Abstract
Lanosterol 14α-demethylase (CYP51F1) from Candida albicans is known to be an essential enzyme in fungal sterol biosynthesis. Wild-type CYP51F1 and several of its mutants were heterologously expressed in Escherichia coli, purified, and characterized. It exhibited a typical reduced CO-difference spectrum with a maximum at 446 nm. Reconstitution of CYP51F1 with NADPH-P450 reductase gave a system that successfully converted lanosterol to its demethylated product. Titration of the purified enzyme with lanosterol produced a typical type I spectral change with Kd = 6.7 μM. The azole antifungal agents econazole, fluconazole, ketoconazole, and itraconazole bound tightly to CYP51F1 with Kd values between 0.06 – 0.42 μM. The CYP51F1 mutations F105L, D116E, Y132H, and R467K frequently identified in clinical isolates were examined to determine their effect on azole drug binding affinity. The azole Kd values of the purified F105L, D116E, and R467K mutants were little altered. A homology model of C. albicans CYP51F1 suggested that Tyr132 in the BC loop is located close to the heme in the active site, providing a rationale for the modified heme environment caused by the Y132H substitution. Taken together, functional expression and characterization of CYP51F1 provide a starting basis for design of agents effective against C. albicans infections.
Keywords: P450, CYP51, lanosterol, azole, Candida albicans
Introduction
Cytochrome P450s (P450s, CYPs) are heme-thiolate-containing enzymes that catalyze reactions involving both endogenous and exogenous substrates [1, 2]. These enzymes are found throughout nature and in microorganisms play important roles, particularly in the biosynthesis of antibiotics and other biologically active molecules [3]. Genomic analysis of Candida albicans suggests it contains 10 putative P450 enzymes, including CYP51, CYP52 and CYP61. It also encodes one NADPH-P450 reductase (NPR) that can deliver electrons to both P450 enzymes and heme oxygenase (http://www.candidagenome.org/) [4].
C. albicans is a well known pathogenic fungus of opportunistic oral and vaginal infections in humans [5, 6]. Due to the recent increase in immunocompromised patients caused by HIV, cancer chemotherapy, and organ/bone marrow transplantation, systemic infections of C. albicans remain a serious clinical problem [7, 8]. A frequent occurrence of numeric and structural chromosomal shuffling in the C. albicans genome and an ability to switch its colony phenotype are its best-known adaptation characteristics [9].
The CYP51 family is very special, as there is a strict functional conservation of enzyme activity among its members in all biological kingdoms except for some nematodes and insects that lack CYP51 and therefore obtain sterols from their diet [10, 11]. They all catalyze the same three sequential oxidative steps that remove the 14α-methyl group from post-squalene sterol precursors [10]. Azole antifungal agents are widely used in the treatment of fungal infections and fungal CYP51 enzymes are considered as their primary target [12]. However, the mechanism of antifungal action at the molecular level has not yet been fully clarified. Azole inhibition of CYP51 in fungi causes the accumulation of membrane-disrupting methylated ergosterol precursors that prevent fungal growth [13]. Azole drugs bind as a sixth ligand to the heme iron atom of CYP51 to give a Type II spectrum with a maximum at 430 nm and a minimum at 410 nm [14]. This interaction involves coordination of the iron with N3 of an imidazole ring or N4 of a triazole ring in the azoles.
Azole-resistant strains have emerged as a serious problem in the clinical use of azole drugs in the treatment to C. albicans infections. Amino acid substitutions in CYP51F1 gene sequences have been found in azole-resistant C. albicans isolates [14, 15]. Interestingly, in the studies of isolates from patients during the emergence of resistance, increased transcriptional levels of CYP51F1 and the drug transporters CDR and MDR1 were observed in addition to the mutations in the CYP51F1 amino acid sequence [16, 17]. Variations in the CYP51F1 gene are thought to be associated with resistance to azole antifungal agents, but few detailed biochemical studies at the protein level have so far addressed this problem.
In this study, we have successfully cloned, overexpressed, and purified CYP51F1 and several of its variants from C. albicans and characterized these proteins in terms of their spectroscopic and catalytic properties.
Materials and Methods
Chemicals and Enzymes
Lanosterol, econazole, fluconazole, itraconazole, ketoconazole, sodium dithionite, glucose-6-phosphate, glucose-6-phosphate dehydrogenase, and NADP+ were purchased from Sigma (St. Louis, MO) or Aldrich Chemical Co. (Milwaukee, WI). Other chemicals were of the highest grade commercially available. Escherichia coli DH5α cells were purchased from Invitrogen (Carlsbad, CA). Recombinant rat NPR was expressed in E. coli and purified as previously reported [18].
Construction of Expression Plasmids and Site-directed Mutagenesis
The general approach has been described previously [19, 20]. The genomic DNA from C. albicans was kindly provided by Professor Won Ki Huh at Seoul National University. The open reading frame for CYP51F1, and an added 6×His-C-terminal tag, were amplified using PCR with forward and reverse primers (5′-AAACAGGATCCATCGATGCTTAGGAGGTCATATGGCTATTGTTGAAAC-′3, 5′-ATTATTTCTAGACCGGAAGCTTTTAGTGATGGTGATGGTGATGAAACATACA AGTTTCTCT-′3) and the amplified PCR fragment was cloned into the pCW(Ori+) vector using the BamHI and XbaI restriction sites. The cloned vectors were verified by nucleotide sequencing analysis and restriction digestion. C. albicans translates CTG as Ser instead of Leu, the amino acid for which CTG is a universal codon [21]. The open reading frame of CYP51F1 contains a CTG encoding a Ser that must be corrected by site-directed mutagenesis to enable expression of the correct recombinant protein in E. coli. Site-directed mutagenesis to change Leu263 to a Ser was carried out using Quick-Change mutagenesis (Stratagene, La Jolla, CA) as previously described [20]. Four CYP51F1 variant clones containing F105L, Y132H, D116E, and R467K substitutions were constructed by site-directed mutagenesis.
Enzyme Expression and Purification
Expression and purification of the CYP51F1 enzyme were carried out as previously described with some modifications [22]. Briefly, the E. coli strains transformed with pCW(Ori+) vectors were inoculated into TB medium containing 100 μg/ml ampicillin and 1.0 mM IPTG. The expression cultures were grown at 37 °C for 3 h and then at 28 °C with shaking at 200 rpm for 24 h in 1 liter Fernbach flasks. Bacterial inner membrane fractions containing CYP51F1 were isolated and prepared from 1 liter TB (with ampicillin, 100 μg/ml) expression cultures of E. coli DH5α. Purification of CYP51F1 enzyme using a Ni2+-nitrilotriacetate column was as previously described [22, 23]. Briefly, membranes were solubilized at 4 °C overnight in 100 mM potassium phosphate buffer (pH 7.4) containing 20 % (w/v) glycerol, 0.5 M NaCl, 10 mM β-mercaptoethanol, and 1.5 % (w/v) CHAPS. The solubilized fraction was then loaded onto a Ni2+-nitrilotriacetate column (Qiagen, Valencia, CA) and the purified protein was obtained with the elution buffer containing 400 mM imidazole. Eluted fraction containing P450 was dialyzed at 4 °C against 100 mM potassium phosphate buffer (pH 7.4) containing 20 % (v/v) glycerol, 0.1 mM EDTA.
Spectroscopic Characterization
The CO-P450 complexes were generated by passing CO gas through solutions of the P450. Then, sodium dithionite was added to reduce the purified ferric CYP51F1 enzymes. UV-visible spectra were collected on a CARY 100 Varian spectrophotometer in 100 mM potassium phosphate buffer (pH 7.4) at room temperature.
Spectral Binding Titrations
Purified CYP51F1 wild-type and variant enzymes were diluted to 2 μM in 100 mM potassium phosphate buffer (pH 7.4) and divided between two glass cuvettes. Spectra (350 – 500 nm) were recorded with subsequent additions of substrate (lanosterol) or azole compounds (in methanol stock) using a CARY 100 Varian spectrophotometer [24]. The difference in absorbance between the wavelength maximum and minimum was plotted versus the substrate concentration [25, 26]. The data were fit to the typical hyperbolic equation for lanosterol and the quadratic equation for azole drugs using nonlinear regression analysis with Graph-Pad Prism software (Graph-Pad, San Diego, CA).
Lanosterol Demethylation Assay
Lanosterol demethylation by CYP51F1 was determined using a P450/NPR/phospholipid reconstituted system. The reaction mixture included 100 pmol purified P450 enzyme, 200 pmol rat NPR, and DLPC (45 μM) in 0.50 ml of 100 mM potassium phosphate buffer (pH 7.4), along with lanosterol (200 μM). An NADPH-generating system [26] was used to start reactions. Incubations were generally done for 30 min at 37 °C and terminated by addition of l ml of CH2Cl2. The CH2C2 extract was dried under N2 and then converted to trimethylsilyl derivatives by incubation with 50 μl of N,O-bis(trimethylsilyl)trifluoroacetamide at 70 °C for 10–15 min. The derivatized samples were allowed to cool, vortexed, and transferred to sealed Teflon-capped glass vials for either manual or autoinjection into the GC-MS. Analyses were performed on an Agilent 6850 gas chromatograph coupled to an Agilent 5973 Network Mass Selective Detector using electron ionization (EI) as previously described [27].
Homology Modeling
A molecular model of C. albicans CYP51F1 structure was constructed using the X-ray crystal structure of M. tuberculosis CYP51 (PDB 1EA1) as a template. The coordinates of the model were obtained using the SWISS-MODEL software from the Swiss Institute of Bioinformatics (http://swissmodel.expasy.org/).
Results
Sequence Alignment with Other P450s
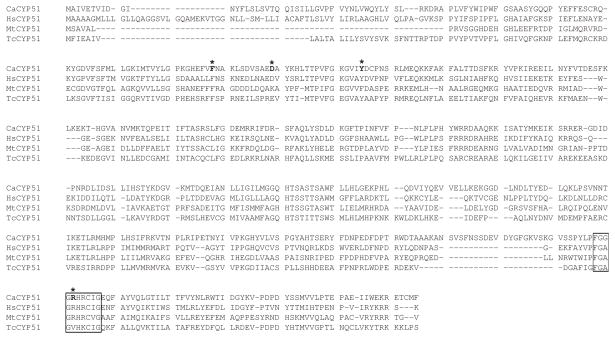
The ERG11 gene from C. albicans (Genebank: X13296, orf19_922) encodes CYP51F1 (lanosterol 14α-demethylase), a member of the cytochrome P450 family that functions in ergosterol biosynthesis. Phylogenetically, CYP51 enzymes are highly conserved throughout nature from bacteria, through fungi, to humans. Amino acid sequence alignment of CYP51F1 from C. albicans with CYP51 of other species reveals high sequence similarities with scores from 85 to 87% identity (Fig. 1) [28]. Good sequence alignment with human, Mycobacterium tuberculosis, and Trypanosoma cruzi CYP51 suggests that the gene product ERG11 from C. albicans is likely to have lanosterol 14α-demethylase catalytic activity.
Fig. 1.
Sequence alignment of C. albicans CYP51F1 with other CYP51 enzymes. The amino acid sequences were aligned using the software T-Coffee (http://www.tcoffee.org). The residues corresponding to the conserved heme binding sites in the CYP51 family are shown with boxes. The alignment scores of C. albicans CYP51F1 to human, Mycobacterium tuberculosis, and Trypanosoma cruzi CYP51’s were 86, 87, and 85 respectively.
Expression and Purification of CYP51F1
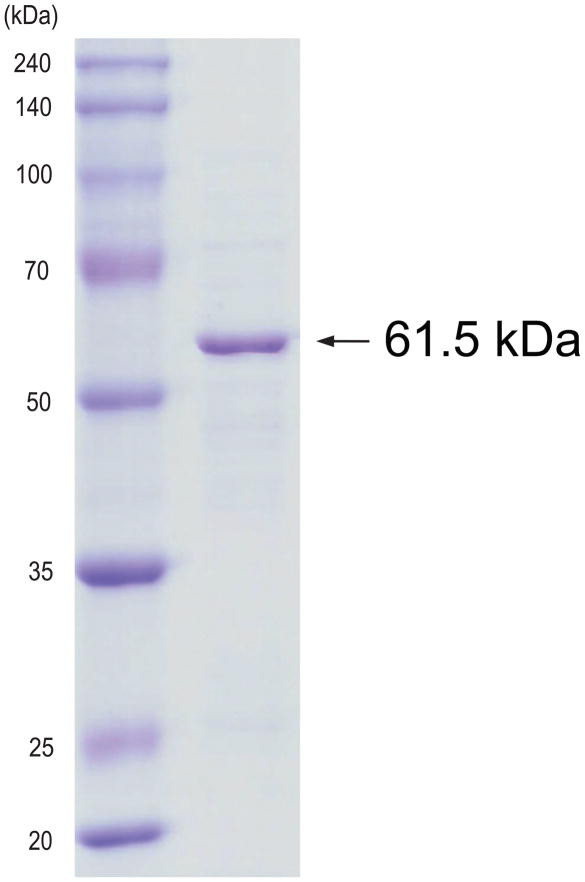
The reduced CO-binding spectrum in the whole cell culture showed the typical P450 expression level of ~50 nmol P450 holoenzyme per liter of culture medium (data not shown). After ultracentrifugation, the expressed P450 protein was observed in the membrane fraction. CHAPS successfully solubilized the membrane-bound P450 protein and the solubilized protein was purified on a Ni-column. The resulting protein migrated on SDS-PAGE as a single band at 61.5 kDa, as expected for the open reading frame of the CYP51F1 gene with a His-tag (Fig. 2). Protein sequencing of the purified protein by mass-spectrometry indicated that the purified protein was the expected CYP51F1 from C. albicans (Supplementary Fig. 1).
Fig. 2.
SDS-PAGE of the purified CYP51F1 protein.
Spectral Properties of CYP51F1
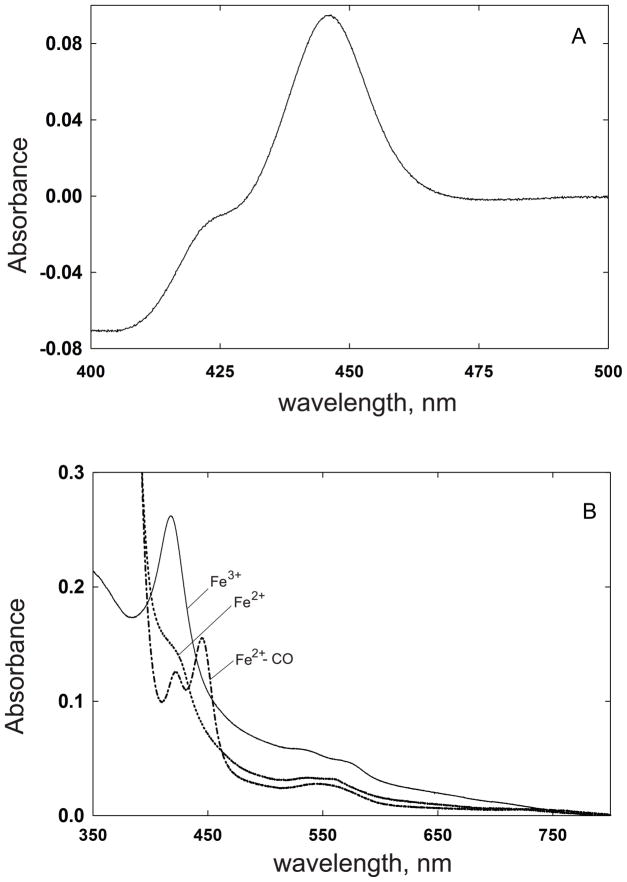
The reduced CO difference spectrum of purified CYP51F1 had a maximum absorption at 446 nm (Fig. 3A). Examination of the absolute spectrum showed that the ferric form of the protein was in the low spin state with a Soret band at 419 nm, while the ferrous protein had a broad absorption peak around 420 nm (Fig. 3B). The smaller α-, and β-bands of the ferric P450 were at 568 and 536 nm, respectively (Fig. 3B). On reduction with sodium dithionite, the α-, and β-bands shifted to 559 and 530 nm respectively.
Fig. 3.
Spectral analysis of the purified CYP51F1 protein. (A) The Fe2+·CO vs Fe2+ difference spectrum; (B) The absolute spectra of the Fe3+, Fe2+, and Fe2+·CO forms were recorded.
Catalytic Activities of CYP51F1
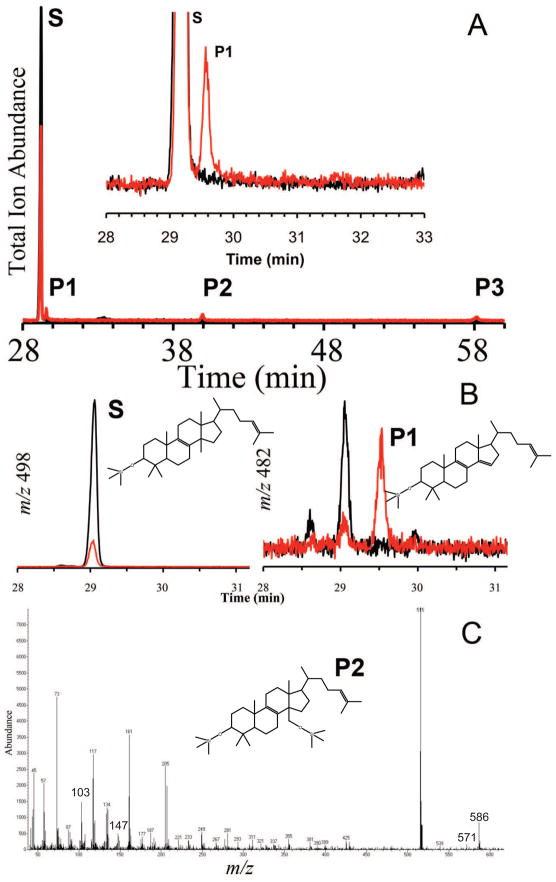
Purified CYP51F1 reconstituted with rat NPR supported the catalytic turnover of lanosterol to yield three GC-MS detectable products (Fig. 4A). The first product peak (P1) in the chromatogram was determined as the final 14α-demethylated metabolite (FF-MAS, follicular fluid-meiosis activating steroid) of lanosterol, although it was partially masked by the tail of the lanosterol peak. Selected ion monitoring (SIM) scans for m/z = 498 (lanosterol TMS ether) and also m/z = 482 (FF-MAS TMS ether) confirmed that peak P1 did in fact correspond to FF-MAS (Fig. 4B). The second product peak (P2) was consistent with the product of oxidation of the methyl to the alcohol, the expected first step of lanosterol demethylation (Fig. 4C). Its fragmentation pattern was consistent with the TMS-ether. The third product (P3) could not be identified. It had a mass 44 Da higher than that of the alcohol but did not fit with any of the intermediates expected in the multi-step demethylation mechanism.
Fig. 4.
GC-MS of products formed in the reaction of CYP51F1 with lanosterol. (A) GC-MS total ion trace of the reaction products, with the inset showing an expanded view of the substrate (lanosterol) and FF-MAS (P1); (B) CYP51F1 reactions and controls were also analyzed in selected ion monitoring (SIM) mode. The SIM spectra for m/z 498 and m/z 482 correspond to lanosterol and FF-MAS, respectively. For each SIM spectrum the RED trace corresponds to CYP51F1 incubations while the black spectrum represents control reactions. (C) elecron impact mass-spectrum of hydroxylanosterol TMS-ether (P2). The molecular ion [M]+• is found at m/z 586, M – CH3 at m/z 571, and the signature ion at m/z 103 corresponds to loss of CH2OTMS from a primary alcohol.
Binding of Lanosterol and Azole Agents to CYP51F1 Wild type and Variants
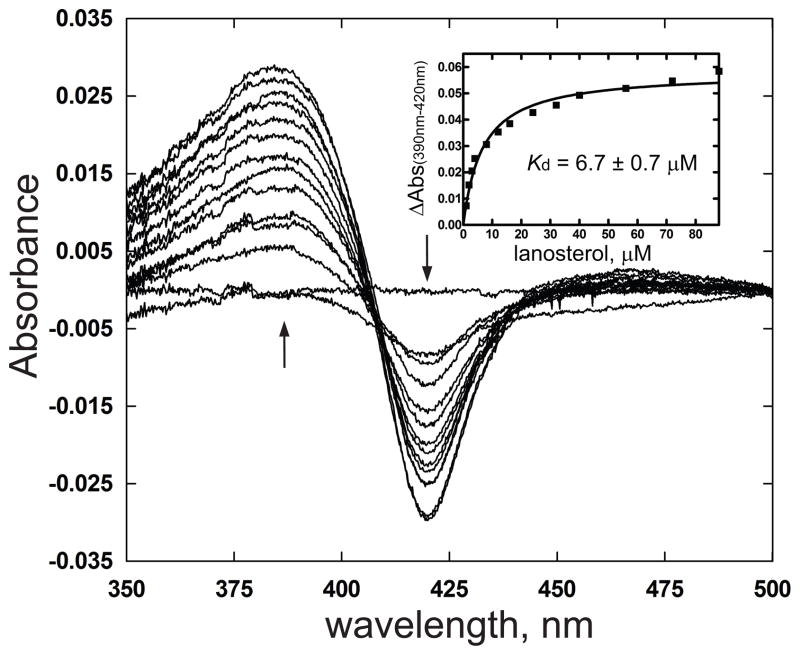
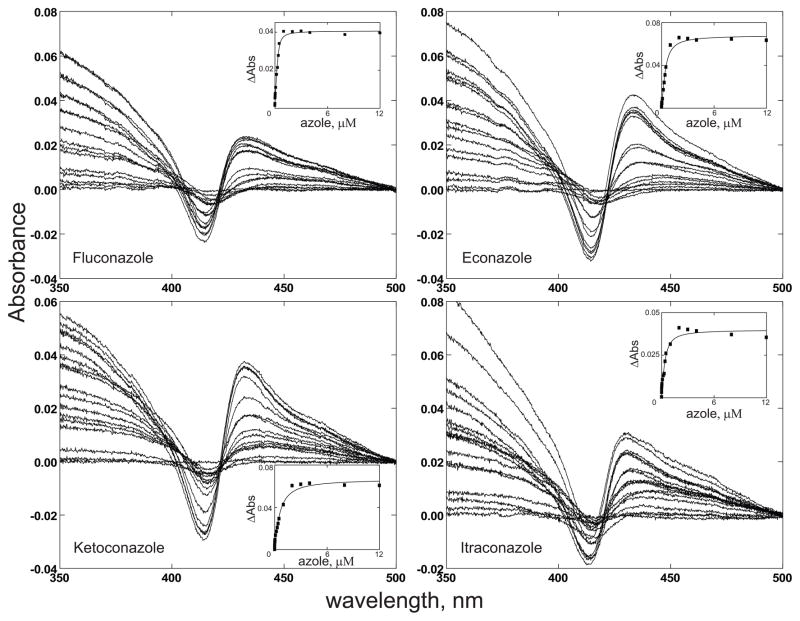
Titration of purified CYP51F1 with lanosterol showed a typical type I spectral change with an increase at 390 nm and decrease at 420 nm, consistent with displacement of the iron-bound water molecule by the lanosterol to give a low-spin hexa coordinated P450 heme (Fig. 5). The calculated Kd value is 6.7 ± 0.7 μM (Fig. 5). Furthermore, purified CYP51F1 produced a typical type II spectral change, with an increase at 430 nm and a decrease at 410 nm, on binding of azole drugs (Fig. 6), as expected for the binding of a strong ligand to the heme iron atom [29]. The calculated Kd values for the azole drugs are between 0.06 and 0.42 μM (Table 1).
Fig. 5.
Titration of CYP51F1 with lanosterol. Increasing concentrations of lanosterol were added to both the sample and reference cuvettes. The inset shows the plot of ΔA390–420nm vs concentration of lanosterol.
Fig. 6.
Titration of CYP51F1 with azole compounds. Purified CYP51F1 (2 μM) was dissolved in 100 mM potassium phosphate buffer (pH 7.4) and the absorption spectrum of the ferric form (350–500 nm) was recorded with subsequent additions of the azole compound fluconazole, econazole, ketoconazole, or itraconazole. The binding affinities of the azoles calculated from the plots are shown in Table 1.
TABLE I.
Binding affinities of azole agents to the C. albicans CYP51F1 wild-type and mutant proteins
| Azole Agents |
Kd (μM) |
||||
|---|---|---|---|---|---|
| Wild type | F105L | D116E | Y132H | R467K | |
| Econazole | 0.20 ± 0.04 | 0.10 ± 0.03 | 0.14 ± 0.03 | 0.42 ± 0.06 | 0.13 ± 0.03 |
| Fluconazole | 0.06 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.15 ± 0.06 | 0.06 ± 0.01 |
| Itraconazole | 0.19 ± 0.06 | 0.16 ± 0.04 | 0.15 ± 0.03 | NDa | 0.18 ± 0.04 |
| Ketoconazole | 0.42 ± 0.08 | 0.17 ± 0.04 | 0.43 ± 0.08 | 0.54 ± 0.07 | 0.66 ± 0.13 |
not determined
Mutations that occur frequently in the CYP51F1 gene (F105L, D116E, Y132H, and R467K) in clinically isolated azole-resistant C. albicans strains were analyzed to determine their effect on the binding affinity of azole drugs [15]. Recombinant mutant enzymes were expressed and purified as already described. There were no notable differences in the reduced-CO difference and absolute spectra (data not shown). Three of the mutant proteins bound azole agents to give typical type II spectra (Supplementary Figs 2~5). However, binding of azoles to the Y132H mutant showed the expected decrease at 410 nm but little increase at 430 nm (Supplementary Fig 4). Unexpectedly, the Kd values for the binding of azoles to the purified F105L, D116E, and R467K mutants were little altered (Table 1). The binding of lanosterol to the purified F105L, D116E, and R467K mutants occurred with a type I spectral change and binding affinities similar to those for the wild-type enzyme (Supplementary Fig 6). Titration of the Y132H mutant with lanosterol gave the expected decrease at 425 nm, but the distinct increase in the peak at 390 nm was not observed.
Discussion
We previously reported purification and characterization of CYP52A1, the Alk8 gene product from C. albicans [30], that oxidizes dodecanoic acid to the ω-hydroxylated metabolite as a major product and the (ω-1)-hydroxylated compound as a minor one. A functional heme oxygenase from C. albicans, the product of the CaHmx1 gene, has also been characterized [20]. Recently, the NPR of C. albicans, coded by the NCP1 gene, was characterized [31]. The purified C. albicans NPR was shown to be an orthologous reductase that supports both the cytochrome P450 and heme oxygenase activities. Our present biochemical characterization of CYP51F1 from C. albicans complements these earlier studies and broadens our understanding of the critical system of catalytic hemoproteins in C. albicans.
The ERG11 gene in C. albicans encodes CYP51F1, a putative sterol 14α-demethylase, that is required for the biosynthesis of ergosterol. Indeed, purified CYP51F1 converts lanosterol to its demethylated product through initial formation of the alcohol metabolite, but the conversion rate is relatively low (Fig. 4). The aldehyde intermediate expected in the oxidation sequence was not detected, possibly because it is too rapidly oxidized to the acid intermediate. One caveat in the assignment of oxidized products is that the identification of the site of hydroxylation is still putative since it was based only on mass spectral evidence. However, the mass spectrometric evidence is commonly used for these types of metabolites when the metabolite standards are not available [27, 32, 33]. HPLC analysis of the enzyme reactions were performed to obtain the enzyme oxidation rate. A new peak appeared in the chromatogram, assuming the putative lanosterol oxidation product (Supplementary Fig. 8A). But, the identification of the product mass was failed due to no ionization in LC-mass spectrometry. The rate of oxidation of lanosterol based on this HPLC analysis was estimated with assuming the new peak as the lanosterol oxidation product. The calculated kcat value was approximately 0.19 min−1 with a Km value of 20 μM for the wild type CYP51F1 enzyme (Supplementary Fig. 8B). In addition, the putative oxidation product peak disappeared when the lanosterol oxidation reaction was performed with azole compounds, suggesting that the oxidation activity of purified enzyme was inhibited by azole drugs (Supplementary Fig. 8A).
Site-specific mutations in the CYP51F1 gene have been considered as one of the main mechanisms for azole resistance in clinically isolated C. albicans strains. So far, about 140 mutations have been reported in the literature, but few of them have been specifically associated with azole resistance [34]. Multiple mechanisms of resistance have been proposed, including a reduction of intracellular azole concentration or an increased CYP51 expression [14]. Changes in CYP51F1 expression have been detected in resistant strains, but their role in resistance is unclear.
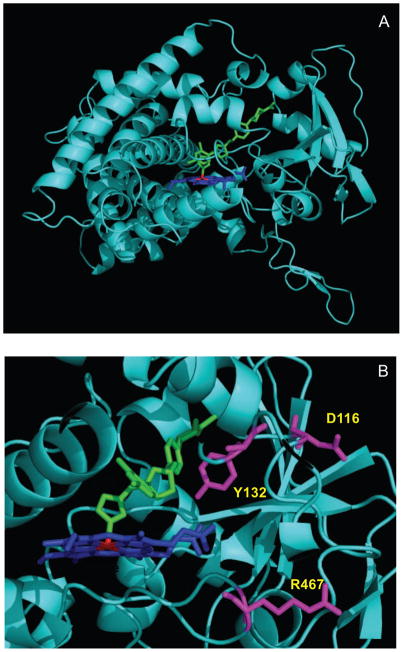
Based on a homology model of C. albicans CYP51F1, we postulate that Tyr132 in the BC loop is located close to the heme in the active site (Fig. 7) and mutations of this residue are likely to modify the active site architecture and heme environment. Kudo et al. reported that an Y132H substitution modified the heme environment but fluconazole resistance was predominantly associated with the double Y132H/F145L mutations [35]. Unusual spectra were obtained on binding of lanosterol or azole drugs to the Y132H mutant in this study. This observation suggests that modification of the heme environment by the substitution Y132H may be a prerequisite for the effective resistance to azole drugs conferred by multiple mutations.
Fig. 7.
Positions of the mutated amino acid residues in CYP51F1. (A) Ribbon diagram of the C. albicans CYP51F1 model using the X-ray crystal structure of M. tuberculosis CYP51 (PDB 1EA1) as a template; (B) The positions of the Y132, D116, and R476 mutations are indicated with colors. Heme and fluconazole are shown in blue and green, respectively.
Interestingly, our study indicated that the azole resistance observed in clinical isolates of C. albicans does not necessarily result only from a lowering of the azole binding affinity of CYP51F1. Thus, at least the four purified mutant enzymes in this study did not show a direct correlation between azole affinity and resistance. Recently, Warrilow et al. also reported that the azole resistance conferred by a I471T mutation of CYP51F1 is not due to significantly alteration of the enzyme’s affinity for azole compounds [36]. They postulated that the increased resistance to azoles conferred by the I471T substitution reflected an enhanced ability of the I471T mutant to remain functionally active at high azole concentrations [36]. Therefore, the mutant enzymes may acquire azole resistance through a combination of mechanisms, not simply by a decreased azole binding affinity. However, C. albicans mutant CYP51F1 enzymes may produce enough ergosterol to maintain cell viability, even if at a very low rate of turnover, despite the presence of a high azole concentration. Measurements of the amount of lanosterol produced by purified mutant enzymes in the presence of high concentrations of azole drugs may provide a clue relative to this postulated mechanism of azole resistance and is currently under investigation.
In conclusion, we have overexpressed, purified, and characterized a critical P450 enzyme from the pathogenic fungus C. albicans. The purified CYP51F1 catalyzes lanosterol demethylation and, interestingly, four of its mutants from azole resistant strains retain essentially unchanged binding affinities for azole drugs. Structure-function studies of this C. albicans CYP51F1 enzyme will provide the basis for the improved design of azole drugs against this pathogen.
Supplementary Material
Protein sequence analysis of N-terminus of purified CYP51F1 using mass-spectrometry.
Titration of purified F105L mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified D116E mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified Y132H mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified R467K mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified mutant proteins (F105L, D116E, Y132H, and R467K) with lanosterol.
Additional spectral analysis of the purified CYP51F1 protein. (A) The absolute spectra of the Fe3+ (—), and Fe3+·lanosterol (----); (B) Second-derivative spectra of purified CYP51F1 Fe3+ form in the absence of lanosterol.
HPLC analysis of the reactions of the purified CYP51F1 protein with lanosterol as substrate. (A) HPLC chromatograms of the oxidation reaction product, Econazole (50 μM) was added to inhibit the reaction as inhibitor; (B) Steady-state kinetic analysis of lanosterol oxidation by CYP51F1. Each point presented is a mean ± SD of duplicate assays.
Acknowledgments
This study was supported by a grant to D.K. from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A084005), and by National Institutes of Health Grant GM25515 to P.R.O.M.
The abbreviations used
- P450 (or CYP)
cytochrome P450
- PCR
polymerase chain reaction
- TB
terrific broth
- DTT
dithiothreitol
- NPR
NADPH-P450 reductase
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
Footnotes
CYP51F1 from Candida albicans is the ERG11 gene (Genebank: X13296, orf19_922) encoding lanosterol 14α-demethylase and it has an amino acid sequence similarity score of 86% with that of human CYP51A1 (NM_000786) lanosterol 14α-demethylase (http://drnelson.uthsc.edu/cytochromeP450.html).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guengerich FP, Tang Z, Salamanca-Pinzon SG, Cheng Q. Mol Interv. 2010;10:153–163. doi: 10.1124/mi.10.3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz de Montellano PR. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Plenum Press; New York: 2005. [Google Scholar]
- 3.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 4.d’Enfert C, Goyard S, Rodriguez-Arnaveilhe S, Frangeul L, Jones L, Tekaia F, Bader O, Albrecht A, Castillo L, Dominguez A, Ernst JF, Fradin C, Gaillardin C, Garcia-Sanchez S, de Groot P, Hube B, Klis FM, Krishnamurthy S, Kunze D, Lopez MC, Mavor A, Martin N, Moszer I, Onesime D, Perez Martin J, Sentandreu R, Valentin E, Brown AJ. Nucleic Acids Res. 2005;33:D353–D357. doi: 10.1093/nar/gki124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odds FC. Crit Rev Microbiol. 1987;15:1–5. doi: 10.3109/10408418709104444. [DOI] [PubMed] [Google Scholar]
- 6.Pendrak ML, Chao MP, Yan SS, Roberts DD. J Biol Chem. 2004;279:3426–3433. doi: 10.1074/jbc.M311550200. [DOI] [PubMed] [Google Scholar]
- 7.Fidel PL., Jr Oral Dis. 2002;8(Suppl 2):69–75. doi: 10.1034/j.1601-0825.2002.00015.x. [DOI] [PubMed] [Google Scholar]
- 8.Kullberg BJ, Oude Lashof AM. Eur J Med Res. 2002;7:183–191. [PubMed] [Google Scholar]
- 9.Calderone R, Odds FC, Boekhout T. FEMS Yeast Res. 2009;9:971–972. doi: 10.1111/j.1567-1364.2009.00585.x. [DOI] [PubMed] [Google Scholar]
- 10.Lepesheva GI, Hargrove TY, Kleshchenko Y, Nes WD, Villalta F, Waterman MR. Lipids. 2008;43:1117–1125. doi: 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strushkevich N, Usanov SA, Park HW. J Mol Biol. 2010;397:1067–1078. doi: 10.1016/j.jmb.2010.01.075. [DOI] [PubMed] [Google Scholar]
- 12.Koltin Y, Hitchcock CA. Curr Opin Chem Biol. 1997;1:176–182. doi: 10.1016/s1367-5931(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 13.Georgopapadakou NH, Walsh TJ. Antimicrob Agents Chemother. 1996;40:279–291. doi: 10.1128/aac.40.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly SL, Kelly DE, Jackson CJ, Warrilow AGS, Lamb DC. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Plenum Press; New York: 2005. pp. 585–617. [Google Scholar]
- 15.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. Microbiology. 1999;145(Pt 10):2701–2713. doi: 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 16.Sanglard D, Ischer F, Monod M, Bille J. Microbiology. 1997;143(Pt 2):405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 17.Franz R, Kelly SL, Lamb DC, Kelly DE, Ruhnke M, Morschhauser J. Antimicrob Agents Chemother. 1998;42:3065–3072. doi: 10.1128/aac.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. Arch Biochem Biophys. 1998;350:324–332. doi: 10.1006/abbi.1997.0534. [DOI] [PubMed] [Google Scholar]
- 19.Kim D, Guengerich FP. Biochemistry. 2004;43:981–988. doi: 10.1021/bi035593f. [DOI] [PubMed] [Google Scholar]
- 20.Kim D, Yukl ET, Moenne-Loccoz P, Ortiz de Montellano PR. Biochemistry. 2006;45:14772–14780. doi: 10.1021/bi061429r. [DOI] [PubMed] [Google Scholar]
- 21.Ohama T, Suzuki T, Mori M, Osawa S, Ueda T, Watanabe K, Nakase T. Nucleic Acids Res. 1993;21:4039–4045. doi: 10.1093/nar/21.17.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Wu ZL, Guengerich FP. J Biol Chem. 2005;280:40319–40327. doi: 10.1074/jbc.M508171200. [DOI] [PubMed] [Google Scholar]
- 23.Yun CH, Miller GP, Guengerich FP. Biochemistry. 2000;39:11319–11329. doi: 10.1021/bi000869u. [DOI] [PubMed] [Google Scholar]
- 24.Yun CH, Kim KH, Calcutt MW, Guengerich FP. J Biol Chem. 2005;280:12279–12291. doi: 10.1074/jbc.M411019200. [DOI] [PubMed] [Google Scholar]
- 25.Schenkman JB, Remmer H, Estabrook RW. Mol Pharmacol. 1967;3:113–123. [PubMed] [Google Scholar]
- 26.Guengerich FP. In: Principles and Methods of Toxicology. Hayes AW, editor. Taylor & Francis; Philadelphia: 2001. pp. 1625–1687. [Google Scholar]
- 27.He X, Cryle MJ, De Voss JJ, Ortiz de Montellano PR. J Biol Chem. 2005;280:22697–22705. doi: 10.1074/jbc.M502632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notredame C, Higgins DG, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 29.Correia MA, Ortiz de Montellano PR. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Plenum Press; New York: 2005. pp. 247–322. [Google Scholar]
- 30.Kim D, Cryle MJ, De Voss JJ, Ortiz de Montellano PR. Arch Biochem Biophys. 2007;464:213–220. doi: 10.1016/j.abb.2007.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park HG, Lim YR, Eun CY, Han S, Han JS, Cho KS, Chun YJ, Kim D. Biochem Biophys Res Commun. 2010;396:534–538. doi: 10.1016/j.bbrc.2010.04.138. [DOI] [PubMed] [Google Scholar]
- 32.Trösken ER, Straube E, Lutz WK, Völkel W, Patten C. J Am Soc Mass Spectrom. 2004;15:1216–1221. doi: 10.1016/j.jasms.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 33.Acimovic J, Lövgren-Sandblom A, Monostory K, Rozman D, Golicnik M, Lutjohann D, Björkhem I. J Chromatog B. 2009;877:2081–2086. doi: 10.1016/j.jchromb.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 34.Morio F, Loge C, Besse B, Hennequin C, Le Pape P. Diagn Microbiol Infect Dis. 2010;66:373–384. doi: 10.1016/j.diagmicrobio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Kudo M, Ohi M, Aoyama Y, Nitahara Y, Chung SK, Yoshida Y. J Biochem. 2005;137:625–632. doi: 10.1093/jb/mvi073. [DOI] [PubMed] [Google Scholar]
- 36.Warrilow AG, Martel CM, Parker JE, Melo N, Lamb DC, Nes WD, Kelly DE, Kelly SL. Antimicrob Agents Chemother. 2010;54:4235–4245. doi: 10.1128/AAC.00587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein sequence analysis of N-terminus of purified CYP51F1 using mass-spectrometry.
Titration of purified F105L mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified D116E mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified Y132H mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified R467K mutant protein with azole compounds. The binding affinities of the azoles calculated from the plots are shown in Table 1.
Titration of purified mutant proteins (F105L, D116E, Y132H, and R467K) with lanosterol.
Additional spectral analysis of the purified CYP51F1 protein. (A) The absolute spectra of the Fe3+ (—), and Fe3+·lanosterol (----); (B) Second-derivative spectra of purified CYP51F1 Fe3+ form in the absence of lanosterol.
HPLC analysis of the reactions of the purified CYP51F1 protein with lanosterol as substrate. (A) HPLC chromatograms of the oxidation reaction product, Econazole (50 μM) was added to inhibit the reaction as inhibitor; (B) Steady-state kinetic analysis of lanosterol oxidation by CYP51F1. Each point presented is a mean ± SD of duplicate assays.