Summary
Protein-protein interactions play an important role in many virus-encoded functions and in virus-host interactions. While a “classical” yeast two-hybrid system (Y2H) is one of the most common techniques to detect such interactions, it has a number of limitations, including a requirement for the proteins of interest to be relocated to the nucleus. Modified Y2H, such as the Sos recruitment system (SRS), which detect interactions occurring in the cytoplasm rather than the nucleus, allow proteins from viruses replicating in the cytoplasm to be tested in a more natural context. In this study, a SRS was used to detect interactions involving proteins from vesicular stomatitis virus (VSV), a prototypic non-segmented negative strand RNA (NNS) virus. All five full-length VSV proteins, as well as several truncated proteins, were screened against each other. Using the SRS, most interactions demonstrated previously involving VSV phosphoprotein, nucleocapsid (N) and large polymerase proteins were confirmed independently, while difficulties were encountered using the membrane associated matrix and glycoproteins. A human cDNA library was also screened against VSV N protein and one cellular protein, SFRS18, was identified which interacted with N in this context. The system presented can be redesigned easily for studies in other less tractable NNS viruses.
Keywords: protein interaction, cytoplasmic yeast two hybrid, vesicular stomatitis virus, negative strand RNA virus, host protein
1. Introduction
Protein-protein interactions are essential for many biological functions and are also involved in host-pathogen interplay. Many steps of virus replication (e.g., virus genome replication and transcription, virion assembly) involve protein complexes comprised of virus-encoded proteins. Furthermore, all viruses depend heavily on host cell functions for their replication, while, at the same time, cellular innate immune components respond to and combat the invading virus. Although direct contacts between viral and host cell components are not necessary for all aspects of these processes, a direct, physical interaction between a viral and host protein may indicate these proteins are influencing each other in a way that affects viral replication.
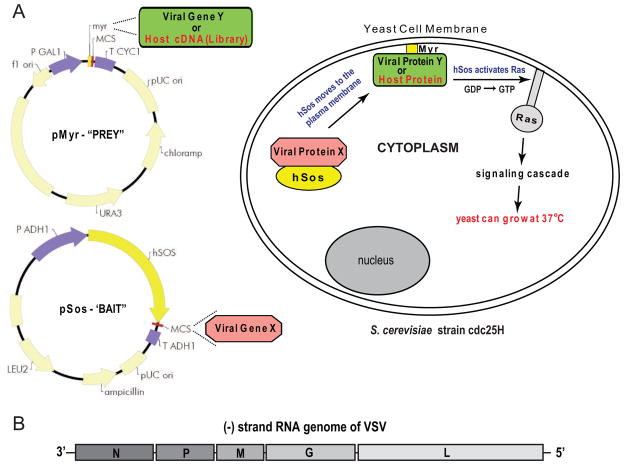
Multiple biochemical and cell based methods have been developed for the detection of protein-protein interactions [reviewed recently in (Guan and Kiss-Toth, 2008; Lalonde et al., 2008; Mendez-Rios and Uetz, 2010; Miernyk and Thelen, 2008)]. A “classical” yeast two-hybrid (Y2H) assay (Fields and Song, 1989) using a protein-protein interaction to reconstitute a functional transcription factor has been, and continues to be, one of the most common techniques used to detect protein-protein interactions. The yeast Saccharomyces cerevisiae can be grown quickly, easily and inexpensively and outcomes can be measured readily through use of reporter genes such as β-galactosidase. These features make the Y2H amenable to high-throughput applications such as cDNA library screens and mapping whole organism interactomes [reviewed in (Parrish, Gulyas, and Finley, 2006)], including those of viruses (Flajolet et al., 2000; Fossum et al., 2009; McCraith et al., 2000; Uetz et al., 2006) and viruses and their host (Calderwood et al., 2007; de Chassey et al., 2008). However, the classical Y2H system also has a number of limitations, including a large number of false positives due to cryptic transcription activation domains and a requirement for the proteins of interest to be relocated to the nucleus (Causier and Davies, 2002). Y2H variants [reviewed in (Bao et al., 2009; Suter, Kittanakom, and Stagljar, 2008)] have been developed to address many of these limitations including systems designed to analyze protein-protein interactions in a cytoplasmic context. Among these is the Sos recruitment system (SRS) first described by Aronheim and colleagues (1997). SRS utilizes a S. cerevisiae strain cdc25H with a point mutation in the CDC25 gene making it temperature sensitive, and takes advantage of the fact that this defect can be complemented by its human homolog, son of sevenless homolog 1 (Sos1), but only when that protein is recruited to the cellular membrane. In this system, one protein (a “prey”) is fused to a myristoylation signal (Myr), causing it to be directed to the inner surface of the cellular membrane where it has the potential to interact with and recruit a second protein (a “bait”) fused to a fragment of the Sos protein. Thus, if there is an interaction, growth can occur at the non-permissive temperature (Fig. 1A). For viruses with a cytoplasmic site of replication, it has the advantage of allowing proteins to be tested in the natural replication environment.
Figure 1.
A. Overview of the Cytotrap yeast two-hybrid system based in the Sos recruitment system. Viral protein baits were cloned into the pSos plasmids and used to screen viral or HeLa cell baits cloned into the pMyr plasmid (pSos and pMyr diagrams are from the Stratagene Cytotrap instruction manual). Protein-protein interactions complement a temperature-sensitive mutation, allowing yeast to grow at the non-permissive temperature. B. Schematic diagram of the VSV genome.
The use of the SRS for detecting protein interactions involving a nonsegmented negative-strand RNA (NNS) virus was evaluated for the first time using vesicular stomatitis virus (VSV, Family Rhabdoviridae). VSV is a prototypic NNS virus, with its replication occurring exclusively in the cytoplasm, and it has a small genome, encoding only five proteins (Fig. 1B). In this study, all five VSV proteins, including some protein fragments, were screened against each other using the SRS. Results of this assay were compared to those achieved previously using a classical Y2H or other methods of detecting protein-protein interaction. Furthermore, potentially functional viral baits were identified for use in screening a human cDNA library for viral-host protein interactions and the results of one such screen are presented.
Since NNS viruses (all belong to the order Mononegavirales) share a similar genome structure and virus replication scheme (Lamb and Parks, 2007; Lyles, 2007), these studies on VSV should be relevant to other members of this order. The system presented can be redesigned easily for studies on protein-protein interactions in other less tractable NNS viruses (such as Ebola, Nipah and rabies), for which the use of mammalian experimental systems is more challenging due to obvious biosafety issues.
2. Materials and Methods
2.1 Plasmids
The pSos and pMyr vectors, used for expression of Sos1 and Myr fusions respectively, provided in the CytoTrap Two-Hybrid System kit (Stratagene) were amplified by transformation into chemically competent DH5α Escherichia coli cells and subsequent purification using a HiSpeed Maxi Prep kit (Qiagen). All other control plasmids contained in the kit were used as supplied.
Insert sequences were PCR amplified from the plasmid pVSVFL(+)g.1 which contains a complete cDNA copy of the VSV (Indiana strain; IND) antigenome (Lawson et al., 1995/5/9). Primers (Table 1) were designed to be complimentary to the target sequence as well as introduce the desired restriction sites. PCR was conducted using the Phusion high-fidelity DNA polymerase (Finnzymes) in accordance with the manufacturer’s instructions. PCR products and vectors were then digested with the indicated restriction enzymes (New England Biolabs), gel purified, ligated utilizing T4 DNA ligase (Promega) and transformed into chemically competent JM109 E. coli with selection for successful transformants using media containing 100μg/ml ampicillin or 30μg/ml chloramphenicol for pSos and pMyr constructs, respectfully. Transformants were amplified and plasmids extracted with commercial kits in accordance with manufacturer instructions. Plasmid inserts were sequenced using the primers indicated in Table 1 to confirm a correct sequence and in-frame insertion.
Table 1.
List of primers used to perform PCR-based cloning and mutagenesis and plasmid sequencing
| Primera | Restriction Enzymeb | Usec |
|---|---|---|
| 5′-agtaggatccccatgagttccttaaagaagattc-3′ | BamHI | pSos-Mwt (+) |
| 5′-agtagcggccgctttgaagtggctgatagaatcc-3′ | NotI | pSos-Mwt (-) |
| 5′-agtaccatggccatggataatctcacaaaagttcg-3′ | NcoI | pSos-Pwt, pSos-PDomainI (+) |
| 5′-agtagtcgaccagagaatatttgactctcg-3′ | SalI | pSos-Pwt (-) |
| 5′-agtagtcgacccactgggatttctgctctcc-3′ | SalI | pSos-PDomainI (-) |
| 5′-agtaccatggccatgtctgttacagtcaagaga-3′ | NcoI | pSos-Nwt, pSos-NΔ10 (+) |
| 5′-agtagtcgactttgtcaaattctgacttagc-3′ | SalI | pSos-Nwt (-) |
| 5′-agtagtcgacaattgtcttctctcttaggcc-3′ | SalI | pSos-NΔ10 (-) |
| 5′agtaggatccccaagttcaccatagtttttccac-3′ | BamHI | pSos-Gwt(mature) (+) |
| 5′-agtagcggccgcctttccaagtcggttcatctc-3′ | NotI | pSos-Gwt(mature) (-) |
| 5′-agtaacgcgtgatggaagtccacgattttgag-3′ | MluI | pSos-Lwt, pSos-L1-1338 (+) |
| 5′-agtagcggccgcatctctccaagagttttcc-3′ | NotI | pSos-Lwt (-) |
| 5′-agtagcggccgcttgtccccacgaaccttcc-3′ | NotI | pSos-L1-1338 (-) |
| 5′-atctgaaggagatgagtgca-3′ | BamHId | pSos-L1139-2127 (+) |
| 5′-agtaacgcgtatctctccaagagttttcc-3′ | MluI | pSos-L1139-2127 (-) |
| 5′-agtagaattcatgagttccttaaagaagattc-3′ | EcoRI | pMyr-Mwt (+) |
| 5′-agtactcgagtttgaagtggctgatagaatcc-3′ | XhoI | pMyr-Mwt (−) |
| 5′-agtagaattcatggataatctcacaaaagttcg-3′ | EcoRI | pMyr-Pwt (+) |
| 5′-agtactcgagcagagaatatttgactctcg-3′ | XhoI | pMyr-Pwt (−) |
| 5′-agtagaattcatgtctgttacagtcaagag-3′ | EcoRI | pMyr-Nwt (+) |
| 5′-agtactcgagtttgtcaaattctgacttagc-3′ | XhoI | pMyr-Nwt (−) |
| 5′-agtagaattcaagttcaccatagtttttccac-3′ | EcoRI | pMyr-Gwt(mature) (+) |
| 5′-agtactcgagctttccaagtcggttcatctc-3′ | XhoI | pMyr-Gwt(mature) (−) |
| 5′-agtacccgggccatggaagtccacgattttgag-3′ | SmaI | pMyr-Lwt (+) |
| 5′-agtactcgagatctctccaagagttttcc-3′ | XhoI | pMyr-Lwt (−) |
| 5′-aatgctgctatacgatcgctcggtctgaaggggaaaggtaagaaatc-3′ | pSos-M2-7 mutagenesis (+) | |
| 5′-cgatcgtatagcagcattcatggggatcctactataactaattttccttgg-3′ | pSos-M2-7 mutagenesis (−) | |
| 5′-aatgctgctatacgatcgggtaagaaatctaagaaattagggatcgcacc-3′ | pSos-M8-13 mutagenesis (+) | |
| 5′-cgatcgtatagcagcattaatcttctttaaggaactcatggggatcc-3′ | pSos-M8-13 mutagenesis (−) | |
| 5′-gagcaaataccaagtcgccagaag-3′ | Overlap PCR (+) | |
| 5′-ccaagaccaggtaccatg-3′ | pSos insert sequencing (+) | |
| 5′-gccagggttttcccagt-3′ | pSos insert sequencing, overlap PCR (−) | |
| 5′-actactagcagctgtaatac-3′ | pMyr insert sequencing (+) | |
| 5′-cgtgaatgtaagcgtgacat-3′ | pMyr insert sequencing (−) | |
| 5′-ggcctttaaacatagggacg-3′ | L insert sequencing (−) | |
| 5′-ttttgaaatacccgacttac-3′ | L insert sequencing (+) | |
| 5′-cccagtgttccgagttatgg-3′ | L insert sequencing (+) | |
| 5′-atctgaaggagatgagtgca-3′ | L insert sequencing (+) | |
| 5′-gggtctaaaacatctgaatctaca-3′ | L insert sequencing (+) | |
| 5′-cctctatctatacaaggtcgtat-3′ | L insert sequencing (+) | |
| 5′-agatgctagagatgcctcca-3′ | L insert sequencing (+) |
nucleotides complimentary to viral sequence shown in bold
restriction site introduced for cloning
(+)=forward primer, (−)=reverse primer
present naturally within viral gene
Site directed mutagenesis of the M gene was carried out using an overlap extension PCR strategy as in (Higuchi, Krummel, and Saiki, 1988). Mutagenic primers containing the 5’-aatgctgctatacgatcg-3’ positive strand DNA sequence encoding the NAAIRS amino acid sequence (Armbruster et al., 2001) are shown in Table 1. This sequence can participate in either α-helices or β-sheets (Wilson et al., 1985), minimizing the disruption this substitution causes to the overall protein structure. The first round of PCR was conducted as above using a mutagenic primer paired with the appropriate flanking primer. Following gel purification, the PCR products were mixed and used as the template for a second round of PCR in combination with the two flanking primers. These PCR products were then cloned into pSos as above.
2.2 Media
Minimal synthetic defined (SD) media using either glucose (Fisher Chemical) (SD/Glucose) or a combination of galactose (Sigma-Aldrich, contains ≤0.01% glucose) and raffinose (Acros) (SD/Galactose) as a carbon source were used for all SRS screens. The latter allowed, while the former repressed expression of the Myr fusion proteins that are under the control of an inducible GAL1 promoter. Sos fusions are under the control of a constitutive ADH1 promoter so that expression occurs with both media types. Omission of uracil (-U) and/or leucine (-L) from the media allowed for selection of yeast transformed with pMyr and pSos constructs, respectively.
2.3 Fusion protein expression
pMyr constructs were transformed individually into cdc25H(α) cells using a lithium acetate method in accordance with the CytoTrap kit instructions. Transformants were selected by plating on SD/Glucose(-U) and incubating at room temperature (RT; 20-24°C). Individual colonies were then inoculated into liquid SD/Galactose(-U) and incubated at RT for 2–3 days with rotation. Prior to protein extraction, yeast were pelleted at 900 × g for 2 minutes at RT.
In the same manner, pSos constructions were co-transformed with pMyr-Sos binding protein (SB). SB interacts directly with Sos1, allowing this construct to serve as a positive control for growth at the non-permissive temperature when paired with any pSos plasmid. Co-transformed yeast were selected by plating on SD/Glucose(-UL) and incubated at RT. Isolated colonies were then transferred to SD/Galactose(-UL) plates and incubated at 34°C for 7 days.
To extract protein, yeast pellets or colonies were resuspended in a buffer containing 50mM Tris (pH 8.0), 10mM MgCl2, 1mM EGTA, 2mM EDTA (pH 8.0), 20% glycerol and 15mM KCl and disrupted mechanically using acid washed glass beads. An equal volume of a second buffer was added to give a final concentration of 1% SDS, 45mM HEPES (pH7.5) and 15mM DTT and the samples were heated at 95ºC for 10 minutes. Insoluble material was pelleted at 16,000 × g for 3 minutes at 4ºC and the resulting supernatant combined with an equal volume of 2x SDS-PAGE sample buffer. VSV virions purified as in (Kalvodova et al., 2009) were disrupted in RIPA buffer [25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% Sodium deoxycholate and 0.1% SDS ] and used as a control where indicated. For western blot analysis, the resulting protein samples were separated by electrophoresis on 10% (Myr fusions) or 7.5% (Sos1 fusions) SDS-PAGE gels and electroblotted to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked using 5% non-fat powdered milk in TBS-T [0.5 M NaCl, 20 mM Tris (pH 7.5), 0.1% Tween 20], which was also used for antibody dilutions. Membranes were first incubated with 1:1000 mouse monoclonal anti-Sos antibodies (clone 25; BD Biosciences), 1:2000 rabbit polyclonal anti-VSV L antibodies (raised against the N-terminal half of the L protein fused to TrpE; “anti-VSV-L1-2”) or 1:5000 rabbit polyclonal anti-VSV antibodies (raised against VSV virions). Detection was with 1:5000 goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) using the Enhanced Chemiluminescence Plus (ECL+) protein detection system (GE Healthcare).
2.4 Screen of viral-viral protein interactions
Specific plasmid pairings were used to transform cdc25H(α) cells and cotransformants were selected on SD/Glucose (-UL) plates incubated at RT. At 8 days post transformation (dpt), five isolated colonies from each pairing were resuspended separately in 20μl sterile water in a 96-well plate. These yeast suspensions were then transferred to SD/Glucose (-UL) and SD/Galactose (-UL) single-well plates in 96-well format using a pin-replicator and incubated at the indicated temperatures (RT, 30°C, 34°C and/or 37°C). Plates were evaluated for growth at 3, 7 and 14 days after transfer.
2.5 Screen of a HeLa cDNA library using pSos-Nwt
The CytoTrap XR HeLa cDNA Library (Stratagene) was amplified in accordance with the manufacturer’s instructions. Cdc25H(α) yeast were transformed with 40 μg of library plasmids and 40 μg of pSos-Nwt (wt = wild-type), plated onto 150 mm SD/Glucose (-UL) plates and incubated at RT to select for cotransformants. At 4 dpt, yeast colonies were replica plated onto SD/Galactose (-UL) plates using velvet and incubated at 34°C. Colonies appearing on these plates at 4, 8, 11, 14 and 18 days after replica plating were picked manually and transferred to single-well SD/Glucose (-UL) plates in a 96-well arrangement, to repress Myr fusion protein expression, and incubated at RT. These putative-positives were then further screened following the scheme laid out by (Aronheim et al., 1997). Briefly, once good growth was observed, colonies were transferred to a SD/Glucose (-UL) and a SD/Galactose (-UL) 150mm plate using a pin-replicator and incubated at 34°C. Colonies showing growth on SD/Galactose (-UL) but not SD/Glucose (-UL) at 34°C were rescreened to confirm this growth pattern. Plasmid DNA was isolated from putative-positive yeast colonies using hot phenol extraction as in (Leeds et al., 1991) and transformed into chemically competent JM109 E. coli which were plated onto LB agar plates with 30μg/ml chloramphenicol to select for bacteria transformed with the pMyr constructs. Plasmids were extracted using the PureYield plasmid miniprep system (Promega). These plasmids were then sequenced using the same primers as for the viral inserts and transformed into cdc25Hα along with pSos or pSos-Nwt and screened as in section 2.4.
3. Results
3.1 VSV fusion constructs
The full-length open reading frames of the VSV matrix (Mwt), phosphoprotein (Pwt), nucleocapsid (Nwt) and large polymerase (Lwt) proteins were cloned into both the pSos and pMyr vectors. The sequence encoding the mature form of the glycoprotein [Gwt(mature)], lacking the N-terminal 16 amino acids (aa) which are cleaved off following insertion of the nascent peptide chain into the endoplasmic reticulum (Irving et al., 1979), was also cloned into both vectors. Also cloned into pSos were: aa 1-138 of P encompassing Domain I of the P protein (PdomainI) (Chen, Ogino, and Banerjee, 2006); N protein with the C-terminal 10 aa deleted (NΔ10); overlapping fragments of the L protein encoding aa 1-1338 (L1-1338) or aa 1139-2127 (L1139-2127); and M protein with aa 2-7 (M2-7) or aa 8-13 (M8-13) mutated to NAAIRS.
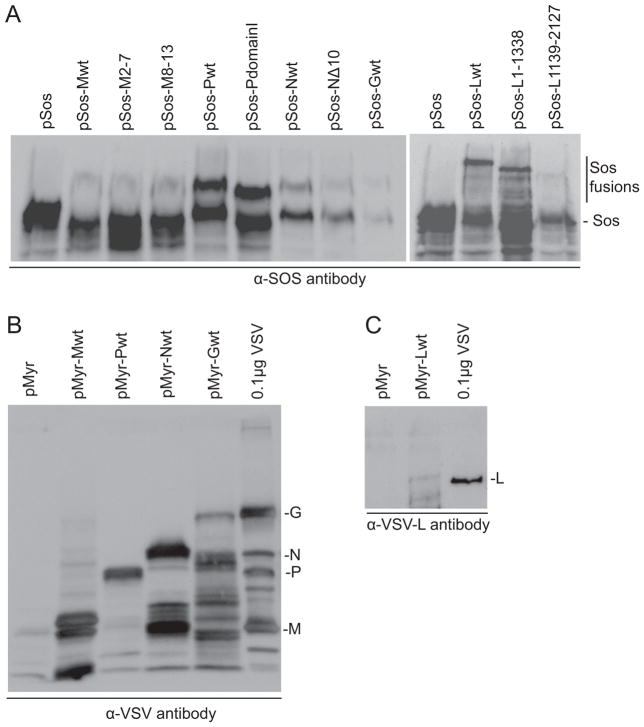
Expression of the VSV fusion proteins was confirmed by western blot (Figure 2). Some variation in protein expression was seen, with Pwt, PdomainI, Lwt and L1-1138 in particular being detected at higher levels (compare products of interest to non-specific bands in Figure 2), indicating there may be variability in expression and/or stability between some of these fusion proteins. However, all viral fusion proteins were detected and many of the proteins detected at lower levels participated in protein-protein interactions within the SRS, suggesting lower expression is not necessarily detrimental. Expression of pMyr constructs transformed individually into yeast was detected after induction of protein expression by incubation in liquid SD/Galactose(-U) at RT. As expected, all proteins were similar in size to their virion encoded counterparts as Myr adds only 15 aa while only 16 aa are lost from the mature form of G (Fig. 2B and C). This also suggests P is being phosphorylated, as the absence of phosphorylation would cause a marked protein mobility shift (Barik and Banerjee, 1991/4; Barik and Banerjee, 1992/7/15). Interestingly, expression of Sos-fusion proteins, encoded by either viral constructs or kit provided control plasmids, could not be detected using similar transformations of pSos plasmids alone and incubation in liquid SD/Glucose(-L) at RT despite gene expression being under the control of the constitutive ADH1 promoter (data not shown). However, when yeast were co-transformed with pMyr-SB and a pSos construct and incubated on SD/Galactose(-UL) plates at 34°C, the fusion proteins could be detected using an antibody against Sos1 (Fig. 2A). As the Sos1 protein fragment used in these fusions is itself quite large (1067 aa), little size variation could be seen between most of the viral fusion proteins although all were significantly larger than the Sos fragment expressed alone (Fig. 2A).
Figure 2.
Verification of fusion protein expression. Total protein was extracted from yeast (A) transformed with the indicated plasmid plus pMyr-SB then incubated on SD/Galactose(-UL) agar at 34ºC or (B and C) transformed with the indicated plasmids then incubated in SD/Galactose(-U) broth at RT. Immunoblots were performed following protein separation on 7.5% (A) or 10% (B and C) SDS-PAGE gels using anti-Sos, -VSV or −L antibodies as indicated. The expected position of the proteins is indicated to the right of the blots
3.2 Optimization of screening conditions
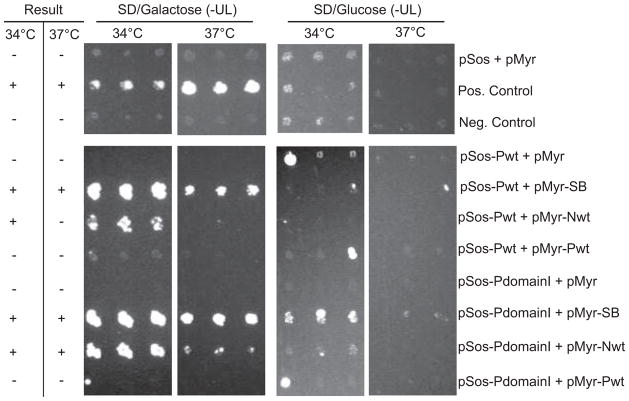
Initial experiments using a subset of the viral fusions paired with each other at the recommended non-permissive temperature of 37°C, failed to detect several interactions shown previously. Therefore, using the same subset of pairings, the incubation temperature was varied from the recommended non-permissive temperature of 37°C in an attempt to increase sensitivity. At 30°C, significant growth was seen for all plasmid pairings (including negative controls) on SD/Glucose(-UL) as well as SD/Galactose(-UL) media making this condition unsuitable for screening purposes (data not shown). In contrast, by using 34°C as the non-permissive temperature, an increase in the number of pairings showing growth on SD/Galactose(-UL) media was observed, as well as improved growth for most of the interactions detected previously, without a significant increase in growth on SD/Glucose(-UL) media. Examples of this observation are shown in Figure 3.
Figure 3.
Temperature optimization. Yeasts were co-transformed with the indicated plasmids, grown initially on SD/Glucose(-UL) at RT, then replica plated to created matching plates for comparison of growth at 34 and 37ºC. Growth on SD/Galactose(-UL) but not SD/Glucose(-UL) is indicative of a protein-protein interaction. For each plasmid pairing, results for 3 of 5 independent transformants are shown. +, protein-protein interaction; -, no interaction.
3.3 Screen for viral-viral protein interactions
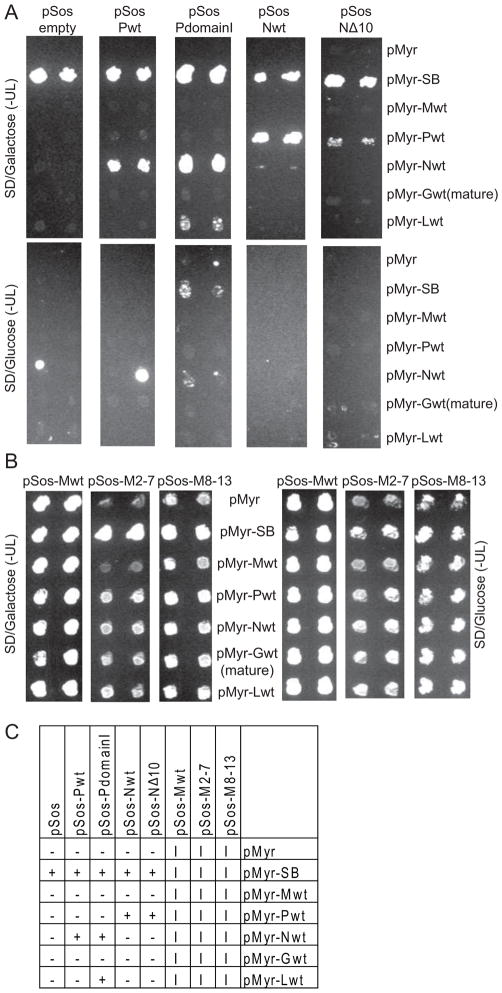
Using 34°C as the non-permissive temperature, a full screen of all possible pairing of the viral constructs against each other as well as the empty vector (pSos or pMyr) and pMyr-SB controls was undertaken. As expected, there was no growth at the non-permissive temperature when the pMyr viral constructs were paired with the “empty” pSos plasmid while all pSos constructs paired with the pMyr-SB positive control showed good growth on SD/Galactose media. There also was no growth at the non-permissive temperature for pSos empty vector or pSos viral fusions paired with pMyr empty vector except for Sos1 fusions with the M proteins. In these cases, growth at the non-permissive temperature was seen for all pairings regardless of the pMyr construct and even occurred on SD/Glucose media, where the Myr fusion protein is not expressed (Figure 4B). As Sos1 must be present at the cellular membrane for growth to occur at the non-permissive temperature, this indicates that M is trafficking to the membrane, thereby relocating the Sos1 protein it is fused to andallowing activation of the Ras signaling cascade in the absence of pMyr protein expression. Replacing aa 2-7 (M2-7) or 8-13 (M8-13) of the M protein with the aa NAAIRS, mutations that have been shown previously to reduce membrane affinity in eukaryotic cells while retaining at least some of the capabilities of M to bind VSV nucleocapsids (Dancho et al., 2009), allowed differences in growth to be distinguished between the positive and negative controls (Fig. 4B). However, background remained high enough to make these mutant pSos-M fusions unsuitable for screening against other viral proteins. Growth on SD/Galactose only at the non-permissive temperature did indicate interactions between pSos-Pwt and pMyr-Nwt, pSos-PdomainI and pMyr-Nwt, pSos-PdomainI and pMyr-Lwt, pSos-Nwt and pMyr-Pwt and pSos-NΔ10 and pMyr-Pwt (Figure 4A). Interactions between pSos-Gwt(mature), pSos-Lwt, pSos-L1-1338 and pSos-L1172-2127 and any of the pMyr viral constructs were not observed (data not shown).
Figure 4.
Screen for viral-viral protein interactions. Yeasts were cotransformed with the indicated plasmids, grown initially on SD/Glucose(-UL) at RT, then replica plated as shown with incubation at the non-permissive temperature of 34ºC. In (A), growth on SD/Galactose(-UL) but not SD/Glucose(-UL) is indicative of a protein-protein interaction. In (B), growth was seen on both media types for all pairings, including pMyr empty vector, indicating the Sos fusions are trafficking to the cell membrane. For each plasmid pairing, results for 2 of 5 independent transformants are shown. (C) Summary of results: +, protein-protein interaction; -, no interaction; I, growth independent of pMyr expression.
3.4 Screen of a HeLa cDNA library for interactions with VSV-Nwt
Approximately 2.0 × 106 yeasts cotransformed with pSos-Nwt and a pMyr plasmid from a HeLa cDNA library were obtained and screened. After replica plating onto SD/Galactose(-UL) media at the non-permissive temperature, 2449 colonies formed. In addition to well defined, independent colonies, a number of tight clusters of small colonies were observed. In all cases tested, yeasts in these clusters contained the same pMyr plasmid (data not shown) and are treated as a single colony. These colonies were further screened to eliminate yeast capable of growth on both glucose and galactose containing media at the non-permissive temperature, meaning that growth was independent of the Myr fusion protein expression, most likely due to a reversion in the CDC25 mutant gene allowing temperature-independent growth. 2359 colonies were eliminated in the first screen and another 65 in a repeat of the screen, leaving 25 colonies where growth was dependent on Myr fusion protein expression. When pMyr plasmids isolated from these colonies were transformed back into yeast with either pSos or pSos-Nwt, 24 showed growth at the non-permissive temperature when paired with either plasmid, indicated that growth is due to properties of the Myr fusion protein rather than an interaction with the VSV N protein. The identities of these proteins, as determined by plasmid insert sequencing combined with PCR screening, are given in Table 2. One insert associated with non-specific growth was unable to be sequenced using either primer. The plasmid containing the cDNA for splicing factor, arginine/serine-rich 18 (SFRS18; NCBI Entrez Gene ID: 25957) was the only plasmid that allowed for growth only when paired with pSos-Nwt, indicating a specific association with N. Growth at the non-permissive temperature for colonies transformed with pSos-Nwt and pMyr-SFRS18 was very weak but reproducible (data not shown).
Table 2.
Identity of inserts from pMyr plasmids causing non-specific growth at the non-permissive temperature
| Protein | Gene | NCBI Entrez Gene ID | # of Colonies | Day(s) Picked |
|---|---|---|---|---|
| Natriuretic peptide receptor A/guanylate cyclase A | NPR1 | 4881 | 11 | 4,8,11 |
| Son of sevenless homolog 1 | SOS1 | 6654 | 5 | 4 |
| v-Ha-ras Harvey rat sarcoma viral oncogene homolog | HRAS | 3265 | 2 | 4 |
| Related RAS viral (r-ras) oncogene homolog 2 | RRAS2 | 22800 | 2 | 8 |
| Poly (ADP-ribose) polymerase family, member 14 | PARP14 | 54625 | 2 | 8 |
| Poly (ADP-ribose) polymerase family, member 10 | PARP10 | 84875 | 1 | 18 |
| No sequence obtained | N/A | N/A | 1 | 11 |
4. Discussion
In this study, the SRS system was used to identify interactions of VSV proteins with each other and with host proteins. SRS has been used previously to test interactions of a specific viral protein with other viral or host proteins (Chomchan, Li, and Shirako, 2003; Frischmuth et al., 2004; Kim, Kim, and Paek, 2006; Takemoto and Hibi, 2005; Yamanaka et al., 2000); or in the case of hepatitis B virus X protein (Barak, Aronheim, and Shaul, 2001; Shamay et al., 2002), tobacco mosaic virus movement protein (Kragler et al., 2003), Papaya ringspot virus helper component-proteinase (Shen et al., 2010) and influenza A virus protein NS1 (Zhao et al., 2009) to screen a host protein library. In this report, a detailed feasibility study was conducted to uncover the strengths and weaknesses of the SRS for defining whole virus interactomes and finding host protein partners for a viral protein. The SRS system has never been used before for VSV or any other NNS virus.
Results of the screen of the viral protein pairings were mixed when compared to previous studies of protein-protein interactions among VSV proteins. Overall, it showed a propensity of the SRS to false-negatives rather than false-positives, at least for the proteins tested, as not all interactions shown previously were detected but no unexpected interactions were seen. This may be one reason why relaxing the selective conditions by lowering the non-permissive temperature 34°C allowed for identification of a greater number of protein-protein interactions.
For VSV proteins that are entirely cytoplasmic (N, P and L), a number of documented interactions were confirmed while some were not. P is capable of binding to both free and RNA-bound N protein although different regions of the P protein are involved in those interactions. Binding RNP has been shown to involve P aa from positions 213–263 (Gill, Chattopadhyay, and Banerjee, 1986/12; Green and Luo, 2009). In contrast, using a mammalian two-hybrid assay, both the C-terminal 10 aa of P and residues within Domain I (aa 1–138), have been shown to be important for binding of P to free N protein, although differences in the relative importance of these domains were seen between the New Jersey and IND strains of VSV. For both strains, phosphorylation of P was not required for binding to N. In this same system, removal of the C-terminal 10 aa of the N protein (equivalent to NΔ10) almost abolished completely the interaction with P (Takacs and Banerjee, 1995/4/20; Takacs, Das, and Banerjee, 1993/11/1). Consistent with these reports for free N, using the SRS Pwt and Nwt were capable of interacting with each other regardless of what vector they were cloned in. Furthermore, Pdomain I was capable of binding Nwt. Binding between Pwt and NΔ10 was also detected, although growth of these colonies was reduced compared to the other N-P combinations, suggesting this deletion weakens the interaction (Fig. 4A). Mammalian two-hybrid has also shown that the N-terminal region of P, particularly Domain I, is also important for binding of P to the L protein, with phosphorylation of P increasing the efficiency of that binding (Takacs and Banerjee, 1995/4/20). In this study, an interaction between PdomainI and Lwt but not Pwt and Lwt was detected, possibly due to conformational differences between the PdomainI and Pwt fusion proteins or to differences in phosphorylation state or multimerization. P also self associates to form dimers. Using a traditional yeast two-hybrid assay, aa 161–210 within the hinge region of P has been shown to be important for this interaction (Chen et al., 2006) and phosphorylation of P appears to be required (Gao and Lenard, 1995/3/15). Using the SRS, no interaction was shown between Pwt proteins. PdomainI would not be expected to participate in P dimerization.
Transformation with a plasmid encoding the VSV M protein fused to Sos1 resulted in growth at the non-permissive temperature when paired with any pMyr construct, including the empty vector, and even occurred when Myr fusion protein expression was repressed by growth on SD/Glucose(-UL) media (Figure 4B). This indicates that in this case, Sos1 relocalization from the cytoplasm to the plasma membrane, a requirement for growth at the non-permissive temperature, is not due to a protein-protein interaction but to M protein trafficking to the yeast membrane. This observation is consistent with the previous studies in mammalian cells, demonstrating that M protein expressed on its own can traffic to the cellular membrane and cause budding of vesicles from the cell surface (Chong and Rose, 1993; Justice et al., 1995). Mutations in M shown to reduce membrane trafficking in mammalian cells, reduced yeast growth at the non-permissive temperature, confirming the observed phenotype is influenced by properties of the M protein and demonstrating that these same mutations also reduce membrane trafficking of M in yeast. While further mutation or deletion of residues involved in membrane association may make M a suitable bait for some applications, these mutations have also been shown to reduce dramatically the association of M with RNP (Dancho et al., 2009) and were not made. Due to this property, the pSos-M plasmids could not be used to screen for protein-protein interactions. This precluded testing for the interaction of M with itself shown previously (Ge et al., 2010; Graham et al., 2008). However, pMyr-Mwt could be used to test for interactions with other viral proteins as this fusion is designed to go to the membrane, but no interactions were detected despite the fact that M has been shown previously to bind RNPs (Chong and Rose, 1993) and G trimers (Lyles, McKenzie, and Parce, 1992) in vitro. In addition to the potential causes of false negatives common to all proteins used in a Y2H assay, M has been shown to have a cooperative mechanism of binding to the nucleocapsid (Flood and Lyles, 1999; Lyles and McKenzie, 1998) which may have contributed to the failure to detect any interactions with these proteins in this assay.
The signal sequence of the G protein was removed prior to cloning into the expression vectors to prevent possible membrane insertion. Unlike for M, the Sos1-G fusion, did not cause growth in the absence of a pMyr construct, indicating it did not traffic to the membrane despite G being an integral membrane protein. However, no interactions were seen between either G fusion and any of the other viral proteins. In addition to its interaction with M, G has been shown previously to form homotrimers (Doms et al., 1987; Roche et al., 2006; Roche et al., 2007). G undergoes extensive post-translational modification some of which are necessary for proper protein folding and trimer formation (Machamer, Florkiewicz, and Rose, 1985; Machamer and Rose, 1988). It is possible that in this system, one or more of those modifications is lacking.
Based on the results from the screen of viral protein interactions, pSos-Nwt, -Pwt and -PdomainI were identified as potential baits to use in the screening of a HeLa cDNA library. Results of the screen using pSos-Nwt are presented, while future screens using the other two baits are planned. The identification of potential cellular proteins interacting with the VSV N protein was somewhat encumbered by the large number of colonies with a reversion in the CDC25 gene (2424 out of 2449). That number may have been exacerbated by use of the longest incubation time prior to replica plating recommended by the manufacturer in an attempt to maximize detection of interactions. A further difficulty was the large numbers of cellular cDNAs encoding proteins capable of rescuing growth even in the absence of an interaction with a Sos fusion protein (24 out of 25), although development of primers capable of amplifying some of the most common of these inserts by PCR aided the screening process (data not shown). The identity of these inserts (Table 2) is largely consistent with the functions of these proteins and previous findings. Trafficking of Sos1 directly to the membrane by means of a Myr tag avoids the need to recruit it via a protein-protein interaction, while mammalian members of the Ras family of proteins have been shown to circumvent the yeast Ras signaling system (Aronheim et al., 1997). Natriuretic peptide receptor A/guanylate cyclase A (NPR1) may allow the initial activation steps to be bypassed as cGMP has been shown to be as effective cAMP in activating S. cerevisiae cAMP-dependent protein kinase (Cytrynska et al., 1999), a downstream effector of the Ras signaling pathway (Broach and Deschenes, 1990). Why expression of two members of the poly (ADP-ribose) polymerase family leads to growth at the non-permissive temperature is less apparent. While to the authors’ best knowledge, PARP activity has not been demonstrated in yeast, many members of this family, including PARP10, have been shown to be involved in cell cycle regulation in other eukaryotic systems (Chou, Chou, and Lee, 2006; Yu et al., 2005). Colonies containing PARP10 or PARP14 were some of the last to be isolated during the initial screen and showed slower growth during rescreening compared to most of the other non-specific activators (data not shown), indicating the mechanism responsible for growth is likely inefficient.
The only protein found to interact specifically with Nwt in yeast was SFRS18 (also known as SRrp130). This serine/arginine-rich (SRr) protein is poorly characterized, but has been shown to colocalize with pinin in nuclear speckles and may be involved in mRNA splicing (Zimowska et al., 2003). Interestingly, several other SRr proteins have been shown to shuttle continuously between the nucleus and the cytoplasm, suggesting that at least some SR proteins may function in cytoplasmic processes (Long and Caceres, 2009). Moreover, the Swedish human protein atlas project (www.proteinatlas.org) (Berglund et al., 2008) indicates cytoplasmic localization of SFRS18 in many human tissues, suggesting an N-SFRS18 interaction in VSV-infected cells is at least hypothetically possible, although this would need to be confirmed experimentally. The failure to detect a larger number of proteins interacting with Nwt is not necessarily an indictment of the SRS as it is unclear how many cellular proteins interact with the N protein. Furthermore, when N is expressed in E. coli in the absence of P, it tends to form aggregates (Das and Banerjee, 1993/3). If the same happens in yeast, it may hinder detection of interactions involving N.
Using VSV as a representative NNS many, although not all, of the interactions previously shown to occur between the viral proteins were confirmed independently, particularly those involving N, P and L, which are strictly cytoplasmic. This trend is likely to hold for other viruses used with the SRS system in the future. One cellular protein that interacts with the VSV N protein in the context of SRS was identified, as well as two additional baits for use in future screens of host proteins.
Acknowledgments
The authors are grateful to Dr. Sue Moyer (University of Florida College of Medicine) for providing VSV reagents for this project. The authors also thank Joshua Stokell, Robert Brown, Sean Vidulich, James Wang, and Natascha Moestl for providing technical assistance. This work was supported by NIH Grant 5R03AI078122 (to V. Z. G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Molecular & Cellular Biology. 2001;21:7775–86. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Molecular & Cellular Biology. 1997;17:3094–102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Redondo C, Findlay JB, Walker JH, Ponnambalam S. Deciphering soluble and membrane protein function using yeast systems (Review) Mol Membr Biol. 2009;26:127–35. doi: 10.1080/09687680802637652. [DOI] [PubMed] [Google Scholar]
- Barak O, Aronheim A, Shaul Y. HBV X protein targets HIV Tat-binding protein 1. Virology. 2001;283:110–20. doi: 10.1006/viro.2001.0883. [DOI] [PubMed] [Google Scholar]
- Barik S, Banerjee AK. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J Virol. 1991 Apr;65:1719–1726. doi: 10.1128/jvi.65.4.1719-1726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S, Banerjee AK. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc Natl Acad Sci U S A. 1992/7/15;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernerus H, Nilsson P, Lundberg E, Sivertsson A, Navani S, Wester K, Kampf C, Hober S, Ponten F, Uhlen M. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–27. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- Broach JR, Deschenes RJ. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A. 2007;104:7606–11. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Davies B. Analysing protein-protein interactions with the yeast two-hybrid system. Plant Molecular Biology. 2002;50:855–70. doi: 10.1023/a:1021214007897. [DOI] [PubMed] [Google Scholar]
- Chen M, Ogino T, Banerjee AK. Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J Virol. 2006;80:9511–8. doi: 10.1128/JVI.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomchan P, Li SF, Shirako Y. Rice grassy stunt tenuivirus nonstructural protein p5 interacts with itself to form oligomeric complexes in vitro and in vivo. J Virol. 2003;77:769–75. doi: 10.1128/JVI.77.1.769-775.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong LD, Rose JK. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J Virol. 1993;67:407–14. doi: 10.1128/jvi.67.1.407-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HY, Chou HT, Lee SC. CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10) J Biol Chem. 2006;281:15201–7. doi: 10.1074/jbc.M506745200. [DOI] [PubMed] [Google Scholar]
- Cytrynska M, Wojda I, Frajnt M, Jakubowicz T. PKA from Saccharomyces cerevisiae can be activated by cyclic AMP and cyclic GMP. Can J Microbiol. 1999;45:31–7. [PubMed] [Google Scholar]
- Dancho B, McKenzie MO, Connor JH, Lyles DS. Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral nucleocapsids. J Biol Chem. 2009;284:4500–9. doi: 10.1074/jbc.M808136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Banerjee AK. Expression of the vesicular stomatitis virus nucleocapsid protein gene in Escherichia coli: analysis of its biological activity in vitro. Virology. 1993 Mar;193:340–347. doi: 10.1006/viro.1993.1130. [DOI] [PubMed] [Google Scholar]
- de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugue S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, Andre P, Rabourdin-Combe C, Lotteau V. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms RW, Keller DS, Helenius A, Balch WE. Role for adenosine triphosphate in regulating the assembly and transport of vesicular stomatitis virus G protein trimers. Journal of Cell Biology. 1987;105:1957–69. doi: 10.1083/jcb.105.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Flajolet M, Rotondo G, Daviet L, Bergametti F, Inchauspe G, Tiollais P, Transy C, Legrain P. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–79. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- Flood EA, Lyles DS. Assembly of nucleocapsids with cytosolic and membrane-derived matrix proteins of vesicular stomatitis virus. Virology. 1999;261:295–308. doi: 10.1006/viro.1999.9856. [DOI] [PubMed] [Google Scholar]
- Fossum E, Friedel CC, Rajagopala SV, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S, Bandyopadhyay S, Rose D, von Brunn A, Uhlmann M, Zeretzke C, Dong YA, Boulet H, Koegl M, Bailer SM, Koszinowski U, Ideker T, Uetz P, Zimmer R, Haas J. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 2009;5:e1000570. doi: 10.1371/journal.ppat.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmuth S, Kleinow T, Aberle HJ, Wege C, Hulser D, Jeske H. Yeast two-hybrid systems confirm the membrane- association and oligomerization of BC1 but do not detect an interaction of the movement proteins BC1 and BV1 of Abutilon mosaic geminivirus. Arch Virol. 2004;149:2349–64. doi: 10.1007/s00705-004-0381-0. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lenard J. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 1995/3/15;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327:689–93. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DS, Chattopadhyay D, Banerjee AK. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc Natl Acad Sci U SA. 1986/12;83:8873–8877. doi: 10.1073/pnas.83.23.8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SC, Assenberg R, Delmas O, Verma A, Gholami A, Talbi C, Owens RJ, Stuart DI, Grimes JM, Bourhy H. Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog. 2008;4:e1000251. doi: 10.1371/journal.ppat.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci U S A. 2009;106:11713–8. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Kiss-Toth E. Advanced technologies for studies on protein interactomes. Adv Biochem Eng Biotechnol. 2008;110:1–24. doi: 10.1007/10_2007_092. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–67. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving RA, Toneguzzo F, Rhee SH, Hofmann T, Ghosh HP. Synthesis and assembly of membrane glycoproteins: presence of leader peptide in nonglycosylated precursor of membrane glycoprotein of vesicular stomatitis virus. Proc Natl Acad Sci U S A. 1979;76:570–4. doi: 10.1073/pnas.76.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice PA, Sun W, Li Y, Ye Z, Grigera PR, Wagner RR. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–60. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, Simons K. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J Virol. 2009;83:7996–8003. doi: 10.1128/JVI.00635-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kim HR, Paek KH. Arabidopsis tonoplast proteins TIP1 and TIP2 interact with the cucumber mosaic virus 1a replication protein. J Gen Virol. 2006;87:3425–31. doi: 10.1099/vir.0.82252-0. [DOI] [PubMed] [Google Scholar]
- Kragler F, Curin M, Trutnyeva K, Gansch A, Waigmann E. MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol. 2003;132:1870–83. doi: 10.1104/pp.103.022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Ehrhardt DW, Loque D, Chen J, Rhee SY, Frommer WB. Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 2008;53:610–35. doi: 10.1111/j.1365-313X.2007.03332.x. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, et al., editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1449–1496. [Google Scholar]
- Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A. 1995/5/9;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes and Development. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lyles D, Rupprecht CE. Rhabdoviridae. In: Knipe DM, Howley PM, et al., editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1363–1408. [Google Scholar]
- Lyles DS, McKenzie M, Parce JW. Subunit interactions of vesicular stomatitis virus envelope glycoprotein stabilized by binding to viral matrix protein. J Virol. 1992;66:349–58. doi: 10.1128/jvi.66.1.349-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles DS, McKenzie MO. Reversible and irreversible steps in assembly and disassembly of vesicular stomatitis virus: equilibria and kinetics of dissociation of nucleocapsid-M protein complexes assembled in vivo. Biochemistry. 1998;37:439–50. doi: 10.1021/bi971812j. [DOI] [PubMed] [Google Scholar]
- Machamer CE, Florkiewicz RZ, Rose JK. A single N-linked oligosaccharide at either of the two normal sites is sufficient for transport of vesicular stomatitis virus G protein to the cell surface. Molecular & Cellular Biology. 1985;5:3074–83. doi: 10.1128/mcb.5.11.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE, Rose JK. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J Biol Chem. 1988;263:5955–60. [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci U S A. 2000;97:4879–84. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Rios J, Uetz P. Global approaches to study protein-protein interactions among viruses and hosts. Future Microbiol. 2010;5:289–301. doi: 10.2217/fmb.10.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA, Thelen JJ. Biochemical approaches for discovering protein-protein interactions. Plant J. 2008;53:597–609. doi: 10.1111/j.1365-313X.2007.03316.x. [DOI] [PubMed] [Google Scholar]
- Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol. 2006;17:387–93. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–91. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–8. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Shamay M, Barak O, Doitsh G, Ben-Dor I, Shaul Y. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J Biol Chem. 2002;277:9982–8. doi: 10.1074/jbc.M111354200. [DOI] [PubMed] [Google Scholar]
- Shen W, Yan P, Gao L, Pan X, Wu J, Zhou P. Helper component-proteinase (HC-Pro) protein of Papaya ringspot virus interacts with papaya calreticulin. Mol Plant Pathol. 2010;11:335–46. doi: 10.1111/j.1364-3703.2009.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Kittanakom S, Stagljar I. Two-hybrid technologies in proteomics research. Curr Opin Biotechnol. 2008;19:316–23. doi: 10.1016/j.copbio.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Takacs AM, Banerjee AK. Efficient interaction of the vesicular stomatitis virus P protein with the L protein or the N protein in cells expressing the recombinant proteins. Virology. 1995 Apr 20;208:821–826. doi: 10.1006/viro.1995.1219. [DOI] [PubMed] [Google Scholar]
- Takacs AM, Das T, Banerjee AK. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc Natl Acad Sci U S A. 1993/11/1;90:10375–10379. doi: 10.1073/pnas.90.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto Y, Hibi T. Self-interaction of ORF II protein through the leucine zipper is essential for Soybean chlorotic mottle virus infectivity. Virology. 2005;332:199–205. doi: 10.1016/j.virol.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–42. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Haft DH, Getzoff ED, Tainer JA, Lerner RA, Brenner S. Identical short peptide sequences in unrelated proteins can have different conformations: a testing ground for theories of immune recognition. Proc Natl Acad Sci U S A. 1985;82:5255–9. doi: 10.1073/pnas.82.16.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, Dean C, Naito S, Ishikawa M. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci U S A. 2000;97:10107–12. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, Mehraein Y, Mertsching J, Kraft R, Austen M, Luscher-Firzlaff J, Luscher B. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–93. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- Zhao L, Xu L, Zhou X, Zhu Q, Yang Z, Zhang C, Zhu X, Yu M, Zhang Y, Zhao X, Huang P. Interaction of influenza virus NS1 protein with growth arrest-specific protein 8. Virol J. 2009;6:218. doi: 10.1186/1743-422X-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G, Shi J, Munguba G, Jackson MR, Alpatov R, Simmons MN, Shi Y, Sugrue SP. Pinin/DRS/memA interacts with SRp75, SRm300 and SRrp130 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2003;44:4715–23. doi: 10.1167/iovs.03-0240. [DOI] [PubMed] [Google Scholar]