Abstract
The Sox family of transcriptional regulators has been implicated in the control of a broad array of developmental processes. One member of this family SOX9 was first identified as a candidate gene for campomelic dysplasia (CD), a human syndrome affecting skeletal and testis development. In these patients most endochondral bones of the face fail to develop resulting in multiple defects such as micrognathia, cleft palate and facial dysmorphia. In this review we describe Sox9 expression during embryonic development and summarize loss of function experiments in frog, fish and mouse embryos highlighting the role of Sox9 in regulating morphogenesis of the face. We also discuss the mutations in and around SOX9 responsible for craniofacial defects in CD patients.
Keywords: Craniofacial, Cartilage, Neural crest, Sox9, Campomelic dysplasia
Introduction
The Sox (Sry HMG-box) gene family of transcriptional regulators was discovered in the early 1990's through the cloning of Sry, the mammalian testis-determining gene (Lovell-Badge, 2010). Sox genes are part of a larger family of high-mobility group (HMG) proteins, defined by their electrophoretic mobility on SDS-PAGE. Sox proteins bind DNA by means of their HMG domain, allowing them to function as transcription factors. This domain is highly conserved among Sox factors, which typically recognize a similar motif on the DNA: (A/T)(A/T)CAA(A/T)G. All Sox proteins bind DNA with low affinity, and are thought to require cofactors to stabilize their interactions with the DNA and mediate their transcriptional activity (Kamachi et al., 2000; Wilson and Koopman, 2002). Because most Sox factors are expressed in more than one cell lineage, cell type-specific cofactors are also required to confer specificity for various promoters (Kamachi et al., 2000; Wilson and Koopman, 2002). According to their sequence homologies within and outside the HMG domain, Sox proteins have been sorted into ten groups, A through J, with Sry being allocated to the SoxA group (Bowles et al., 2000; Schepers et al., 2002).
Sox9 belongs to the SoxE group, which also comprises Sox8 and Sox10. SoxE proteins are critical for the development of neural crest progenitors (Hong and Saint-Jeannet, 2005; Haldin and LaBonne, 2010). In all vertebrates one or more SoxE proteins are implicated in the specification, multipotency and survival of neural crest cells. Later in embryogenesis, individual SoxE proteins direct the development of distinct neural crest derivatives, including melanocytes, chondrocytes and sensory neurons (Hong and Saint-Jeannet, 2005; Haldin and LaBonne, 2010). SoxE factors are also essential for many aspects of peripheral and central nervous system development where they play a major role in oligodendrocyte and Schwann cell formation (Wegner, 2008; Stolt and Wegner, 2010). SOX9 was first identified as a candidate gene for campomelic dysplasia (CD), a condition characterized by skeletal defects, cranial dismorphology and sex reversal (Wagner et al., 1994; Foster et al., 1994). Sox9 turns out to be a master regulator of cartilage development. It has essential roles in the specification and differentiation of mesenchymal cells toward the chondrogenic lineage in all developing skeletal elements. Sox9 binds to and controls the activity of chondrocyte-specific enhancers of several genes critical for chondrocyte differentiation, including type-II collagen, Col2a1, the major matrix protein of the mature cartilage (Lefebvre and de Combrugghe, 1998; Lefebvre and Smits, 2005).
Here we summarize the expression pattern of Sox9 as it relates to craniofacial development and review some of the key features of Sox9 function during morphogenesis of the face. We also discuss the mutations in or near SOX9 causative of the craniofacial defects in CD patients.
Developmental expression of Sox9
During embryonic development in all species examined Sox9 is expressed in a broad array of tissues including the gonad, otic vesicle, lung, notochord, neural tube, pancreas and cardiac cushions (Wright et al., 1995; Ng et al., 1997; Zhao et al., 1997; Chiang et al., 2001; Spokony et al., 2002). The multi-lineage expression of Sox9 suggests the existence of complex regulatory mechanisms to control its tissue-specific expression during embryogenesis (Bagheri-Fam et al., 2006). More relevant to the focus of this review, Sox9 is also detected in neural crest progenitors at the lateral edge of the neural plate and is maintained in cranial neural crest cells as they populate the pharyngeal arches in the head region (Hong and Saint-Jeannet, 2005).
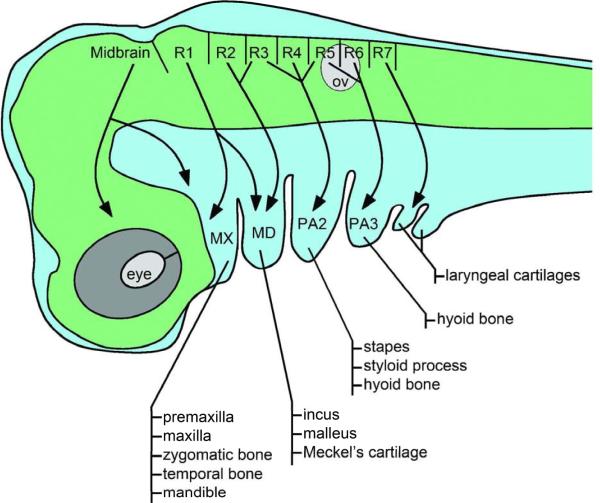
The neural crest is a multipotent population of migratory cells unique to the vertebrate embryo with the remarkable ability to give rise to a broad range of derivatives. The premigratory neural crest can be divided into five distinct and partially overlapping domains along the antero-posterior axis, the cranial, cardiac, vagal, trunk and sacral neural crest, each forming a specific set of derivatives in the periphery (LeDouarin and Kalcheim, 1999). Cranial neural crest cells delaminate from the posterior midbrain and individual rhombomeres in the hindbrain, and migrate into the pharyngeal arches to give rise to skeletal elements of the face, and contribute to the pharyngeal glands (thymus, thyroid and parathyroid) and portions of the cardiovascular system (Fig 1; LeDouarin and Kalcheim, 1999). In each pharyngeal arch the neural crest will contribute to a specific set of skeletal elements. The first pharyngeal arch has two components, the maxillary prominence forming the premaxilla, maxilla, zygomatic bone and temporal bone; and the mandibular prominence forming the middle ear bones, malleus and incus, and Meckel's cartilage. The second pharyngeal arch gives rise to the stapes, styloid process and lesser horn of the hyoid, while the greater horn of the hyoid is derived from the third pharyngeal arch. The elements derived from the most caudal pharyngeal arches (4 to 6) form the laryngeal cartilages (Fig 1; Minoux and Rijli, 2010).
Figure 1. Pattern of cranial neural cell migration and their skeletal derivatives.
In the mammalian embryo neural crest cells delaminate from the posterior midbrain and individual rhombomeres (R1–R7) in the hindbrain, and migrate into the pharyngeal arches (PA). Neural crest cells migrate in a stereotypical pattern based on their origin in the hindbrain (arrows). In each arch the neural crest contributes a specific set of skeletal elements as indicated. The first pharyngeal arch has two parts the maxillary (MX) and mandibular (MD) prominences. The most caudal pharyngeal arches form laryngeal cartilages. Lateral view, anterior to left, dorsal to top. ov, otic vesicle.
In zebrafish two orthologs of sox9 have been identified: sox9a and sox9b (Chiang et al., 2001). sox9b is expressed earlier than sox9a in progenitors of the neural crest (Li et al., 2002; Yan et al., 2002; 2005). Both genes are co-expressed in the premigratory trunk and cranial neural crest, though sox9b is expressed at higher levels than sox9a. Sox9b expression is downregulated in neural crest cells shortly after they initiate their migration towards the pharyngeal arches (Li et al., 2002; Yan et al., 2005). While sox9b expression is reactivated once neural crest cells reach the pharyngeal arches, sox9a is expressed at higher levels than sox9b in these structures (Yan et al., 2005). Early on sox9a is detected in the pre-condensed mesenchyme of the pharyngeal arch primordial, whereas sox9b expression in the mesenchyme of the pharyngeal arch starts just before hatching (Chiang et al., 2001; Yan et al., 2002). The pectoral fin rudiments (a cranial neural crest derivative) also express sox9b (Chiang et al., 2001). Craniofacial cartilages begin to condensate and differentiate between day 2 and day 3. sox9a and sox9b expression persists during chondrogenesis in essentially all the elements derived from the pharyngeal arches and in the neurocranium (Chiang et al., 2001).
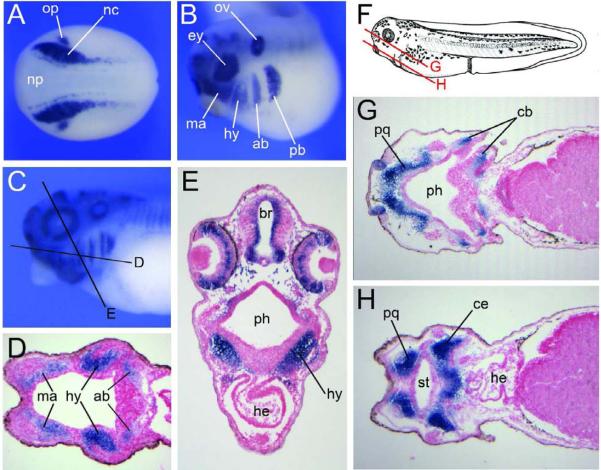
In the frog, Xenopus laevis, Sox9 is also first detected in the neural crest forming regions at the end of gastrulation (Fig 2; Spokony et al., 2002). As the neural tube closes, Sox9 expression persists in both the cranial and trunk neural crest. At the early tailbud stage as neural crest cells start to migrate in the cranial region, Sox9 is strongly expressed in all four streams of migrating cranial neural crest, the mandibular, hyoid, anterior and posterior branchial neural crest, as well as at the dorsal midline in the trunk neural crest (Fig 2; Spokony et al., 2002). Sox9 expression is subsequently downregulated in the trunk as cranial neural crest cells accumulate in the pharyngeal arches. Sox9 expression persists in the mesenchyme of the pharyngeal arch primordia until the swimming tadpole stage (Fig 2; Spokony et al., 2002; Kerney et al., 2007). Later in development Sox9 is mostly restricted to the condensing mesenchyme corresponding to the future palatoquadrate, ceratohyal, ceratobranchial, and Meckel's cartilages (Fig 2; Sadaghiani and Thiebaud, 1987; Gross and Hanken, 2008; Kerney et al., 2007).
Figure 2. Developmental expression of Xenopus Sox9 in the neural crest lineage.
(A) By in situ hybridization, at the end of gastrulation Sox9 is detected in neural crest progenitors (nc) at the lateral edge of the neural plate (np), and in the presumptive otic placode (op). Dorsal view, anterior to left. (B) At the tailbud stage Sox9 is detected in the four streams of cranial neural crest migrating towards the pharyngeal arches, the mandibular (ma), hyoid (hy), anterior branchial (ab) and posterior branchial (pb) neural crest. Other domains of expression include the developing eye (ey) and the otic vesicle (ov). Lateral view, anterior to left, dorsal to top. (C) Sox9 expression in the head region of a stage 35 embryo (Nieuwkoop and Faber, 1967). Lateral view, anterior to left, dorsal to top. The black lines indicate the level of the sections shown in the subsequent panels. (D–E) Sections showing Sox9 expression in the mesenchyme of the pharyngeal arches. (F) Diagram of a stage 40 embryo, after Nieuwkoop and Faber (1967). Lateral view, anterior to left, dorsal to top. The red lines indicate the level of the sections shown in the subsequent panels. (G–H) Sox9 is detected in all differentiating cartilage elements, including the palatoquadrate (pq), ceratobranchial (cb), ceratohyal (ce) and Meckel's cartilage (not shown). br, brain; he, heart; ph, pharynx; st, future stomodeum.
Sox9 expression in the mouse neural crest is first detected around E8.5 at the dorsal tip of the closing neural tube in the hindbrain region, as well as in the neural folds in the trunk region (Ng et al., 1997; Zhao et al., 1997). In the mouse, neural crest cells start to migrate before the neural tube is fully closed, and Sox9 is detected in cranial neural crest cells as they migrate away from the neuroepithelium. At E9.5, Sox9 is expressed in the mesenchyme of the frontal nasal mass and in the first, second and third pharyngeal arches (Wright et al., 1995, Ng et al., 1997; Zhao et al., 1997). Sox9 expression is subsequently associated with pre-chondrogenic mesenchyme condensed into cartilage primordia throughout the developing skeleton. In E14.5 embryos, in which most skeletal structures have formed, Sox9 is abundantly expressed at the base of the skull, in cartilage precursors of the supraoccipital bones, nasal, hyoid and Meckel's cartilages (Wright et al., 1995; Ng et al., 1997; Zhao et al., 1997).
It is important to mention that in all species examined, once cranial neural crest cells have settled in the pharyngeal arches, Sox9 is co-expressed with Col2a1 in the pharyngeal arch mesenchyme (Chiang et al., 2001; Kerney et al., 2007, Ng et al., 1997; Zhao et al., 1997), consistent with the predicted function of Sox9 in the regulation of chondrocyte development.
Sox9 function in craniofacial development
As discussed above, the expression of Sox9 in the developing neural crest and pharyngeal arch mesenchyme has been well conserved during evolution suggesting a conserved function for Sox9 in craniofacial development across species. Loss-of-function experiments have been especially critical to understand the specific requirement for Sox9 in the morphogenesis of the face. Here we summarize some of these findings in frog, fish and mouse embryos.
In Xenopus embryos, Sox9 knockdown by means of morpholino antisense oligonucleotides resulted in embryos exhibiting a specific loss of early neural crest markers, including Snail2, FoxD3 and Sox10 (Spokony et al., 2002; Aoki et al., 2003; Lee et al., 2004). Later in development, the loss of neural crest progenitors correlated with a number of defects in skeletal elements derived from the cranial neural crest while the development of melanocytes, a trunk neural crest derivative, was largely unaffected. Craniofacial defects included a complete loss of Meckel's cartilage and defective ceratohyal and ceratobranchial cartilages (Spokony et al., 2002), elements derived from the mandibular, hyoid and branchial neural crest, respectively (Sadaghiani and Thiebaud, 1987; Gross and Hanken, 2008). The basihyal, a mesoderm-derived cartilage that forms at the midline (Sadaghiani and Thiebaud, 1987; Gross and Hanken, 2008), was unaffected in these embryos (Spokony et al., 2002).
These defects were interpreted as the result of a specific depletion of the cranial neural crest progenitor pool, consistent with a role of Sox9 in neural crest specification. Yet, it is likely that Sox9 may also have a later role in the differentiation of neural crest cells once they reach the pharyngeal arches, a possibility that could not be evaluated with this type of knockdown approach. In another set of experiments embryos were injected with an inducible inhibitory mutant of Sox9 activated at the neurula stage to bypass the early requirement for Sox9 in neural crest specification. In these embryos the pattern of cranial neural crest migration was unperturbed suggesting that Sox9 is dispensable for neural crest cells migration in the frog (Lee et al., 2004). However the craniofacial phenotype of these embryos has not been analyzed.
The situation is a little different in zebrafish due to the presence of two sox9 orthologs, which carry distinct but complementary functions. In the sox9a zebrafish mutants, jellyfish, neural crest specification and migration occur normally, however these embryos have reduced expression of col2a1 and lack all cranial cartilage, with the exception of a few cells of the ceratohyal (Yan et al., 2002). sox9a is not required for neural crest specification or pharyngeal cartilage condensation; but appears to play a later role in the subsequent differentiation of these cells by regulating the stacking of chondrocytes and the shaping of individual cartilages (Yan et al., 2002). The analysis of the phenotype of sox9b morpholino knockdown and mutants revealed distinct roles for sox9a and sox9b in neural crest and cartilage development (Yan et al., 2005). sox9b mutant have reduced first and second pharyngeal arch cartilages. In sox9b homozygous mutants chondrocytes stack normally but are reduced in number, suggesting a specific role of sox9b is regulating survival of these precursors (Yan et al., 2005). Each single mutant retains different amounts of pharyngeal cartilages, the double mutants are completely lacking cranial cartilages (Yan et al., 2005).
sox9b is expressed earlier than sox9a in neural crest progenitors and appears to carry a function in neural crest specification similar to the one described for Xenopus Sox9 (Spokony et al., 2002). sox9b mutant embryos show reduced expression of the early neural crest markers snail1b, foxd3 and sox10 at the neural plate border. Moreover, in overexpression studies, sox9a and sox9b have a positive regulatory influence on each other's expression and cause ectopic expression of several early neural crest markers consistent with an early function of sox9b in neural crest specification (Yan et al., 2005).
Sox9 homozygous mutant mouse embryos die midway through gestation, too early to evaluate any cranial malformation (Bi et al., 1999). Sox9 heterozygous mutants die after birth with major skeletal defects, characterized by hypoplasia of all bones formed by endochondral ossification of cartilage, including a shortened jaw and cleft palate reminiscent of the malformations seen in CD patients (Bi et al., 2001). The skeleton of the body forms either by endochondral or intramembranous bone formation. Endochondral bone formation is the process by which bone develops from cartilage, while in intramembranous ossification mesenchymal cells differentiate directly into osteoblasts. The neural crest-derived craniofacial skeleton develops using both mechanisms.
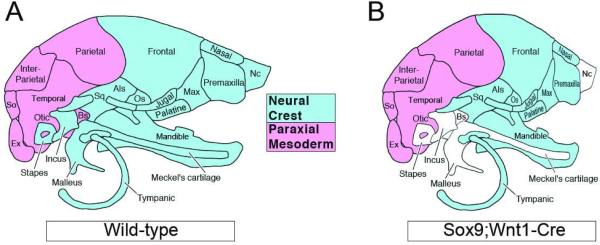
To evaluate the role of Sox9 in the neural crest lineage, Sox9 was inactivated in neural crest cells by means of a Wnt1-Cre transgene (Mori-Akiyama et al., 2003). Sox9;Wnt1-Cre embryos died at birth from respiratory distress because of a large cleft palate. Newborn mutants exhibited craniofacial deformities characterized by a domed skull, a short snout, and short mandibles. Skeletal preparations of mutant skulls showed that all of the cartilages and endochondral bones including the basisphenoid and presphenoid, were missing. Among the first pharyngeal arch-derived skeletal elements of the mutants, all intramembranous bones were conserved. In contrast, the endochondral bones, such as malleus and incus, and the nasal capsule were totally absent. Meckel's cartilage was also completely absent, and the mandibles were smaller than those of the wild type embryos. The skeletal elements derived from the second and third branchial arches, such as the stapes, the hyoid bone and the styloid process were missing in the mutant embryos (Fig 3). These defects result not from abnormal migration or increased apoptosis in the cranial neural crest compartment, but rather from an inability of the post-migrating cranial neural crest cells to differentiate into chondrocytes (Mori-Akiyama et al., 2003). Early on, the expression of Foxd3, Snail and Sox10 was unaffected in the premigratory neural crest of mutant embryos suggesting that neural crest specification is initiated normally in the absence of Sox9 (Cheung et al., 2005), unlike what has been reported in zebrafish and Xenopus (Spokony et al., 2002; Yan et al., 2005). This difference in the early requirement for Sox9 in neural crest specification may reflect species-specific differences in the relative importance of the three SoxE proteins during development of the neural crest lineage (Hong and Saint-Jeannet, 2005; Haldin and LaBonne, 2010).
Figure 3. Diagram illustrating the craniofacial defects observed in Sox9;Wnt1-Cre mouse embryos.
(A) Diagram showing the paraxial mesoderm (red) and neural crest (blue) contribution to the mouse head skeleton. Lateral view, anterior to right. Als, Alisphenoid; Bs, Basisphenoid; Ex, Exooccipital; Nc, Nasal capsule; Os, Orbitosphenoid; So, Supraoccipital; Sq, Squamosal (modified from Noden and Schneider, 2006). (B) Sox9;Wnt1-Cre mouse embryos have a domed skull, a short snout and short mandibles. In these animals the missing skeletal elements of neural crest origin are depicted in white.
Cleft palate is a major defect observed in Sox9 mutant mouse embryos (Bi et al., 2001; Mori-Akiyama et al., 2003). In mutant animals the development of the palate is arrested after the initial outgrowth of the palatal shelves. Sox9 is normally expressed within the subepithelial mesenchyme of the palatal shelves before and during fusion, and therefore is likely to be required for the growth and fusion process of the palatal shelves (Yamashiro et al., 2004; Nie, 2006). Sox9 mutants also show delayed posterior frontal suture closure. The posterior frontal suture is a mesenchymal structure derived from the cranial neural crest, and suture closure is a process occurring through endochondral ossification. In wild-type embryos, Sox9 expression is typically upregulated within the suture mesenchyme prior to suture closure (Sahar et al., 2005).
In summary, these studies show that during craniofacial development Sox9 is required in neural crest cells that will generate both the chondrocyte and osteoblast lineages participating in endochondral bone formation. However, Sox9 is dispensable for the osteoblast lineage in intramembranous skeletal elements derived from the neural crest. Additionally, in fish and frogs, Sox9 appears to have also an early function in neural crest specification.
SOX9 mutations and the craniofacial features of CD patients
Campomelic dysplasia (CD; OMIM # 114290) is a rare autosomal dominant skeletal dysmorphology syndrome characterized by congenital bowing of the long bones (campomelia), hypoplastic scapulae, eleven pairs of ribs, pelvic and spinal malformations, clubbed feet, facial dysmorphia and Pierre Robin sequence, which includes micrognathia (small lower jaw), macroglossia (enlarged tongue), and cleft palate (Maroteaux et al., 1971; Mansour et al., 1995). Campomelia is absent in about 10% of the cases, and the disorder is then referred to as acampomelic campomelic dysplasia, or ACD (Houston et al., 1983; Mansour et al., 1995; Moog et al., 2001). Other features include laryngotracheomalacia (softening of the laryngo-tracheal cartilages) with respiratory deficiencies, ambiguous genitalia or male to female sex reversal in 75% of XY CD patients, hearing impairment, and a variety of congenital heart defects have been described in a small number of cases (Houston et al., 1983).
CD can be identified on prenatal ultrasound examinations based on the shortness and bowing of the long bones (Olney et al., 1999), however ACD is often undetectable. Newborns with CD usually die in the first year of life due to respiratory distress. However there are a few reports of affected infants surviving the neonatal period. In a study published in 2002 (Mansour et al., 2002), all five surviving patients analyzed displayed distinctive facial anomalies such as macrocephaly, depressed nasal bridge, hypertelorism (increased distance between the eyes), elongated philtrum and micrognathia with or without cleft palate. The clinical problems of these individuals were primarily of a respiratory nature, including recurrent apnoea sometimes requiring tracheotomy. Later in life, complications included conductive hearing loss and developmental delay (Mansour et al., 2002). Because most patients present the symptoms as a result of de novo mutations, parents are typically not affected. There are few examples in which both the parent and child are presenting the symptoms, but in all cases the parent is only mildly affected and diagnosed only after the birth of a severely affected child (Mansour et al., 2002; Savarirayan et al., 2003; Leipoldt et al., 2007).
CD is due to haploinsufficiency of the transcription factor SOX9 on chromosome 17q24.3. SOX9 was first identified as the candidate gene for CD in patients with reciprocal de novo translocations upstream of the SOX9 coding sequence (Young et al., 1992; Tommerup et al., 1993; Wagner et al., 1994; Foster et al., 1994). Subsequently, the characterization of de novo heterozygous loss-of-function mutations within the SOX9 coding region were also linked to CD cases (Wagner et al., 1994; Foster et al., 1994; Kwok et al., 1995; Meyer et al., 1997). Nonsense, missense and frameshift mutations in the SOX9 coding region represent approximately 90% of CD cases. The remaining cases are the result of SOX9 deletions, translocations, or inversions upstream of SOX9. Nonsense and frameshift mutations of SOX9 are distributed over the entire coding region, while missense mutations are clustered in the HMG domain, interfering with DNA binding (Meyer et al., 1997, McDowall et al., 1999) or in the dimerization domain, preventing dimer formation (Bernard et al., 2003, Sock et al., 2003). Nonsense and most frameshift mutations at the C-terminus result in a SOX9 protein with truncation of the transactivation domain. In mutations at the N-terminus, typically part or the entire HMG domain is missing. The corresponding mutant proteins lacking both the transactivation domain and the HMG domain constitute loss-of-function alleles, while mutant proteins retaining the HMG domain may function as dominant-negative alleles (McDowall et al., 1999; Preiss et al., 2001).
Transcriptional regulation of SOX9 involves a ~1 Mb cis-regulatory domain upstream of SOX9. Translocation and inversion breakpoints that interrupt this upstream sequence are all unique but fall within the same breakpoint clusters: a proximal cluster and a distal cluster located ~400 kb apart (Leipoldt et al., 2007). In long-term survivors of CD de novo translocations or inversions with breakpoints upstream of SOX9 are more likely to be seen than mutations in the SOX9 coding region, suggesting that they are generally less severe than the intragenic mutations (Pfeifer et al 1999, Leipoldt et al 2007). Recently, lesions have been described that map upstream of the 1 Mb regulatory domain and downstream of SOX9 in patients with isolated Pierre Robin sequence, a craniofacial disorder characterized by micrognathia, cleft palate and macroglossia (Jackobsen et al., 2007, Benko et al., 2009). Pierre Robin sequence is also one of the more consistent features of CD and ACD. The identification of mutations on either side of SOX9 associated with craniofacial defects seen in CD patients indicates that SOX9 regulatory sequences are covering a much larger genomic region than initially suspected. Further these observations support the existence of tissue specific enhancers acting over a long distance to regulate SOX9 expression during craniofacial development. The loss or dysregulation of these regulatory regions around SOX9 may account for the spectrum of craniofacial defects described in CD and ACD. A couple of excellent reviews have recently discussed these findings (Gordon et al., 2009; Amiel et al., 2010).
Acknowledgements
We thank Allison Williams for comments on the manuscript, and for assistance in the preparation of Figure 2. We apologize to colleagues whose work is not cited here, due to space limitations. Work in J-P S-J's lab is supported by grants from the National Institutes of Health (RO1-DE014212 and RO1-DC07175).
References
- Amiel J, Benko S, Gordon CT, Lyonnet S. Disruption of long-distance highly conserved noncoding elements in neurocristopathies. Ann N Y Acad Sci. 2010;1214:34–46. doi: 10.1111/j.1749-6632.2010.05878.x. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink EK, Lee Y-H, Credidio C, Saint-Jeannet J-P. Sox10 regulates the development of the neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–97. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, McBride D, Golzio C, Fisher M, Perry P, Abadie V, Ayuso C, Holder-Espinasse M, Kilpatrick N, Lees MM, Picard A, Temple IK, Thomas P, Vazquez MP, Vekemans M, Crollius HR, Hastie ND, Munnich A, Etchevers HC, Pelet A, Farlie PG, Fitzpatrick DR, Lyonnet S. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet. 2003;12:1755–1765. doi: 10.1093/hmg/ddg182. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–55. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Chiang EF, Pai CI, Wyatt M, Yan YL, Postlethwait J, Chung B. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kowk G, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, Brook JD, Schafer AJ. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- Gordon CT, Tan TY, Benko S, Fitzatrick D, Lyonnet S, Farlie PG. Long-range regulation of the SOX9 locus in development and disease. J Med Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- Gross JB, Hanken J. Segmentation of the vertebrate skull: neural-crest derivation of adult cartilages in the clawed frog, Xenopus laevis. Int Comp Biol. 2008;48:681–696. doi: 10.1093/icb/icn077. [DOI] [PubMed] [Google Scholar]
- Haldin CE, Labonne C. SoxE factors as multifunctional neural crest regulatory factors. Int J Bioch Cell Biol. 2010;42:441–444. doi: 10.1016/j.biocel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C-S, Saint-Jeannet J-P. Sox proteins and neural crest development. Sem. Cell Dev Biol. 2005;16:694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Houston CS, Opitz JM, Spranger JW, Macpherson RI, Reed MH, Gilbert EF, Herrmann J, Schinzel A. The campomelic Syndrome: Review, report of 17 cases, and follow-up on the currently 17-year-old boy first reported by Maroteaux et al. in 1971. Amer J Med Genet. 1983;15:3–28. doi: 10.1002/ajmg.1320150103. [DOI] [PubMed] [Google Scholar]
- Jakobsen LP, Ullmann R, Christensen SB, Jensen KE, Molsted K, Henriksen KF, Hansen C, Knudsen MA, Larsen LA, Tommerup N, Tumer Z. Pierre Robin sequence may be caused by dysregulation of SOX9 and KCNJ2. J Med Genet. 2007;44:381–386. doi: 10.1136/jmg.2006.046177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Kerney R, Gross JB, Hanken J. Runx2 is essential for larval hyobranchial cartilage formation in Xenopus laevis. Dev Dyn. 2007;236:1650–1662. doi: 10.1002/dvdy.21175. [DOI] [PubMed] [Google Scholar]
- Kwok C, Weller PA, Guioli S, Foster JW, Mansour S, Zuffardi O, Punnett HH, Dominguez-Steglich MA, Brook JD, Young ID, et al. Mutations in SOX9, the gene responsible for campomelic dysplasia and autosomal sex reversal. Am J Hum Genet. 1995;57:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- LeDouarin NM, Kalcheim C. The Neural Crest. Second edition Cambridge Univ. Press; London: 1999. [Google Scholar]
- Lee YH, Aoki Y, Hong CS, Saint-Germain N, Credidio C, Saint-Jeannet JP. Early requirement of the transcriptional activator Sox9 for neural crest specification in Xenopus. Dev Biol. 2004;275:93–103. doi: 10.1016/j.ydbio.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, de Crombrugghe B. Towards understanding Sox9 function in chondrocyte differentiation. Matrix Biology. 1998;16:529–540. doi: 10.1016/s0945-053x(98)90065-8. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res. 2005;75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- Leipoldt M, Erdel M, Bien-Willner G, Smyk M, Theurl M, Yatsenko S, Lupski J, Lane A, Shanske A, Stankiewicz P, Scherer G. Two novel translocation breakpoints upstream of SOX9 define borders of the proximal and distal breakpoint cluster region in campomelic dysplasia. Clin Genet. 2007;71:67–75. doi: 10.1111/j.1399-0004.2007.00736.x. [DOI] [PubMed] [Google Scholar]
- Li M, Zhao C, Wang Y, Zhao Z, Meng A. Zebrafish sox9b is an early neural crest marker. Dev Genes Evol. 2002;212:203–206. doi: 10.1007/s00427-002-0235-2. [DOI] [PubMed] [Google Scholar]
- Lovell-Badge R. The early history of the Sox genes. Int J Bioch Cell Biol. 2010;42:378–380. doi: 10.1016/j.biocel.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Mansour S, Hall CM, Pembrey ME, Young ID. A clinical and genetic study of campomelic dysplasia. J Med Genet. 1995;32:415–420. doi: 10.1136/jmg.32.6.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S, Offiah AC, McDowall S, Sim P, Tolmie J, Hall C. The phenotype of survivors of campomelic dysplasia. J Med Genet. 2002;39:597–602. doi: 10.1136/jmg.39.8.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroteaux P, Spranger J, Opitz JM, Kucera J, Lowry RB, Schimke RN, Kagan SM. Le syndrome campomelique. Presse Med. 1971;79:1157–1162. [PubMed] [Google Scholar]
- McDowall S, Argentaro A, Ranganathan S, Weller P, Mertin S, Mansour S, Tolmie J, Harley V. Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J Biol Chem. 1999;274:24023–24030. doi: 10.1074/jbc.274.34.24023. [DOI] [PubMed] [Google Scholar]
- Meyer J, Sudbeck P, Held M, Wagner T, Schmitz ML, Bricarelli FD, Eggermont E, Friedrich U, Haas OA, Kobelt A, Leroy JG, Van Maldergem L, Michel E, Mitulla B, Pfeiffer RA, Schinzel A, Schmidt H, Scherer G. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum Mol Genet. 1997;6:91–98. doi: 10.1093/hmg/6.1.91. [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Moog U, Jansen NJG, Scherer G, Schrander-Stumpel CTRM. Acampomelic campomelic syndrome. Am J Med Gen. 2001;104:239–245. [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci USA. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- Nie X. Sox9 mRNA expression in the developing palate and craniofacial muscles and skeletons. Acta Odontol Scand. 2006;64:97–103. doi: 10.1080/00016350500420089. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North Holland Publishing Company; Amsterdam, The Netherlands: 1967. [Google Scholar]
- Noden DM, Schneider RA. Neural crest cells and the community of plan for craniofacial development: Historical debates and current perspectives. Adv Exp Med Biol. 2006;589:1–23. doi: 10.1007/978-0-387-46954-6_1. [DOI] [PubMed] [Google Scholar]
- Olney PN, Kean LS, Graham D, Elsas LJ, May KM. Campomelic syndrome and deletion of Sox9. Am J Med Genet. 1999;84:20–24. [PubMed] [Google Scholar]
- Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, Birren B, Korniszewsky L, Back E, Scherer G. Campomelic dysplasia translocation breakpoints are scattered over 1Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet. 1999;65:111–124. doi: 10.1086/302455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss S, Argentaro A, Clayton A, John A, Jans DA, Ogata T, Nagai T, Barroso I, Schafer AJ, Harley VR. Compound effects of point mutations causing campomelic dysplasia/autosmal sex reversal upon SOX9 structure, nuclear transport, DNA binding, and transcriptional activation. J Biol Chem. 2001;276:27864–27872. doi: 10.1074/jbc.M101278200. [DOI] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Sahar DE, Longaker MT, Quarto N. Sox9 neural crest determinant gene controls patterning and closure of the posterior frontal cranial suture. Dev Biol. 2005;280:244–261. doi: 10.1016/j.ydbio.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Savarirayan R, Robertson SP, Bankier A, Rogers JG. Variable expression of campomelic dysplasia in a father and his 46, XY daughter. Ped Path Mol Med. 2003;22:37–46. doi: 10.1080/pdp.22.1.37.46. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–70. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, Scherer G. Loss of DNAdependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum Mol Genet. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink EK, Saint-Jeannet J-P. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Wegner M. SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol. 2010;42:441–444. doi: 10.1016/j.biocel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Tommerup N, Schempp W, Meinecke P, Pedersen S, Bolund L, Brandt C, Goodpasture C, Guldberg P, Held KR, Reinwein H, Saugstad OD, Scherer G, Skjeldal O, Toder R, Westvik J, van der Hagen CB, Wolf U. Assignment of an autosomal sex reversal locus (SRA1) and campomelic dysplasia (CMPD1) to 17q24.3–q25.1. Nat Genet. 1993;4:170–174. doi: 10.1038/ng0693-170. [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- Yamashiro T, Wang XP, Li Z, Oya S, Aberg T, Fukunaga T, Kamioka H, Speck NA, Takano-Yamamoto T, Thesleff I. Possible roles of Runx1 and Sox9 in incipient intramembranous ossification. J Bone Miner Res. 2004;19:1671–1677. doi: 10.1359/JBMR.040801. [DOI] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Young ID, Zuccollo JM, Maltby EL, Broderick NJ. Campomelic dysplasia associated with a de novo 2q;17q reciprocal translocation. J Med Genet. 1992;29:251–252. doi: 10.1136/jmg.29.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]