Abstract
We investigated phylogenetic affiliation, occurrence of virulence genes and quinolone resistance in 109 integron-containing strains of Escherichia coli isolated from a wastewater treatment plant. Selection for integron-bearing strains caused a shift toward phylogroup D, which was most numerous, followed by A, B1 and B2. Phylogroups D and B2, both of which are reported to include virulent extraintestinal pathotypes, made up 50.5% of all isolates and were present in every stage of wastewater treatment, including final effluent. Diarrheagenic pathotypes made up 21% of the strains. The average virulence factor genes score was low (1.40) and the range was from 0 to 5. Quinolone and fluoroquinolone resistance was observed in 56.0% and 50.4% of the strains, respectively; however, it was not associated with virulence factor score. Although the average virulence factor score was low, 17.4% of strains had three and more virulence genes. They were isolated mostly from raw sewage, but 30% of them were cultured from final effluent. Release of multiresistant integron-bearing E. coli strains with virulence traits into the environment may create potential threat and be of public health concern.
Keywords: Escherichia coli, Antibiotic resistance, Virulence, Wastewater
Introduction
Escherichia coli is a well recognized etiological agent of community-acquired and nosocomial infections. Clinical categories of E. coli include commensal, intestinal pathogenic and extraintestinal pathogenic (ExPEC) strains. Intestinal pathogenic isolates comprise enterohemorrhagic (EHEC), enteropathogenic (EPEC), enteroaggregative (EAEC), enterotoxigenic (ETEC), enteroinvasive (EIEC) and diffusely adherent E. coli (DAEC). ExPEC strains, which are responsible for several infectious diseases, including urinary tract (uropathogenic strains, UPEC), bacteremia and neonatal meningitis (MNEC, meningitidis-associated E. coli), possess virulence factors like adhesins, toxins, iron acquisition systems, polysaccharide coats and invasins (Kaper et al. 2004). The virulence genes in both diarrheagenic and extraintestinal strains are often located within mobile genome elements, e.g. plasmids, transposons, bacteriophages, and pathogenicity islands (Kaper 2005). This phenomenon may contribute to spreading of virulence traits among related bacterial species. It has been shown that pathogenic E. coli may be derived from commensal strains by acquisition of chromosomal or extra-chromosomal virulence operons (Ochman et al. 2000). Phylogenetically, E. coli strains are divided upon amplification of genes chuA coding for the heme transport protein, yjaA, initially identified in the complete genome sequence of E. coli K-12 encoding a hypothetical protein, and a non-coding region TSPE4.C2 (Clermont et al. 2000). The patterns of amplicons creates four groups A, B1, B2 and D. ExPEC strains are derived from group B2 and to a lesser extent D, commensal from A and B1, and diarrheagenic from A, B1 and D (Johnson and Stell 2000; Hamelin et al. 2007). However, the relative predominance of different phylogroups may vary due to geographical location, the site of infection and the level of antibiotic resistance (Duriez et al. 2001; Bukh et al. 2009). Several studies on clinical E. coli have demonstrated relationship between antibiotic resistance and low prevalence of virulence factors and suggested that quinolone resistance may be directly associated with the loss of virulence (Horcajada et al. 2005; Takahashi et al. 2009; Kawamura-Sato et al. 2010). The mechanism underlying this correlation is not clear, yet, but probably a particular genetic background is required for mutations leading to quinolones resistance (Kawamura-Sato et al. 2010). Soto et al. (2006) have shown that quinolones can induce in vitro partial or total loss of a pathogenicity island containing hemolysin and cytotoxic necrotizing factor genes, but other studies have not confirmed that phenomenon (Sabaté et al. 2008).
Wastewater treatment plant creates excellent conditions for dissemination of resistance genes through horizontal gene transfer facilitated by plasmids and transposons (Schlüter et al. 2007). From wastewater treatment plants bacteria can return to aquatic and land habitats, and affect human and animals. Resistance genes are often assembled in integrons, which are natural systems mediating the capture of gene cassettes coding for antibiotic resistance. Integrons are frequently embedded in mobile DNA elements such as transposons and conjugative plasmids that enable them to spread horizontally through bacterial populations (Fluit and Schmitz 2004). Recently, it has occurred that resistance due to slow reversibility can persist long in bacterial communities even if the selective pressure imposed by antibiotic usage is reduced (Andersson and Hughes 2010). Moreover, the integron-mediated resistance, can be silenced in bacterial population at no biological cost (Guerin et al. 2009). Multidrug resistance of Enterobacteriaceae strains is associated with the presence of integrons (Leverstein-van Hall et al. 2003). As the economic and clinical impact of E. coli infections is considerable, monitoring of non-clinical reservoirs of these bacteria with virulence and resistance determinants has become a serious concern worldwide.
The aim of the study was to analyze the distribution of phylogenetic groups and occurrence of virulence genes in relation to quinolone resistance in integron-bearing strains of E. coli isolated from the sewage in wastewater treatment plant of a city with population of about 550 thousand.
Materials and methods
Sampling and E. coli isolation
Sewage samples were taken from a municipal wastewater treatment plant three times: in December 2008, April 2009 and July 2009. The samples were taken from (i) raw sewage in primary sedimentation tank, (ii) aeration tank and (iii) final effluent. The sewage was collected to sterile containers at the depth of 0.3 m and the distance of one meter from the side of the respective sampling sites. Serial decimal dilutions of the samples were prepared in 0.9% NaCl, inoculated onto Brilliance™ E. coli/coliform Selective Agar (Oxoid) and incubated overnight at 44°C. Purple colonies were picked from the medium and used in further experiments. Identification was done with API 20E (bioMérieux).
ERIC PCR typing
The ERIC PCR was done according to Versalovic et al. (1991) with primers ERIC 1 and ERIC 2. The PCR products were separated in 1.5% agarose gel, stained with ethidium bromide and digitalized with Bio-Print v. 99 gel documentation system (Vilbert-Lourmat). The electrophoretic patterns were analyzed by using GelCompar II 3.5 software (Applied Maths). Similarity between fingerprints was calculated with the Dice coefficient. Cluster analysis was performed using the unweighted pair-group method with average linkages (UPGMA).
Identification of integrase genes
Genomic DNA was isolated by the boiling lysate method. Integron classes were determined by multiplex PCR. The sequences of primers targeting intI1, intI2 and intI3 integrase genes were recommended by Dillon et al. (2005). Amplification involved an initial denaturation (94°C, 5 min) followed by 30 cycles of denaturation (94°C, 1 min), annealing (59°C, 1 min) and extension (72°C, 1 min) with a final extension step (72°C, 8 min). Negative control contained sterile deionized water instead of template DNA. To exclude false positive results, the PCR products of the expected sizes (160 bp for intI1, 788 bp for intI2 and 979 bp for intI3) were sequenced in a 3130xl Genetic Analyzer (Applied Biosystems). The amplicon sequences were compared against GenBank database by using BLAST (Basic Local Alignment Search Tool).
Phylogenetic analysis
A, B1, B2 and D phylogenetic groups were determined in triplex PCR method (Clermont et al. 2000) with primers ChuA.1, ChuA.2, YjaA.1, YjaA.2, TspE4C2.1, and TspE4C2.2. The PCR reaction consisted of initial denaturation for 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C followed by a final extension step of 7 min at 72°C.
Detection of genes characteristic of diarrheagenic pathotypes of E. coli
Multiplex PCR assays according to Aranda et al. (2004) were done to detect typical and atypical enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC) enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and Shiga toxin-producing E. coli (STEC) isolates. The targeted genes were: eae and bfpA for EPEC, CVD432 for EAEC, heat-labile (LT) and heat-stable (ST) enterotoxin genes for ETEC, ipaH for EIEC, and stx1 and stx2 for STEC.
Identification of virulence genes
Virulence genes characteristic for extraintestinal E. coli were detected by PCR assays. The following genes were targeted using primers and PCR conditions recommended by Johnson and Stell (2000): fyuA (encoding yersiniabactin receptor), iutA (aerobactin receptor gene), hlyA (haemolysin A gene), papA (P fimbriae subunit gene), sfaS (S fimbriae subunit gene), focG (F1C fimbriae subunit gene), and cnf1 (cytotoxic necrotizing factor gene). VF (virulence factor) gene score was determined as a total number of virulence genes per strain.
Determination of antimicrobial susceptibility including quinolone and fluoroquinolone resistance
Antibiotic resistance patterns were determined by disc diffusion method according to Clinical and Laboratory Standards Institute guidelines (CLSI, 2009). Isolates were tested for susceptibility to antibiotics representing aminoglycosides, amphenicols, carbapenems, penicillins, cephalosphorins, quinolones, fluoroquinolones, sulphonamides, and tetracyclines. Quinolones were represented by nalidixic acid and fluoroquinolones by norfloxacin and ciprofloxacin.
Statistical analysis
Occurrence of virulence factors among phylogenetic groups were compared by using Fisher’s exact test. VF scores were compared with the Mann–Whitney U test (Statistica 9.1, StatSoft). P < 0.05 was considered to indicate statistical significance.
Results and discussion
E. coli isolation and identification of the integrase genes
Altogether, 973 isolates of E. coli were recovered from the wastewater; 109 of them harboured integrons in their genomes and were used in further experiments.
PCR amplification of the integrase genes representative for three classes of integrons revealed the presence of intI1 in 105 strains (10.8%). Two strains (0.2%) had intI2 gene and two isolates (0.2%) harboured both intI1 and intI2. No integron of class 3 was found. Integrons of class 2 were present in strains belonging to phylogroup A (2 strains) and D (2 strains). Seventy-five percent of intI-positive strains originated from raw sewage, 10% from aeration tank and 15% from final effluent. They made 11% of the total number of E. coli strains isolated from the wastewater samples.
Similar values have been reported by da Silva et al. (2007), who noted 10% frequency of occurrence of class 1 integrons among E. coli strains isolated from an urban wastewater treatment plant in Portugal. On the other hand, Moura et al. (2007) have noticed higher incidences of integron carriage among Enterobacteriaceae and Aeromonadaceae originated from a slaughterhouse wastewater treatment plant. The mean percentage of integrase-positive strains reached 35%, with intI1 gene present in 30.7% of strains, intI2 gene in 4.5% and both genes in one isolate (0.3%). Integrons have been also detected in multiresistant E. coli isolated from raw and treated municipal wastewaters used for irrigation in agriculture (Pignato et al. 2009).
The frequencies of integron-positive strains in our study are comparable to those obtained previously for E. coli strains isolated from healthy subjects without recent antibiotic exposure: 11% for class 1 integrase, 3% for class 2 integrase and 0.6% for both class 1 and 2 integrases (Skurnik et al. 2005). Our results are also similar to the incidencies of intI-positive strains isolated from Seine estuary, which reached 11% (Laroche et al. 2009).
The frequencies of integron-carrying E. coli isolated from clinical specimens are noticeably higher and reach as much as 27.6% in bloodstream infections (Daikos et al. 2007) and 44.8% in urine and stool strains (Japoni et al. 2007).
Clonal relatedness of E. coli strains
To exclude the possibility of analyzing strains clonally identical, we performed ERIC PCR typing. The fingerprints of E. coli isolates consisted of 1–21 bands ranging in size from 110 to 3500 bp. None of the strains shared identical ERIC PCR pattern.
Phylogenetic analysis
The most abundant phylogroup among intI-positive E. coli strains was D (36.7%), followed by A (30.3%), B1 (19.3%), and B2 (13.8%) (Table 1). A set of 150 E. coli strains randomly picked from the total pool of isolates (973) showed noticeably different pattern of phylogenetic group distribution: 47.0% of strains belonged to group A, whereas groups D, B2 and B1 were represented by 24.7, 23.3 and 10.0%, respectively. We observed that the selection for integron-harbouring strains caused a shift toward group D and B1 isolates (P < 0.05).
Table 1.
Distribution of genes coding for virulence factors and quinolone resistance among phylogenetic groups of intI-positive E. coli strains isolated from a wastewater treatment plant
| Virulence factor gene | Virulence factor | Prevalence in phylogenetic groups | ||||
|---|---|---|---|---|---|---|
| A (n = 33) | B1 (n = 21) | B2 (n = 15) | D (n = 40) | Total (n = 109) | ||
| eae | Intimin | 0 | 1 (4.8%) | 1 (6.7%) | 1 (2.5) | 3 (2.8%) |
| ST gene* | Heat-stable toxin | 2 (6.1%) | 7 (33.3%) | 0 | 3 (7.5%) | 12 (11.0%) |
| stx1, stx2 | Shiga toxin | 3 (9.1%) | 1 (4.8%) | 2 (13.3%) | 1 (4.8%) | 7 (6.4%) |
| fyuA | Yersiniabactin | 9 (27.3%) | 3 (14.3%) | 7 (46.7%) | 10 (25%) | 29 (26.6%) |
| iutA* | Aerobactin | 10 (30.3%) | 8 (38.0%) | 10 (66.7%) | 17 (52.5%) | 45 (41.3%) |
| hlyA | Haemolysin A | 1 (3.0%) | 1 (4.8%) | 1 (6.7%) | 1 (2.5%) | 4 (3.7%) |
| papA | P fimbriae | 5 (15.2%) | 3 (14.3%) | 2 (13.3%) | 8 (20.0%) | 18 (16.5%) |
| sfaS | S fimbriae | 9 (27.2%) | 4 (19.0%) | 2 (13.3%) | 3 (7.5%) | 18 (16.5%) |
| focG | F1C fimbriae | 2 (6.1%) | 2 (9.5%) | 2 (13.3%) | 4 (7.5%) | 10 (9.2%) |
| cnf1 | CNF | 1 (3.0%) | 1 (4.8%) | 1 (6.7%) | 2 (2.5%) | 5 (4.6%) |
| VF score (mean) | 0–4 (1.24) | 0–4 (1.52) | 0–5 (1.87) | 0–4 (1.28) | 0–5 (1.40) | |
| Quinolone resistance | 19 (57.6%) | 13 (61.9%) | 9 (60.0%) | 20 (50.0%) | 61 (56.0%) | |
| Fluoroquinolone resistance | 14 (42.2%) | 9 (42.9%) | 9 (60.0%) | 23 (57.5%) | 55 (50.4%) | |
* Statistically significant differences among phylogenetic groups (P < 0.05)
The frequencies of phylogroups were different than those reported previously for human, environmental and animal samples. Analysing integron-bearing intestinal E. coli isolates from healthy subjects without antibiotic exposure, Skurnik et al. (2005) have found phylogroup A as the most numerous (44.4%), followed by equal numbers of B1 and D (25.9%) and the least abundant group B2 (3.7%). In our research the intI-positive B2 strains were the least frequent as well, but their frequency was noticeably higher (13.8%).
When it comes to phylogrouping of E. coli strains isolated from wastewater without an a priori integron selection, the proportions were also different. Sabaté et al. (2008) have shown that phylogroup D was the least abundant in human wastewater isolates of E. coli (8%) and not frequent among chicken sewage strains (12%). Majority of the strains belonged to group A (52% of human and 51% of chicken wastewater strains), followed by B1 (20% of human and 5% of chicken wastewater strains) and B2 (20% of human and 33% of chicken wastewater strains). Our results are also in contrast to those observed in random environmental and animal samples of E. coli in the study of Higgins et al. (2007), in which phylogroup B1 was most abundant (31%), followed by A (26%), D (25%) and B2 (17%). Duriez et al. (2001) in an a set of 168 human commensal E. coli strains isolated from the stools of three geographically distinct human populations (France, Croatia, and Mali) have found phylogenetic groups A (40%) and B1 (34%) the most common, followed by group D (15%) and B2 (11%).
Members of group B2 and D possess the chuA gene—these groups are supposed to encompass most of pathogenic E. coli strains and in our research they made up 50.5% of intI-positive strains. It has been suggested that the protein coded by chuA is involved in human host colonization and thus can have impact on pathogenicity (Hoffmann et al. 2001). In our study, the frequency of chuA is higher than that reported by Hoffmann et al. (2001) for strains isolated from environmental samples (30%) and lower than those isolated from human sources (70%).
Diarrheagenic pathotypes of E. coli
Twenty-two strains (20.2%) were classified as diarrheagenic pathotypes: 12 ETEC strains, seven STEC and three EPEC. No EAEC and EIEC strains were found. Diarrheagenic E. coli were found mostly in B1 phylogenetic group (42.9%), but this was due to high number of ETEC isolates. All ETEC strains had only the heat-stable toxin gene. It was identified in 33.3% of strains belonging to B1 group, and in groups D (three strains) and A (two strains). The presence of ST toxin gene was associated with the phylogenetic group affiliation (P < 0.05) (Table 1). Seven STEC isolates harboured stx2 gene, one of them additionally had stx1 gene. Shiga toxin genes stx1 and stx2 were present in 0.9 and 6.4% strains, respectively. The enteropathogenic E. coli isolates were determined as atypical EPEC, as they lacked bfpA gene encoding the bundle-forming pilus subunit. They belonged to groups B2, B1 and D. Atypical EPEC are considered less virulent than typical EPEC. Still, the strains can cause infection among children (Gomes et al. 2004) and both animals and humans can be their reservoirs (Trabulsi et al. 2002).
Distribution of virulence genes characteristic for ExPEC isolates
Genes iutA and fyuA coding for aerobactin and yersiniabactin receptor protein, respectively, are a part of siderophore-mediated systems of iron acquisition. Both aerobactin and yersiniabactin are recognized virulence factors in Enterobacteriaceae. The iutA gene was present in 41.3% of E. coli strains, whereas 26.6% possessed fyuA (Table 1). The cnf1 gene located within PAI II and coding for cytotoxic necrotizing factor was found in five strains (4.6%) belonging to all four phylogroups. Haemolysin A gene, hlyA, associated with pathogenicity islands PAI I and II of uropathogenic E. coli, was present in four (3.7%) strains. P fimbriae subunit gene, papA, also embedded in pathogenicity islands, was observed in 18 strains (16.5%). Similarly, S fimbriae subunit gene, sfaS, located within PAI III, was found in 16.5% strains. Fimbrial F1C subunit gene, focG, was present in 9.2% of the strains.
Only presence of iutA was associated with phylogenetic group affiliation. It was significantly higher among isolates from group B2 (compared versus group A, B1, and groups A, B1 and D taken together). It was also higher among group D strains compared with those from group A and B1 (P < 0.05) (Table 1).
Antimicrobial susceptibility and distribution of quinolone resistance
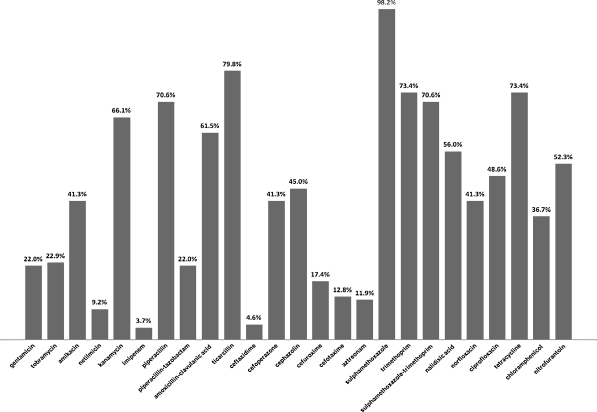
All of the strains were multiresistant, i.e. resistant to three unrelated antimicrobials. The highest percent of resistance was noted for sulfamethoxazole, ticarcillin, tetracycline and trimethoprim. The lowest frequencies of resistant strains were noted for imipenem, ceftazidime and netilmicin (Fig. 1).
Fig. 1.
Frequency of antibiotic resistance in integron-positive E. coli strains isolated from wastewater treatment plant
Resistance to nalidixic acid was observed in 56% of E. coli strains; 50.4% were resistant to fluoroquinolones: ciprofloxacin or norfloxacin (Table 1). The distribution of Q (quinolone) and FQ (fluoroquinolones) resistance within phylogenetic groups was as follow: A: 69.7 and 42.4%, B1: 61.9 and 42.9%, B2: 60 and 60%, and D: 50 and 57.5%, respectively (Table 1). The resistance to quinolones and fluoroquinolones was not associated with phylogenetic group. The frequency of resistance to quinolones was higher in comparison with that reported by Sabaté et al. (2008) for E. coli strains from human wastewater, which were in 24% resistant to quinolones and in 20% to ciprofloxacin. None of their B2 strains has shown quinolone resistance, which is in contrast with the high frequency of Q and FQ resistance in B2 strains in our research.
Association of quinolone resistance with virulence genes
Although integron-positive strains are often multiresistant, only resistance to quinolones has been reported to be associated with decrease in pathogenic potential of E. coli strains (Kawamura-Sato et al. 2010). There is little information about virulence potential of integron-bearing E. coli isolated from wastewater. In our study, the average number of genes associated with diarrheagenic pathotypes and ExPEC per strain was low and reached 1.40. The highest average VF score was found in phylogroup B2 (1.87), whereas the lowest (1.24) was observed in group A; however, the differences in VF scores among phylogenetic groups were not statistically significant (Table 1). In clinical strains of E. coli causing bacteraemia it ranges from 1.9 to 4.5, depending on resistance profiles, and is lower in multiresistant strains (2.8) and in ciprofloxacin-resistant (1.9) (Rijavec et al. 2008). In our study, at low level of VF score, phenotypic resistance to quinolones and fluoroquinolones was not associated with VF score (P > 0.05). Nevertheless, we noticed significantly higher presence of iutA gene in quinolone and fluoroquinolone resistant isolates and lower presence of sfaS gene in fluoroquinolone resistant strains (P < 0.05) (Table 2).
Table 2.
Association between resistance to quinolones and presence of virulence factors coding genes in integron-bearing E. coli strains isolated from a wastewater treatment plant (n = 109)
| Virulence gene | Virulence factor | Quinolone resistant | Fluroquinolone resistant | Susceptible |
|---|---|---|---|---|
| 61 (56.0%) | 55 (50.4%) | 34 (31.2%) | ||
| eae | Intimin | 1 (1.6%) | 0 | 2 (5.9%) |
| ST gene | ST toxin | 8 (13.1%) | 9 (16.4%) | 2 (5.9%) |
| stx1, stx2 | Shiga toxin | 2 (3.3%) | 2 (3.6%) | 4 (11.8%) |
| fyuA | Yersiniabactin | 21 (34.4%) | 15 (27.3%) | 6 (17.6%) |
| iutA | Aerobactin | 30 (49.2%)* | 28 (50.1%)* | 9 (26.5%)* |
| hlyA | Haemolysin A | 2 (3.3%) | 0 | 2 (5.9%) |
| papA | P fimbriae | 10 (16.4%) | 7 (12.8%) | 6 (17.6%) |
| sfaS | S fimbriae | 7 (11.5%) | 4 (7.3%)* | 9 (26.5%)* |
| focG | F1C fimbriae | 7 (11.5%) | 5 (9.1%) | 2 (5.9%) |
| cnf1 | CNF | 3 (4.9%) | 1 (1.8%) | 1 (2.9%) |
| VF score (mean) | 0–5 (1.53) | 0–4 (1.30) | 0–3 (1.24) | |
*Statistically significant differences (P < 0.05)
In conclusion, integron-positive E. coli strains originating from wastewater treatment plant belong mostly to phylogenetic group D, usually connected with extraintestinal pathotypes. Although the average VF score was low, more than one-sixth of the multiresistant strains had three or more virulence genes. They were isolated mostly from raw sewage, but 30% of them were cultured from final effluent which is released to the corresponding river. This may create potential threat of spreading multiresistant, integron-bearing E. coli strains with virulence traits into the environment and be of public health concern. Moreover, some virulence genes are embedded in pathogenicity islands and together with potentially mobile resistance integrons located in plasmids they may be disseminated in the environment via horizontal gene transfer.
Acknowledgment
This work was partially supported by grant NN304 080635 from the Polish Ministry of Science and Higher Education.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Andersson D, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- Aranda KRS, Fagundes-Neto U, Scaletsky ICA. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol. 2004;42:5849–5853. doi: 10.1128/JCM.42.12.5849-5853.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh AS, Schønheyder HC, Emmersen JMG, Søgaard M, Bastholm S, Roslev P. Escherichia coli phylogenetic groups are associated with site of infection and level of antibiotic resistance in community-acquired bacteraemia: a 10 year population-based study in Denmark. J Antimicrob Chemother. 2009;64:163–168. doi: 10.1093/jac/dkp156. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2009) Performance standards for antimicrobial disk susceptibility tests; approved standard—tenth edition. M02-A10, 29(1)
- Da Silva MF, Vaz-Moreira I, Gonzalez-Pajuelo M, Nunes OC, Manaia CM. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an Urban wastewater treatment plant. FEMS Microbiol Ecol. 2007;60:166–176. doi: 10.1111/j.1574-6941.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- Daikos G, Kosmidis C, Tassios P, Petrikkos G, Vasilakopoulou A, Psychogiou M, Stefanou I, Avlami A, Katsilambros N. Enterobacteriaceae bloodstream infections: presence of integrons, risk factors, and outcome. Antimicrob Agents Chemother. 2007;51:2366–2372. doi: 10.1128/AAC.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon B, Thomas L, Mohmand G, Zelynsky A, Iredell J. Multiplex PCR for screening of integrons in bacterial lysates. J Microbiol Methods. 2005;62:221–232. doi: 10.1016/j.mimet.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001;147:1671–1676. doi: 10.1099/00221287-147-6-1671. [DOI] [PubMed] [Google Scholar]
- Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- Gomes TA, Irino K, Girao DM, Girao VB, Guth BE, Vaz TM, Moreira FC, Chinarelli SH, Vieira MA. Emerging enteropathogenic Escherichia coli strains? Emerg Infect Dis. 2004;10:1851–1855. doi: 10.3201/eid1010.031093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbé J, Ploy M, Mazel D. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- Hamelin K, Bruant G, El-Shaarawi A, Hill S, Edge TA, Fairbrother J, Harel J, Maynard C, Masson L, Brousseau R. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit River areas. Appl Environ Microbiol. 2007;73:477–484. doi: 10.1128/AEM.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J, Hohn C, Hornor S, Frana M, Denver M, Joerger R. Genotyping of Escherichia coli from environmental and animal samples. J Microbiol Methods. 2007;70:227–235. doi: 10.1016/j.mimet.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Hoffmann H, Hornef MW, Schubert S, Roggenkamp A. Distribution of the outer membrane haem receptor protein ChuA in environmental and human isolates of Escherichia coli. Int J Med Microbiol. 2001;291:227–230. doi: 10.1078/1438-4221-00123. [DOI] [PubMed] [Google Scholar]
- Horcajada JP, Soto S, Gajewski A, Smithson A, Jimenez de Anta MT, Mensa J, Vila J, Johnson JR. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J Clin Microbiol. 2005;43:2962–2964. doi: 10.1128/JCM.43.6.2962-2964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japoni A, Gudarzi M, Farshad S, Basiri E, Ziyaeyan M, Alborzi A, Rafaatpour N. Assay for integrons and pattern of antibiotic resistance in clinical Escherichia coli strains by PCR-RFLP in Southern Iran. Jpn J Infect Dis. 2007;61:85–88. [PubMed] [Google Scholar]
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- Kaper JB. Pathogenic Escherichia coli. Int J Med Microbial. 2005;295:355–356. doi: 10.1016/j.ijmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kawamura-Sato K, Yoshida R, Shibayama K, Ohta M. Virulence genes, quinolone and fluoroquinolone resistance, and phylogenetic background of uropathogenic Escherichia coli strains isolated in Japan. Jpn J Infect Dis. 2010;63:113–115. [PubMed] [Google Scholar]
- Laroche E, Pawlak B, Berthe T, Skurnik D, Petit F. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France) FEMS Microbiol Ecol. 2009;68:118–130. doi: 10.1111/j.1574-6941.2009.00655.x. [DOI] [PubMed] [Google Scholar]
- Leverstein-van Hall MA, Blok HEM, Rogier A, Donders T, Paauw A, Fluit AC, Verhoef J. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J Infect Dis. 2003;187:251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- Moura A, Henriques I, Ribeiro R, Correia A. Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J Antimicrob Chemother. 2007;60:1243–1250. doi: 10.1093/jac/dkm340. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Pignato S, Coniglio MA, Faro G, Weill FX, Giammanco G. Plasmid-mediated multiple antibiotic resistance of Escherichia coli in crude and treated wastewater used in agriculture. J Water Health. 2009;7:251–258. doi: 10.2166/wh.2009.019. [DOI] [PubMed] [Google Scholar]
- Rijavec M, Müller-Premru M, Zakotnik B, Žgur-Bertok D. Virulence factors and biofilm production among Escherichia coli strains causing bacteraemia of urinary tract origin. J Med Microbiol. 2008;57:1329–1334. doi: 10.1099/jmm.0.2008/002543-0. [DOI] [PubMed] [Google Scholar]
- Sabaté M, Prats G, Moreno E, Ballesté E, Blanch AR, Andreu A. Virulence and antimicrobial resistance profiles among Escherichia coli strains isolated from human and animal wastewater. Res Microbiol. 2008;159:288–293. doi: 10.1016/j.resmic.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Szczepanowski R, Pühler, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plant provides evidence for a widely accessible drug resistance pool. FEMS Microbiol Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- Shigemura K, Arakawa S, Miura T, Nakano Y, Tanaka K, Fujisawa M. Significance of fluoroquinolone-resistant Escherichia coli in urinary tract infections. Jpn J Infect Dis. 2008;61:226–228. [PubMed] [Google Scholar]
- Skurnik D, Le Menac’h A, Zurakowski D, Mazel D, Courvalin P, Denamur E, Andremont A, Ruimy R. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob Agents Chemother. 2005;49:3062–3065. doi: 10.1128/AAC.49.7.3062-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto SM, Jimenez de Anta MT, Vila J. Quinolones induce partial or total loss of pathogenicity island in uropathogenic Escherichia coli by SOS-dependent or -independent pathways, respectively. Antimicrob Agents Chemother. 2006;50:649–653. doi: 10.1128/AAC.50.2.649-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Muratani T, Yasuda M, Takahashi S, Monden K, Ishikawa K, Kiyota H, Arakawa S, Matsumoto T, Shima H, Kurazono H, Yamamoto S. Genetic profiles of fluoroquinolone-resistant Escherichia coli isolates obtained from patients with cystitis: phylogeny, virulence factors, PAIusp subtypes, and mutation patterns. J Clin Microbiol. 2009;47:791–795. doi: 10.1128/JCM.01740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabulsi LR, Keller R, Tardelli TAG. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–513. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]