Subjects were paid for their time and signed a consent form that was approved by, as were all procedures, the Stanford University Institutional Review Board. This protocol conforms to the ethical standards outlined in the Declaration of Helsinki.
3. Results
Of the 46 individuals on whom we were able to obtain genotyping, 11 (24%) were positive for the DQB1*0602 allele and the remaining 35 were negative. Baseline demographic, cognitive, and psychiatric variables were statistically indistinguishable between individuals positive and negative for the allele (Table 1). There was no significant difference in the Respiratory Disturbance Index (screened to be ≤10) between the two groups (Table 2), nor was there a significant difference in the overnight mean oxygen saturation (Table 2). The two groups were also similarly sleepy during the daytime, according to the Epworth Sleepiness Scale (Table 1). According to prospectively recorded sleep logs, the subjects with or without the DQB1*0602 allele were similar for most sleep characteristics, including minutes in bed, minutes asleep, sleep efficiency, minutes of wakefulness after sleep onset, and minutes to fall asleep (Table 2). Despite similar subjective sleep characteristics, DQB1*0602 positive subjects reported that they were significantly less well rested in the morning (3.16 ± 0.858) than those without the allele (3.96 ± 1.02, t-test, P < 0.05). To determine if this difference in subjective assessment of restedness was consistent across similar sleep efficiencies in those positive or negative for the DQB1*0602 allele, we plotted the subjective sleep efficiency value for each night (calculated from the sleep log) against the subjective rating of restedness after that same night. These data were then fit with a linear regression model (P < 0.0001 for both genotypes). We found that at sleep efficiencies lower than 70%, the curves were significantly different from one another (non-overlapping 95% confidence intervals) (Figure 1). Thus, given the same subjective sleep efficiency, when reporting sleeping less than 70% of their time in bed, individuals who were DQB1*0602 positive reported lower restedness ratings than individuals negative for DQB1*0602.
Table 1.
Baseline data for subjects with the DQB1*0602 allele (0602+, n=11) or without the allele (0602-, n=35) are presented as mean ± SD. t-tests or Chi Square tests, as appropriate, were used to determine P-values. Abbreviations as in text. Only 34 of 35 0602- subjects had available data for the EXIT and MEQ; all other data sets are complete.
| 0602+ | 0602- | P-value | |
|---|---|---|---|
| Age (yrs) | 66.3 ± 7.73 | 63.1 ± 7.17 | 0.21 |
| Sex (% female) | 63.6% | 74.3% | 0.70 |
| Education (yrs) | 17.0 ± 1.90 | 16.1 ± 2.47 | 0.26 |
| MMSE | 28.5 ± 1.04 | 28.7 ± 1.38 | 0.53 |
| EXIT | 2.09 ± 1.81 | 3.15 ± 2.19 | 0.16 |
| GDS | 3.82 ± 2.75 | 4.14 ± 3.12 | 0.76 |
| PSWQ | 39.2 ± 8.18 | 47.4 ± 14.8 | 0.09 |
| STAI (Trait) | 35.5 ± 7.83 | 34.4 ± 7.96 | 0.69 |
| ESS | 7.36 ± 3.01 | 6.97 ± 4.44 | 0.79 |
| PCS | 51.9 ± 5.20 | 51.8 ± 8.05 | 0.96 |
| MCS | 53.7 ± 3.90 | 50.3 ± 8.88 | 0.08 |
| MEQ | 67.5 ± 6.06 | 61.6 ± 9.84 | 0.07 |
Table 2.
Basic sleep log-, actigraph-, and polysomnography (PSG)-derived sleep characteristics for subjects with (0602+, n=11) or without (0602-, n=35) the DQB1*0602 allele. Sleep logs and actigraphy were averaged over two weeks (except only one week for “Rested”) of at-home collection, while PSG data were averaged over two nights (one subject from each group had only one usable night of PSG collected). Data were averaged within subjects then between subjects and are presented as mean ± SD. P-values were determined using unpaired t-Tests corrected for multiple comparisons (7 in sleep log data, 5 in actigraphy data, and 18 in PSG data). SE: sleep efficiency; WASO: wake after sleep onset; ISA: intrasleep awakening; NREM: non-rapid eye movement sleep; REM: rapid eye movement sleep; S1: stage 1, NREM; S2: stage 2, NREM; S3/4: combined stages 3 and 4, NREM; RDI: Respiratory Disturbance Index.
| 0602+ | 0602- | P-value | ||
|---|---|---|---|---|
| Sleep logs derived | Time in Bed | 517 ± 40.6 | 502 ± 63.2 | 0.49 |
| Total Sleep Time | 351 ± 52.3 | 341 ± 51.8 | 0.56 | |
| SE | 68.3 ± 8.98 | 68.7 ± 11.5 | 0.92 | |
| WASO | 73.2 ± 25.4 | 66.1 ± 45.0 | 0.62 | |
| Sleep Latency | 31.7 ± 26.9 | 33.0 ± 28.2 | 0.89 | |
| “Rested” | 3.16 ± 0.858 | 3.96 ± 1.02 | 0.02* | |
| Actigraph-derived | Time in Bed | 521 ± 37.1 | 499 ± 60.2 | 0.25 |
| Total Sleep Time | 436 ± 57.0 | 395 ± 54.8 | 0.04 | |
| SE | 83.6 ± 7.35 | 79.4 ± 7.33 | 0.11 | |
| WASO | 54.2 ± 22.5 | 63.7 ± 32.4 | 0.37 | |
| Sleep Latency | 14.2 ± 15.4 | 21.9 ± 14.8 | 0.14 | |
| Polysomnography-derived | Total Sleep Time | 340 ± 70.3 | 353 ± 51.5 | 0.51 |
| Sleep Latency | 18.9 ± 13.9 | 17.4 ± 20.9 | 0.83 | |
| SE | 77.7 ± 15.1 | 82.6 ± 8.06 | 0.33 | |
| WASO | 98.0 ± 63.7 | 74.9 ± 35.6 | 0.27 | |
| ISA > 2 min | 7.00 ± 3.77 | 6.04 ± 2.43 | 0.32 | |
| All transitions | 186 ± 54.4 | 186 ± 45.9 | 0.99 | |
| Transitions, NREM to REM | 20.5 ± 7.75 | 20.1 ± 7.10 | 0.89 | |
| Transitions, REM to NREM | 16.1 ± 6.30 | 15.8 ± 6.29 | 0.88 | |
| REM (min) | 61.2 ± 18.0 | 58.8 ± 17.8 | 0.69 | |
| REM (%) | 17.3 ± 4.08 | 16.3 ± 3.93 | 0.51 | |
| REM latency (min) | 69.1 ± 25.1 | 84.1 ± 37.9 | 0.23 | |
| S1 (min) | 156 ± 55.2 | 178 ± 46.3 | 0.19 | |
| S1 (%) | 45.7 ± 11.9 | 50.6 ± 9.72 | 0.17 | |
| S2 (min) | 112 ± 36.6 | 110 ± 34.2 | 0.85 | |
| S2 (%) | 34.3 ± 12.0 | 31.6 ± 9.51 | 0.44 | |
| S3/4 (min) | 10.0 ± 14.9 | 5.49 ± 8.67 | 0.36 | |
| S3/4 (%) | 2.77 ± 3.96 | 1.50 ± 2.24 | 0.33 | |
| O2 saturation (%) | 87.8 ± 7.82 | 88.5 ± 6.21 | 0.75 | |
| RDI | 3.79 ± 2.30 | 3.14 ± 2.69 | 0.48 |
Figure 1.
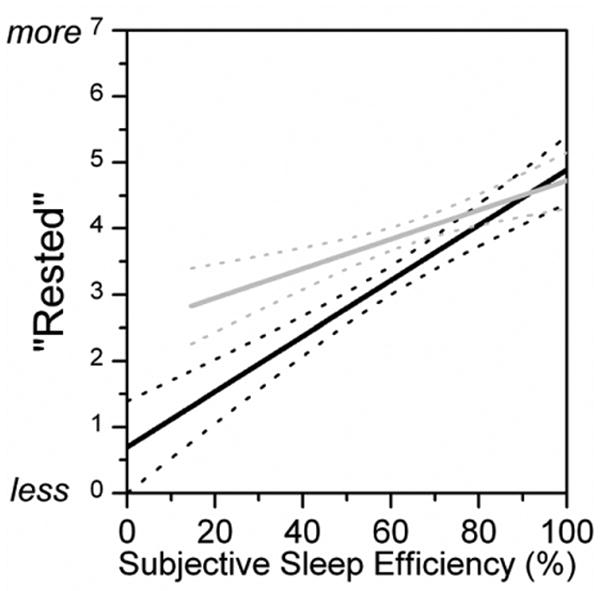
Subjective restedness by sleep efficiency varies by genotype. Subjective sleep efficiency correlates strongly with subjective restedness after a night of sleep and this relationship varies by the presence (black) or absence (grey) of DQB1*0602 such that below 70%, the relationship is different in the two genotypes. Linear regressions shown with 95% confidence intervals.
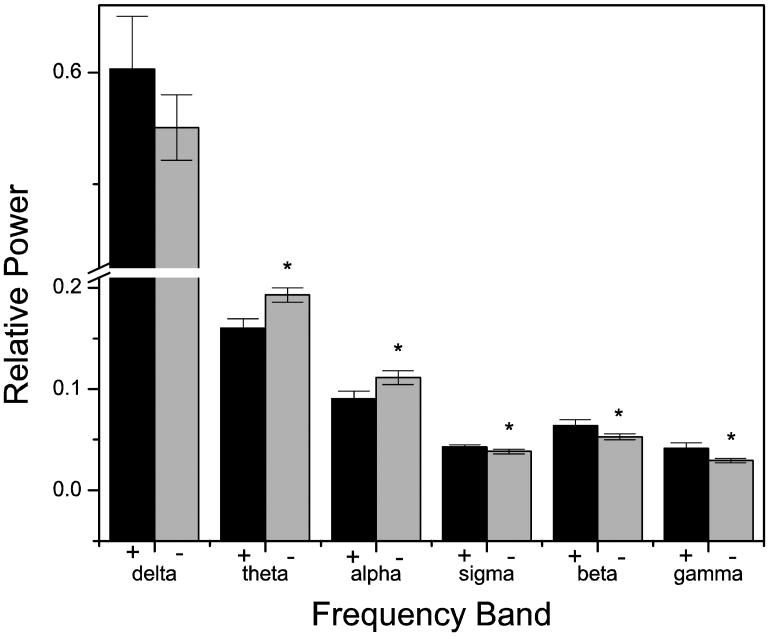
As all standard PSG, sleep log, and actigraphy data indicated that there were no differences between the sleep of individuals with the DQB1*0602 allele and those without (see Table 2), we examined the C3-A2 derivation to see if there might be differences specifically in the REM sleep EEG that would not be noticed with traditional sleep/wake scoring. ANOVA indicated that there was no significant effect of fifth of night (P's>0.07) for any of the six frequency bands, but that there was a significant effect of genotype in the theta (P < 0.0001), alpha (P < 0.005), sigma (P < 0.01), beta (P < 0.0005), and gamma (P < 0.0001) bands; there was no effect of genotype in the delta band (P = 0.10). For visual presentation (Figure 2), as there was no effect of time of night, we collapsed across the entire night averaging all of the data that was derived from REM sleep. As is evident in this figure, there is a frequency shift such that faster frequencies are over-represented in individuals with the DQB1*0602 allele. In REM sleep, there is significantly more relative sigma, beta, and gamma power and significantly less relative theta and alpha power in individuals with the DQB1*0602 allele.
Figure 2.
Frequency shift in REM sleep varies by genotype. Relative power in individuals with (black) or without (grey) the DQB1*0602 allele varies such that those with the allele have more faster frequencies during REM sleep. Data were averaged over two nights within and then between subjects. Average ± SD is shown. The asterisk indicates significance by t-test at P < 0.00833.
While our a priori hypothesis was to examine differences in REM sleep, we also advantageously examined the power spectra in non-REM sleep. As expected during non-REM sleep, each of the six frequency bins had a significant change over the course of the night (P's<0.0002, ANOVA). There were no statistically significant time of night by genotype interactions (P's>0.33, ANOVA) and while trending in the correct direction, with more relative power in the faster frequencies in the subjects with DQB1*0602, the differences by genotype were not consistently different with delta (P = 0.048), sigma (P = 0.078), and gamma (P = 0.027) bands not reaching threshold for statistical significance.
4. Discussion
While in need of replication in a larger data set, our study is the first to show that a genetic polymorphism may help explain both interindividual differences in the perception of sleep quality and basic EEG physiology in older individuals with insomnia. We show that those older individuals with insomnia who are carriers of the DQB1*0602 allele have a shift to faster EEG frequencies during REM sleep and feel less well rested given the same amount of sleep as those who do not carry this allele. The results from non-REM sleep are less consistent, though there may be a similar shift to faster EEG frequencies. These data are consistent with the idea of a general hyperarousal during sleep in individuals with insomnia (Riemann et al., 2010). Alternatively, it may be that neural activity between disparate brain regions during sleep is less well coordinated. If sleep were treated as a local phenomenon (Huber et al., 2004), in those with DQB1*0602 and possibly diminished hypocretin neuron function, local pockets of sleep around the cortex would be less well temporally coordinated with one another (i.e., there would be more intermixed sleep and wake). This could help explain why individuals with same amount of “sleep” (the determination of which is from a single EEG derivation) could have a different sense of refreshment after sleep, with those having more coordinated sleep activity awakening more refreshed. This could also impact particular functions of sleep, such as memory consolidation, in individuals with insomnia (Nissen et al., 2006). This hypothesis would, however, need to be tested in individuals with insomnia using dense EEG arrays.
Unlike previously published findings from a population study (Mignot et al., 1998), we did not detect a difference in REM latency between HLA DQB1*0602 positive and negative individuals. We did observe a trend towards a shorter REM latency in individuals with HLA DQB1*0602 (see Table 2, 15 minutes shorter in DQB1*0602 positive), consistent with the previous study, but the variance is so large that only a difference of 40.7 minutes is detectable in our sample size (cf., an 18 minute difference is reported in the population study of 490 individuals (Mignot et al., 1998)). Thus, a similar difference in REM latency may exist between HLA DQB1*0602 positive and negative older individuals with insomnia, but a larger study would be required to detect such a difference.
In the sleep of young, healthy individuals, it may be difficult to observe phenotypic differences based on non-pathological genotypic differences since sleep is generated by multiple, likely highly overlapping neuromodulatory systems (España and Scammell, 2004). In examining healthy older individuals in whom some of these systems may be damaged and have lower amplitude, one might be able to detect the otherwise compensated changes in sleep physiology. In fact, we have previously shown that in individuals with Alzheimer's disease, CSF hypocretin-1 concentrations are in the “normal” range (i.e., non-narcoleptic), but those at the lower end of normal concentrations nap while those on the higher end of normal do not (Friedman et al., 2007).
Our data agree with that of both Feige et al. (2008) and Perlis et al. (2001) that intrusion of wakefulness into REM may fundamentally alter one's perception of sleep quality. Given the relationship between DQB1*0602 and hypocretin cell loss in narcolepsy, we hypothesize that in older individuals who are DQB1*0602 positive, an alteration of hypocretin cell function is responsible for this state instability, though exploration of this remains for future study.
Acknowledgments
The authors would like to thank Dr. Ling Lin for conducting the genotyping, Beatriz Hernandez for data collection and management, and Dan East for EEG data scoring. Grant support for this study was provided by the National Institutes of Health (R01 AG12914); Department of Veterans Affairs Sierra-Pacific Mental Illness Research, Education, and Clinical Center; Office of the Vice Provost for Undergraduate Education, Stanford University; Human Biology Research Exploration program, Stanford University; and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Ancoli-Israel S, Cole R, Alessi C, Chambers M, Moorcroft W, Pollack CP. The role of actigraphy in the study of sleep and circadian rhythms. Sleep. 2003;26:342–392. doi: 10.1093/sleep/26.3.342. [DOI] [PubMed] [Google Scholar]
- Bourgin P, Zeitzer JM, Mignot E. CSF hypocretin-1 assessment in sleep and neurological disorders. Lancet Neurology. 2008;7:649–662. doi: 10.1016/S1474-4422(08)70140-6. [DOI] [PubMed] [Google Scholar]
- España RA, Scammell TE. Sleep neurobiology for the clinician. Sleep. 2004;27:811–820. [PubMed] [Google Scholar]
- Feige B, Al-Shajlawi A, Nissen C, Voderholzer U, Hornyak M, Spiegelhalder K, Kloepfer C, Perlis M, Riemann D. Does REM sleep contribute to subjective wake time in primary insomnia? A comparison of polysomnographic and subjective sleep in 100 patients. Journal of Sleep Research. 2008;17:180–190. doi: 10.1111/j.1365-2869.2008.00651.x. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Friedman L, Zeitzer JM, Lin L, Hoff D, Mignot E, Peskind ER, Yesavage JA. Hypocretin-1 is inversely associated with wake fragmentation in Alzheimer's disease. Neurology. 2007;68:793–794. doi: 10.1212/01.wnl.0000256731.57544.f9. [DOI] [PubMed] [Google Scholar]
- Horne JA, Östberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. International Journal of Chronobiology. 1976;4:97–110. [PubMed] [Google Scholar]
- Huber R, Ghilardi MF, Massimini M, Tononi G. Local sleep and learning. Nature. 2004;430:78–81. doi: 10.1038/nature02663. [DOI] [PubMed] [Google Scholar]
- Johns MW. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep. 1991;14:540–545. doi: 10.1093/sleep/14.6.540. [DOI] [PubMed] [Google Scholar]
- Kushida CA, Chang A, Gadkary C, Guilleminault C, Carrillo O, Dement WC. Comparison of actigraphic, polysomnographic, and subjective assessment of sleep parameters in sleep-disordered patients. Sleep Medicine. 2001;2:389–396. doi: 10.1016/s1389-9457(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Meyer TJ, Miller ML, Metzger RL, Borkovec TD. Development and validation of the Penn State Worry Questionnaire. Behaviour Research and Therapy. 1990;28:487–495. doi: 10.1016/0005-7967(90)90135-6. [DOI] [PubMed] [Google Scholar]
- Mignot E, Lin L, Finn L, Lopes C, Pluff K, Sundstrom ML, Young T. Correlates of sleep-onset REM periods during the Multiple Sleep Latency Test in community adults. Brain. 2006;129:1609–1623. doi: 10.1093/brain/awl079. [DOI] [PubMed] [Google Scholar]
- Mignot E, Young T, Lin L, Finn L, Palta M. Reduction of REM sleep latency associated with HLA-DQB1*0602 in normal adults. Lancet. 1998;351:727. doi: 10.1016/S0140-6736(05)78496-8. [DOI] [PubMed] [Google Scholar]
- Nissen C, Kloepfer C, Nofzinger EA, Feige B, Voderholzer U, Riemann D. Impaired sleep-related memory consolidation in primary insomnia--a pilot study. Sleep. 2006;29:1068–1073. doi: 10.1093/sleep/29.8.1068. [DOI] [PubMed] [Google Scholar]
- Perlis ML, Smith MT, Andrews PJ, Orff H, Giles DE. Beta/gamma EEG activity in patients with primary and secondary insomnia and good sleep controls. Sleep. 2001;24:110–117. doi: 10.1093/sleep/24.1.110. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen A, Kales A. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects. US Department of Health, Education, and Welfare, Government Printing Office; Washington D.C.: 1968. [DOI] [PubMed] [Google Scholar]
- Riemann D, Spiegelhalder K, Feige B, Voderholzer U, Berger M, Perlis M, Nissen C. The hyperarousal model of insomnia: a review of the concept and its evidence. Sleep Medicine Reviews. 2010;14:19–31. doi: 10.1016/j.smrv.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Royall DR, Mahurin RK, Gray KF. Bedside assessment of executive cognitive impairment: the executive interview. Journal of the American Geriatrics Society. 1992;40:1221–1226. doi: 10.1111/j.1532-5415.1992.tb03646.x. [DOI] [PubMed] [Google Scholar]
- Spielberger C, Gorusch RI, Lushene R, Vagg PR, Jacobs GA. State-Trait Anxiety Inventory for Adults. Consulting Psychologists Press, Inc.; Palo Alto CA: 1983. [Google Scholar]
- Ware JE, Jr, Sherbourne CD. The MOS 36-item short-form health survey (SF-36). I. Conceptual framework and item selection. Medical Care. 1992;30:473–483. [PubMed] [Google Scholar]
- Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, Leirer VO. Development and validation of a geriatric depression screening scale: a preliminary report. Journal of Psychiatric Research. 1982;17:37–49. doi: 10.1016/0022-3956(82)90033-4. [DOI] [PubMed] [Google Scholar]