Abstract
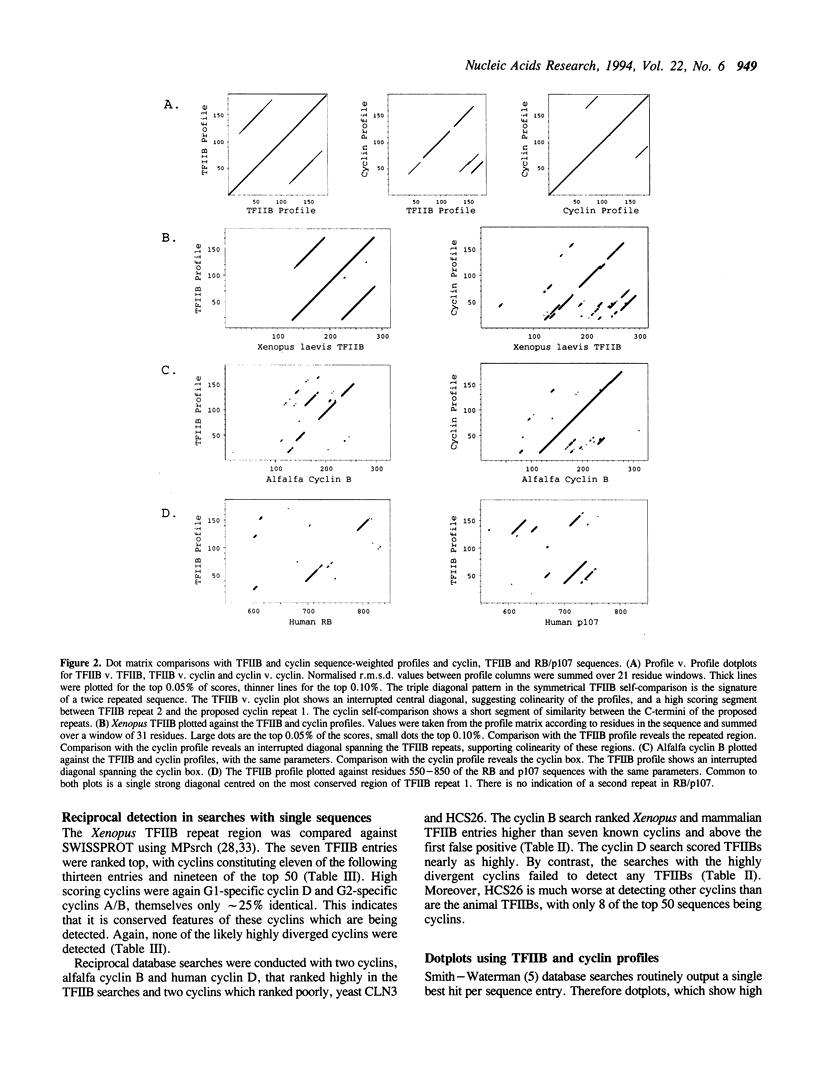
Cyclins, TFIIB and RB play major roles in cell cycle and/or gene regulation. Earlier work has suggested common ancestry for the TFIIB repeats and RB pocket B which share 20% sequence identity. We now report that database searches with profiles based on a multiple alignment of cyclin core regions (the 'cyclin box') detect the TFIIB repeats with equivalent scores to divergent cyclins. Several features of the sequences support the notion of common ancestry: e.g. cyclins A/B, C and D share approximately 20-30% identity but each have approximately 15-20% identity with vertebrate TFIIB, showing that conserved cyclin features underlie the match. These results suggest the presence of a domain superfamily, which we term the TR domain, in nuclear regulatory proteins belonging to the TFIIB, cyclin and RB families, that has been duplicated many times during eukaryotic evolution. The TR domain appears to function in protein-protein interactions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Carroll R. J., Lipman D. J. Weights for data related by a tree. J Mol Biol. 1989 Jun 20;207(4):647–653. doi: 10.1016/0022-2836(89)90234-9. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank, recent developments. Nucleic Acids Res. 1993 Jul 1;21(13):3093–3096. doi: 10.1093/nar/21.13.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara L. R., Adamczewski J. P., Hunt T., La Thangue N. B. Cyclin A and the retinoblastoma gene product complex with a common transcription factor. Nature. 1991 Jul 18;352(6332):249–251. doi: 10.1038/352249a0. [DOI] [PubMed] [Google Scholar]
- Creti R., Londei P., Cammarano P. Complete nucleotide sequence of an archaeal (Pyrococcus woesei) gene encoding a homolog of eukaryotic transcription factor IIB (TFIIB). Nucleic Acids Res. 1993 Jun 25;21(12):2942–2942. doi: 10.1093/nar/21.12.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto S. H., Mudryj M., Pines J., Hunter T., Nevins J. R. A cyclin A-protein kinase complex possesses sequence-specific DNA binding activity: p33cdk2 is a component of the E2F-cyclin A complex. Cell. 1992 Jan 10;68(1):167–176. doi: 10.1016/0092-8674(92)90215-x. [DOI] [PubMed] [Google Scholar]
- Dowdy S. F., Hinds P. W., Louie K., Reed S. I., Arnold A., Weinberg R. A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993 May 7;73(3):499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- Etzold T., Argos P. SRS--an indexing and retrieval tool for flat file data libraries. Comput Appl Biosci. 1993 Feb;9(1):49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- Evans T., Rosenthal E. T., Youngblom J., Distel D., Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983 Jun;33(2):389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Ewen M. E., Faha B., Harlow E., Livingston D. M. Interaction of p107 with cyclin A independent of complex formation with viral oncoproteins. Science. 1992 Jan 3;255(5040):85–87. doi: 10.1126/science.1532457. [DOI] [PubMed] [Google Scholar]
- Faha B., Ewen M. E., Tsai L. H., Livingston D. M., Harlow E. Interaction between human cyclin A and adenovirus E1A-associated p107 protein. Science. 1992 Jan 3;255(5040):87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Flemington E. K., Speck S. H., Kaelin W. G., Jr E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T. J., Rice P. M., Thompson J. D., Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem Sci. 1993 Sep;18(9):331–333. doi: 10.1016/0968-0004(93)90068-x. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Thompson J. D., Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993 Jun 21;324(3):361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Giordano A., Whyte P., Harlow E., Franza B. R., Jr, Beach D., Draetta G. A 60 kd cdc2-associated polypeptide complexes with the E1A proteins in adenovirus-infected cells. Cell. 1989 Sep 8;58(5):981–990. doi: 10.1016/0092-8674(89)90949-5. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Schneider J. W., Condorelli G., Kaushal S., Mahdavi V., Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993 Feb 12;72(3):309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- Hagemeier C., Bannister A. J., Cook A., Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier C., Cook A., Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993 Nov 11;21(22):4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P. A., Gallie B. L., Phillips R. A. The retinoblastoma protein and cell cycle regulation. Trends Genet. 1992 May;8(5):180–185. doi: 10.1016/0168-9525(92)90221-o. [DOI] [PubMed] [Google Scholar]
- Haslam R. J., Koide H. B., Hemmings B. A. Pleckstrin domain homology. Nature. 1993 May 27;363(6427):309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hisatake K., Hasegawa S., Takada R., Nakatani Y., Horikoshi M., Roeder R. G. The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993 Mar 11;362(6416):179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- Kato J., Matsushime H., Hiebert S. W., Ewen M. E., Sherr C. J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993 Mar;7(3):331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Stewart E., Poon R., Adamczewski J. P., Gannon J., Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell. 1992 Nov;3(11):1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Transcriptional regulation by the retinoblastoma protein. Trends Cell Biol. 1993 Jul;3(7):211–213. doi: 10.1016/0962-8924(93)90113-f. [DOI] [PubMed] [Google Scholar]
- Lees E., Faha B., Dulic V., Reed S. I., Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992 Oct;6(10):1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- Levine A. J. The tumor suppressor genes. Annu Rev Biochem. 1993;62:623–651. doi: 10.1146/annurev.bi.62.070193.003203. [DOI] [PubMed] [Google Scholar]
- Lew D. J., I Reed S. A proliferation of cyclins. Trends Cell Biol. 1992 Mar;2(3):77–81. doi: 10.1016/0962-8924(92)90076-y. [DOI] [PubMed] [Google Scholar]
- López-De-León A., Librizzi M., Puglia K., Willis I. M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992 Oct 16;71(2):211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- Mayer B. J., Ren R., Clark K. L., Baltimore D. A putative modular domain present in diverse signaling proteins. Cell. 1993 May 21;73(4):629–630. doi: 10.1016/0092-8674(93)90244-k. [DOI] [PubMed] [Google Scholar]
- Mudryj M., Devoto S. H., Hiebert S. W., Hunter T., Pines J., Nevins J. R. Cell cycle regulation of the E2F transcription factor involves an interaction with cyclin A. Cell. 1991 Jun 28;65(7):1243–1253. doi: 10.1016/0092-8674(91)90019-u. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Gibson T., Rice P., Thompson J., Saraste M. The PH domain: a common piece in the structural patchwork of signalling proteins. Trends Biochem Sci. 1993 Sep;18(9):343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- Na J. G., Hampsey M. The Kluyveromyces gene encoding the general transcription factor IIB: structural analysis and expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1993 Jul 25;21(15):3413–3417. doi: 10.1093/nar/21.15.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J. Earliest phylogenetic branchings: comparing rRNA-based evolutionary trees inferred with various techniques. Cold Spring Harb Symp Quant Biol. 1987;52:825–837. doi: 10.1101/sqb.1987.052.01.090. [DOI] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992 Oct 16;71(2):189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- Pascarella S., Argos P. Analysis of insertions/deletions in protein structures. J Mol Biol. 1992 Mar 20;224(2):461–471. doi: 10.1016/0022-2836(92)91008-d. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Luckey C., Horton L., Esser M., Templeton D. J. Biological function of the retinoblastoma protein requires distinct domains for hyperphosphorylation and transcription factor binding. Mol Cell Biol. 1992 Dec;12(12):5363–5372. doi: 10.1128/mcb.12.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Fuchs R., Higgins D. G., Stoehr P. J., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1993 Jul 1;21(13):2967–2971. doi: 10.1093/nar/21.13.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippmann F., Taylor W. R., Rothbard J. B., Green N. M. A hypothetical model for the peptide binding domain of hsp70 based on the peptide binding domain of HLA. EMBO J. 1991 May;10(5):1053–1059. doi: 10.1002/j.1460-2075.1991.tb08044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. G., Ha I., Maldonado E., Reinberg D., Green M. R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993 Jun 24;363(6431):741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- Ruppert S., Wang E. H., Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993 Mar 11;362(6416):175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shirodkar S., Ewen M., DeCaprio J. A., Morgan J., Livingston D. M., Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992 Jan 10;68(1):157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993 Mar 11;21(5):1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann R. The basic RNA polymerase II transcriptional machinery. Gene Expr. 1992;2(2):81–91. [PMC free article] [PubMed] [Google Scholar]




