Abstract
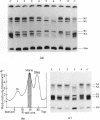
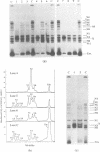
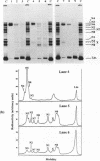
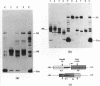
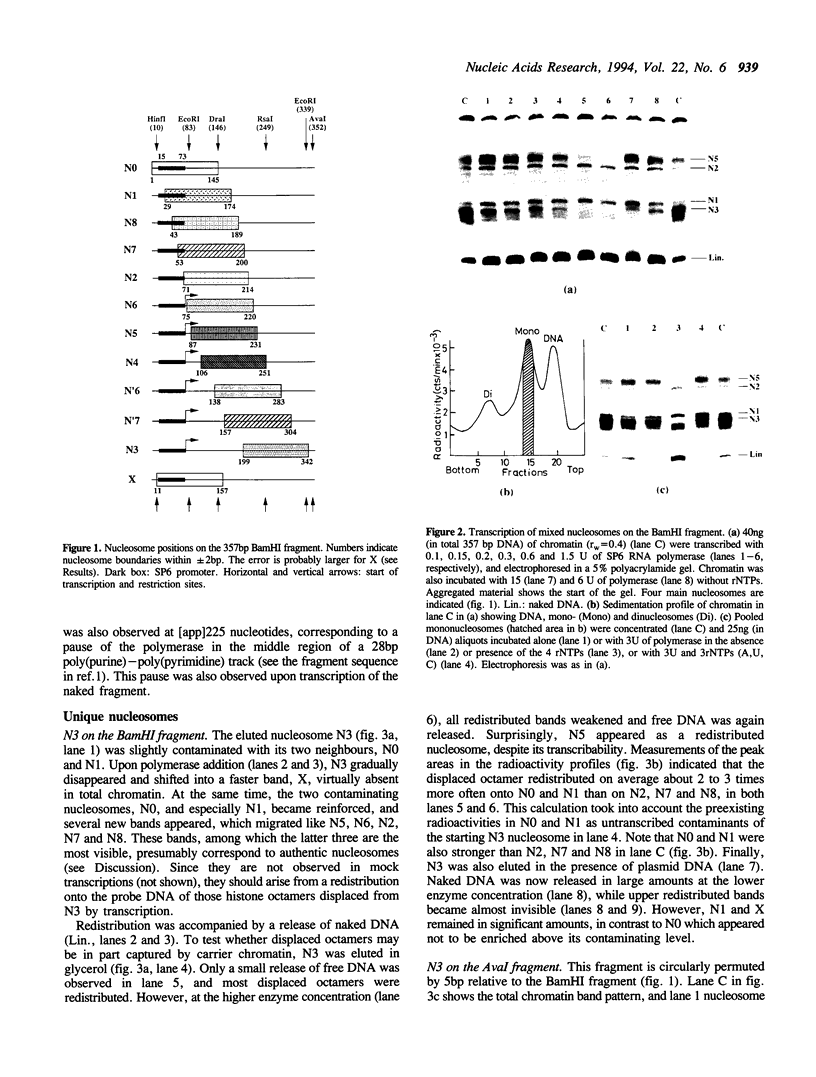
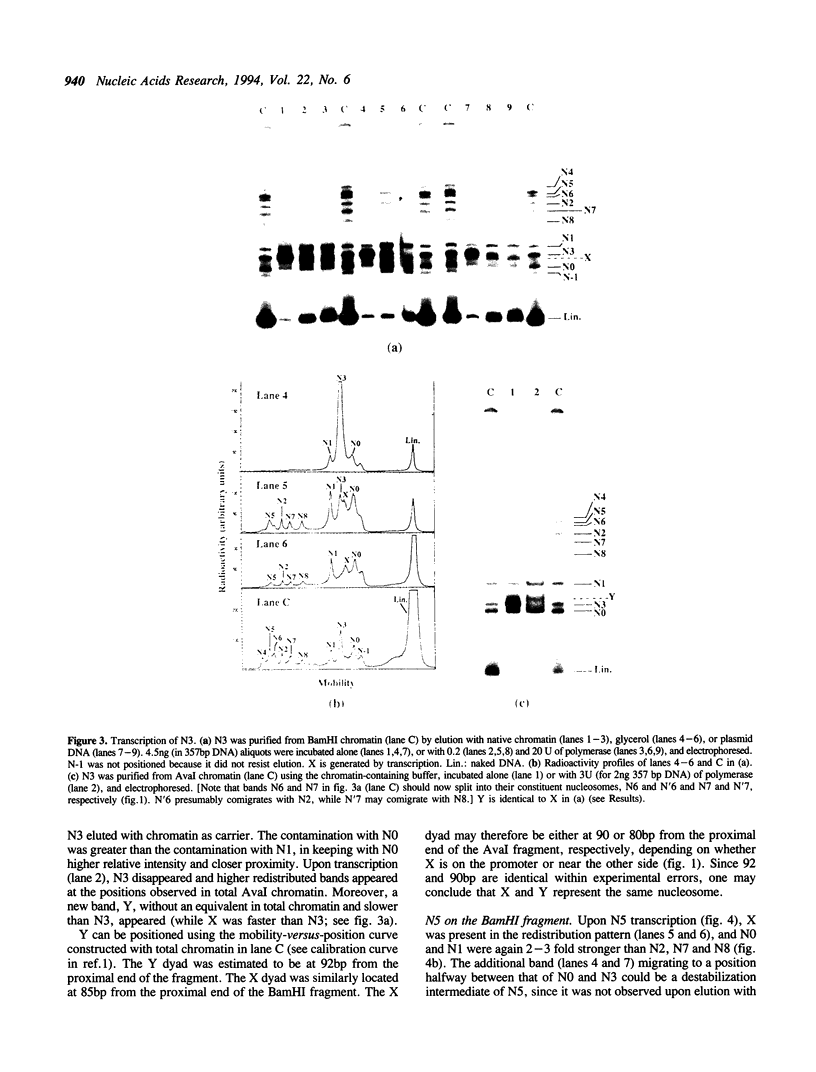
Single nucleosomes were assembled on a 357bp DNA fragment containing a 5S RNA gene from sea urchin and a promoter for SP6 RNA polymerase, and were fractionated as a function of their positions by gel electrophoresis. Transcribed nucleosome positions were detected by observing band disappearance in gels, which in turn provided evidence for the displacement of the histone octamer upon transcription. Differential band disappearance showed that nucleosomes closer to the promoter were harder to transcribe, and transcription was blocked when the nucleosome proximal boundary was at the start site. Nucleosomes located at discrete positions were also eluted from the gel bands and transcribed. In this case, new bands appeared as a consequence of octamer redistribution. Such redistribution occurred over all untranscribed positions, as well as over transcribed positions close enough to the promoter. Similar conclusions were derived from another previously investigated fragment containing a Xenopus 5S RNA gene.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Nucleosome displacement in transcription. Cell. 1993 Feb 12;72(3):305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- Ausio J. Structure and dynamics of transcriptionally active chromatin. J Cell Sci. 1992 May;102(Pt 1):1–5. doi: 10.1242/jcs.102.1.1. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992 Oct 2;71(1):11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- Duband-Goulet I., Carot V., Ulyanov A. V., Douc-Rasy S., Prunell A. Chromatin reconstitution on small DNA rings. IV. DNA supercoiling and nucleosome sequence preference. J Mol Biol. 1992 Apr 20;224(4):981–1001. doi: 10.1016/0022-2836(92)90464-u. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Gould H. J., Cowling G. J., Harborne N. R., Allan J. An examination of models for chromatin transcription. Nucleic Acids Res. 1980 Nov 25;8(22):5255–5266. doi: 10.1093/nar/8.22.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov N., Tsaneva I., Einbinder E., Tsanev R. In vitro transcription through nucleosomes by T7 RNA polymerase. EMBO J. 1992 May;11(5):1941–1947. doi: 10.1002/j.1460-2075.1992.tb05247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Irresistible force meets immovable object: transcription and the nucleosome. Cell. 1991 Nov 29;67(5):833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. On the displacement of histones from DNA by transcription. Cell. 1988 Dec 2;55(5):743–744. doi: 10.1016/0092-8674(88)90128-6. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Meersseman G., Pennings S., Bradbury E. M. Mobile nucleosomes--a general behavior. EMBO J. 1992 Aug;11(8):2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989 Aug;8(8):2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H. Transcribed chromatin. Trends Biochem Sci. 1992 Jan;17(1):23–26. doi: 10.1016/0968-0004(92)90422-6. [DOI] [PubMed] [Google Scholar]
- O'Neill T. E., Roberge M., Bradbury E. M. Nucleosome arrays inhibit both initiation and elongation of transcripts by bacteriophage T7 RNA polymerase. J Mol Biol. 1992 Jan 5;223(1):67–78. doi: 10.1016/0022-2836(92)90716-w. [DOI] [PubMed] [Google Scholar]
- O'Neill T. E., Smith J. G., Bradbury E. M. Histone octamer dissociation is not required for transcript elongation through arrays of nucleosome cores by phage T7 RNA polymerase in vitro. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6203–6207. doi: 10.1073/pnas.90.13.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A. Periodicity of exonuclease III digestion of chromatin and the pitch of deoxyribonucleic acid on the nucleosome. Biochemistry. 1983 Oct 11;22(21):4887–4894. doi: 10.1021/bi00290a004. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Stein A. DNA folding by histones: the kinetics of chromatin core particle reassembly and the interaction of nucleosomes with histones. J Mol Biol. 1979 May 15;130(2):103–134. doi: 10.1016/0022-2836(79)90421-2. [DOI] [PubMed] [Google Scholar]
- Thoma F. Structural changes in nucleosomes during transcription: strip, split or flip? Trends Genet. 1991 Jun;7(6):175–177. doi: 10.1016/0168-9525(91)90429-t. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Eisenberg H. Binding of additional histones to chromatin core particles. Nature. 1978 Jun 8;273(5662):446–448. doi: 10.1038/273446a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. Studies on the mechanism of transcription of nucleosomal complexes. Eur J Biochem. 1980 Jan;103(2):219–226. doi: 10.1111/j.1432-1033.1980.tb04306.x. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Thevenin G., Oudet P., Chambon P. Transcription of in vitro assembled chromatin by Escherichia coli RNA polymerase. J Mol Biol. 1979 Mar 5;128(3):411–440. doi: 10.1016/0022-2836(79)90095-0. [DOI] [PubMed] [Google Scholar]
- Williamson P., Felsenfeld G. Transcription of histone-covered T7 DNA by Escherichia coli RNA polymerase. Biochemistry. 1978 Dec 26;17(26):5695–5705. doi: 10.1021/bi00619a015. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Drew H. R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Duband-Goulet I., Schultz P., Stofer E., Oudet P., Prunell A. Chromatin reconstitution on small DNA rings. III. Histone H5 dependence of DNA supercoiling in the nucleosome. J Mol Biol. 1990 Jul 20;214(2):479–495. doi: 10.1016/0022-2836(90)90195-R. [DOI] [PubMed] [Google Scholar]
- ten Heggeler-Bordier B., Wahli W., Adrian M., Stasiak A., Dubochet J. The apical localization of transcribing RNA polymerases on supercoiled DNA prevents their rotation around the template. EMBO J. 1992 Feb;11(2):667–672. doi: 10.1002/j.1460-2075.1992.tb05098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K. E., Lohr D. E., Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992 Feb 15;267(5):2837–2840. [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]