Abstract
When ruptured, the anterior cruciate ligament (ACL) of the human knee has limited regenerative potential. However, the goal of this report was to show that the cells that migrate out of the human ACL constitute a rich population of progenitor cells and we hypothesize that they display mesenchymal stem cell (MSC) characteristics when compared with adherent cells derived from bone marrow or collagenase digests from ACL. We show that ACL outgrowth cells are adherent, fibroblastic cells with a surface immunophenotype strongly positive for cluster of differentiation (CD)29, CD44, CD49c, CD73, CD90, CD97, CD105, CD146, and CD166, weakly positive for CD106 and CD14, but negative for CD11c, CD31, CD34, CD40, CD45, CD53, CD74, CD133, CD144, and CD163. Staining for STRO-1 was seen by immunohistochemistry but not flow cytometry. Under suitable culture conditions, the ACL outgrowth-derived MSCs differentiated into chondrocytes, osteoblasts, and adipocytes and showed capacity to self-renew in an in vitro assay of ligamentogenesis. MSCs derived from collagenase digests of ACL tissue and human bone marrow were analyzed in parallel and displayed similar, but not identical, properties. In situ staining of the ACL suggests that the MSCs reside both aligned with the collagenous matrix of the ligament and adjacent to small blood vessels. We conclude that the cells that emigrate from damaged ACLs are MSCs and that they have the potential to provide the basis for a superior, biological repair of this ligament.
Introduction
Much of the mechanical stability of the human knee is provided by the anterior cruciate ligament (ACL). With increased participation in sports, the frequency of ACL injuries is rapidly increasing, and over 100,000 patients rupture their ACL each year in the United States.1 Injury to the ACL presents enormous problems for both the patient and the orthopaedic surgeon.
Even with surgical repair, the ruptured ACL will not heal.2 However, if left unattended, it remains symptomatic and considerably increases the likelihood of developing premature osteoarthritis (OA) via exposure of pathologic loads to the cartilaginous joint surfaces in the unstable knee.3 Synthetic ACL substitutes have been evaluated, but these have had very limited clinical success due to mechanical failure and severe inflammatory reactions.4 For these reasons, it is common to surgically reconstruct the ACL using autograft hamstring or patellar tendon, and also allografts.5 Not only are these procedures highly invasive, with a protracted recovery period, but they are also very expensive, costing the U.S. healthcare system ∼$100 million per annum.1 Further, they fail to obviate the development of secondary OA.3
Recent data from Murray and colleagues challenge the dogma that the ACL lacks any intrinsic ability to heal. When the human, ruptured ACL is placed into organ culture, there is a rapid egress of cells.6,7 Under suitable culture conditions, these cells divide and form a collagenous repair tissue that resembles neoligament; indeed, if provided with a suitable scaffold, the cells participate in the successful repair of damaged ACL in animal models.8–10 These findings offer the prospect of developing strategies for the biological repair of the ACL with the potential to be more effective, less invasive, quicker, and more economical than the existing practice of surgical reconstruction. Because the outgrowth cells are central to the development of regenerative approaches to healing ACL ruptures, we have examined their properties in detail and were surprised to find them almost indistinguishable from mesenchymal stem cells (MSCs) derived from human bone marrow. Although the term MSC is controversial, it is used neutrally throughout this article to conform to the abundant literature on the subject.
MSCs are multipotent, fibroblastic cells11,12 first identified in bone marrow.13 Similar cells have since been isolated from an expanding list of connective tissues, including fat,14 muscle,15 skin,16 bone,17 periosteum,18 synovium,19 meniscus,20 cartilage,21,22 intervertebral disc,23 tendon,24 and only recently ligaments.25 Their phenotypic plasticity has generated considerable enthusiasm for using them to repair and regenerate connective tissues either with ex vivo, tissue engineering strategies,26 or by in situ techniques.27,28
Lack of specific markers impedes the detailed study of MSC biology, and many investigators define them operationally on the basis of their ability to differentiate along multiple lineages, particularly those leading to chondrogenesis, osteogenesis, and adipogenesis. Kolf et al.29 recently collated the literature concerning the surface immunophenotype of human MSCs, noting that, according to most authors, cultures of these cells stain positively for CD13, CD29, CD44, CD90, and CD105. Staining for STRO-1 and CD106 is more variable, and CD11b, CD31, CD34, and CD45 are normally absent.
Here we report than human ACL outgrowth cells share very similar properties with MSCs derived from bone marrow and collagenase digests of ACLs, but subtle differences were found that will be discussed in detail further in the article. Moreover, MSCs are common within the body of the ACL, both aligned to its collagen fibers and around blood vessels. The unexpected discovery of a mobile population of MSCs within the ACL offers a new insight into the biology of this ligament and new possibilities for its biological repair.
Materials and Methods
Tissue collection and cell isolation
This study was approved by the Institutional Review Boards of Children's Hospital and Brigham and Women's Hospital, Boston, MA, and the University of Würzburg, Würzburg, Germany. All patients gave informed consent. Human injured ACLs were harvested aseptically from a total of 20 patients (M:F ratio, 5:15), mean age 24.6 years (age range, 18–35 years), undergoing ACL reconstruction following complete ACL rupture after trauma during skiing (n=6), rollerblading (n=1), soccer playing (n=12), or biking (n=1). Surgery was performed within a mean time after trauma of 8 weeks (range, 4–16 weeks).
For comparison, we also retrieved intact ACL cells from 10 patients suffering knee OA (aged 46–68 years), whose intact ACLs were removed during total knee arthroplasty surgery.
Bone marrow cells were harvested from intramedullary reamings from the femoral shafts of 20 patients (M:F ratio, 15:5), mean age 56 years (age range 45–67 years), undergoing total hip replacement surgery after informed consent, as approved by the local Institutional Review Board.
For ACL explant cultures, ACLs were rinsed twice with serum-free Dulbecco's modified Eagle's medium (DMEM)/F-12 media (PAA Laboratories GmbH, Pasching, Austria) containing 2% antibiotic/antimycotic solution (Invitrogen, Carlsbad, CA). The synovial sheath and the ends of the ACL remnants were removed meticulously, and the ACL fascicles were dissected into pieces of about 3 mm3. The ACL fragments were placed in 12-well plates for 2 h at 37°C with a minimal amount of medium to promote adherence, followed by the addition of 1 mL complete medium consisting of DMEM/F-12 supplemented with 10% fetal bovine serum (FBS) (Invitrogen), antibiotics (50 IU penicillin/mL and 50 μg streptomycin/mL; PAA Laboratories), and 50 μg/mL ascorbate 2-phosphate (Sigma, St. Louis, MO) for 3–4 weeks. During this period, cells migrated from the tissue fragments forming a population of outgrowth cells (ACLOUT). Second passage cells were used for all experiments.
For ACL digest cell cultures, cells were isolated by direct digestion of ACL fragments in a buffer containing 0.1% (w/v) collagenase 1 and 3 (Invitrogen). The cells released from the ACL in this fashion (ACLDIG) were plated in monolayer culture in complete DMEM/F-12 medium. Second passage cells were used for all experiments.
Human bone-marrow-derived mesenchymal stromal cells (BMSCs) were isolated on marrow reamings (10–20 mL) harvested from the femoral shaft, which were transferred to 50 mL conical tubes containing 20 mL DMEM/F-12 (PAA Laboratories). The tubes were vortexed to detach marrow cells from the bone plugs and centrifuged (1000 rpm for 5 min) to pellet cells and bone plugs. The supernatant was discarded and the pellets were reconstituted in 10 mL complete medium. After repeated vortexing, the marrow cells were collected by aspiration with 10-mL syringes fitted with 20-gauge needles. The remaining cells in the bone plugs were extracted using the identical procedure for a total of four times until the bone plugs appeared yellowish-white. The collected cells were pelleted (1000 rpm for 5 min), resuspended in the complete medium, counted with a hemocytometer, and plated at a density of 8×107 nucleated cells per 150 cm2 tissue culture flask (TPP, Trasadingen, Switzerland). Nonadherent cells were removed after 2 days, and attached BMSCs were washed with phosphate-buffered saline (PBS) and cultured in the complete medium for 10–14 days with medium changes every 3–4 days. Cells to be analyzed for chondrogenic differentiation were expanded in the presence of 10 ng/mL fibroblast growth factor-2 (PeproTech, Rocky Hill, NJ), which has been shown to maintain the chondrogenic potential of MSCs in monolayer.30 All cells were observed in tissue culture using a light microscope (Model Axioscope 2; Carl Zeiss, Göttingen, Germany) and photographed with a digital camera (AxioCam MRc; Carl Zeiss).
Cell proliferation assay
Cell proliferation was assessed by measuring adenosine 5′ triphosphate (ATP) using the sensitive luminometric Cell Titer-Glo® assay system (Promega, Mannheim, Germany) according to the manufacturer's instructions. Briefly, first passage ACL-outgrowth cells, ACL-digest cells, or BMSCs were seeded at 1000 cells per well in 96-well plates (Thermo Fischer Scientific Nunc, Langenselbold, Germany) and cultured in 100 μL complete DMEM/F-12 mediu per well for 12 days, with medium changes every 2 days. At days 2, 4, 6, 8, 10, and 12, luminescence of n=10 wells per cell type and donor was measured for 0.1 s in a luminometer after an equal volume of Cell Titer-Glo Reagent was added per well followed by 5 min incubation at room temperature. A total of five donors for each cell type were included.
Fluorescence-activated cell sorting
Cells were detached with 10 mM ethylenediaminetetraacetic acid, and suspended in 4°C PBS with sodium azide (to minimize internalization or shedding of surface molecules) to a final concentration of 2×107 cells/mL. Samples were distributed into 96-well V-bottom plates (BD Biosciences, San Jose, CA) and spun at 400 g for 3 min. Supernatants were decanted and the cell pellets washed twice with PBS. After washing, samples were suspended in 100 (microL blocking buffer, and incubated for 30 min at 4°C. Direct single or multicolour immunofluorescent staining was performed as follows: 100 μL antigen-specific fluorescent mAb or an immunoglobulin isotype-matched control were added to samples and incubated for 30 min at 4°C. After incubation, samples were pelleted at 400 g for 3 min and the supernatant decanted. Samples were resuspended in 4°C PBS with sodium azide, and this wash step repeated three times. Samples were analyzed immediately or stored in 2% paraformaldehyde for later analysis. Samples were analyzed using a Cryonics FC 500 flow cytometer (Beckman Coulter, Fullerton, CA). All antibodies (Serotec, Düsseldorf, Germany; BD Biosciences; R&D Systems, Minneapolis, MN; Acris Antibodies GmbH, Hiddenhausen, Germany; Santa Cruz Biotechnology, Santa Cruz, CA) were conjugated to the fluorochromes fluorescein isothiocyanate, phycoerythrin, or allophycocyanin for the following cluster of differentiation (CD) antigens: CD11c, CD14, CD29, CD31, CD34, CD40, CD44, CD45, CD49c, CD53, CD73, CD74, CD90, CD97, CD105, CD106, CD133, CD144, CD146, CD163, and CD166, as well as alkaline phosphatase (ALP), HLA A,B,C, and STRO-1. Nonspecific mAbs for all fluorochromes were used as comparative controls. Labeling, manufacturer, and marker specification for each antibody is listed in Table 1.
Table 1.
Cell Surface Phenotype of ACLOUT, ACLDIG, and Bone Marrow Mesenchymal Stromal Cells
| AB | Label | Manufacturer | Marker specification | ACLOUT | ACLDIG | BMSC |
|---|---|---|---|---|---|---|
| CD11c | PE | Serotec | Cell adhesion (αX integrin), immune cell marker | − | − | + |
| CD14 | FITC | BD Biosciences | Monocyte marker | + | + | + |
| CD29 | PE | Serotec | Cell adhesion (β1 integrin) | +++ | +++ | +++ |
| CD31 | FITC | Serotec | PECAM 1, endothelial and hematopoietic cells | − | − | − |
| CD34 | APC | BD Biosciences | HSC marker, cell adhesion | − | − | − |
| CD40 | FITC | BD Biosciences | Epithelial and monocyte marker | − | − | − |
| CD44 | PE | BD Biosciences | Hyaluronic acid receptor, HCAM | +++ | +++ | +++ |
| CD45 | APC | BD Biosciences | Hematopoetic cells, leukocyte antigen | − | − | − |
| CD49c | PE | BD Biosciences | Cell adhesion, signal transduction | ++ | ++ | ++ |
| CD53 | FITC | Serotec | Osteoblast, osteoclast, signal transduction | − | − | − |
| CD73 | PE | BD Biosciences | Lymphocyte-vascular adhesion protein 2, mesenchymal, epithelial and endothelial cells | +++ | +++ | +++ |
| CD74 | FITC | BD Biosciences | Epithelial and endothelial cells | − | − | − |
| CD90 | APC | BD Biosciences | Thy1, fibroblasts, stromal cells, HSCs | +++ | +++ | +++ |
| CD97 | FITC | BD Biosciences | Monocyte marker | ++ | ++ | + |
| CD105 | PE | R&D Systems | Endoglin, mesenchymal and erythroid progenitors | +++ | +++ | +++ |
| CD106 | APC | BD Biosciences | VCAM-1, Leukocyte adhesion | − | − | ++ |
| CD133 | FITC | R&D Systems | AC133, HSC marker | − | − | − |
| CD144 | FITC | Serotec | Endothelial cells, cell adhesion, β-catenin | − | − | − |
| CD146 | PE | R&D Systems | Endothelial cells, cell adhesion | ++ | ++ | ++ |
| CD163 | FITC | Acris Antibodies | Monocyte marker, endocytosis | − | − | − |
| CD166 | PE | BD Biosciences | Monocyte, fibroblast, epithelial and mesenchymal stem cell marker, cell adhesion | +++ | +++ | +++ |
| ALP | APC | R&D Systems | Alkaline phosphatase | ++ | ++ | ++ |
| HLAA,B,C | PE/ABC | BD Biosciences | MHC I class | +++ | +++ | +++ |
| STRO-1 (Ig M) | PE | Santa Cruz | Mesenchymal stem cells | − | − | + |
| Ig G1 | FITC | Serotec | Isotype control antibody | − | − | − |
| Ig G1 | PE | R&D Systems | Isotype control antibody | − | − | − |
| Ig G1 | APC | BD Biosciences | Isotype control antibody | − | − | − |
| Ig M | PE | Santa Cruz | Isotype control antibody | − | − | − |
, marker expression on >95% of the cells;++, marker expression on 15%–95% of the cells;+, marker expression on 5%–15% of the cells, −, marker expression on <5% of the cells; AB, antibody; ACLDIG, anterior cruciate ligament collagenase digest cells; ACLOUT, anterior cruciate ligament outgrowth cells; ALP, alkaline phosphatase; APC, allophycocyanin; BMSC, bone marrow mesenchymal stromal cells; CD, cluster of differentiation; FITC, fluorescein isothiocyanate; PE, phycoerythrin; HCAM, homing cell adhesion molecule; HSC, hematopoietic stem cell; PECAM, platelet/endothelial cell adhesion molecule 1; Thy1, thymus cell antigen 1; VCAM-1, vascular cell adhesion molecule-1.
Histology and immunohistochemistry
Monolayer cultures were washed three times with PBS and then fixed with 100% methanol cooled to 4°C. Staining of ACL tissue sections or constructs was performed after fixation in 10% buffered formalin for 5 days. The fixed tissues were embedded in paraffin and sectioned at 5 μm. Sections were deparaffinized for 2 min in xylene and rehydrated in graded alcohols. For histological analyses, ACL sections or constructs were stained using hematoxylin and eosin (H&E), Azan, and Masson/Goldner (M/G). For immunohistochemical analyses of ACL tissue sections or monolayer cultures, slides were washed for 20 min in Tris buffered saline, and then incubated in 2% bovine serum albumin (Sigma) and 2.5% normal horse serum. After washing in Tris buffered saline, sections were trypsinized (1 mg/mL) for 30 min at 37°C, and then incubated with 5 μg/mL primary antibody. Based on the fluorescence-activated cell sorting (FACS) data (Table 1), immunostaining for selected markers was performed to locate these cells within the bulk tissue, using primary diluted antibodies for the CD44 (1:200), CD90 (1:25), CD105 (1:50) (all DAKO, Hamburg, Germany), and STRO-1 (1:20) (R&D Systems) antigens. Antibody binding was detected using the Link-Label IHC Detection System (BioGenex, San Ramon, CA), as directed by the supplier, using Fast Red (BioGenex) as substrate chromogen. Sections were then analyzed for red color by light microscopy.
Chondrogenesis
Chondrogenic differentiation was assessed by the pellet culture method as modified recently.31 Cells were suspended to a concentration of 1×106 cell/mL in serum-free DMEM and 200-μL aliquots (2×105 cells) were distributed to a polypropylene, V-bottom 96-well plate (Corning Inc., Corning, NY). The plate was centrifuged at 400 g for 5 min, and the supernatant aspirated and replaced with the chondrogenic medium consisting of DMEM-high glucose (Invitrogen) with 1% antibiotic/antimycotic cocktail, 1% ITS+Premix (BD Biosciences), 40 μg/mL proline, 100 nM dexamethasone, and 50 μg/mL ascorbate-2-phosphate (all from Sigma). To certain aggregates, 10 ng/mL recombinant human transforming growth factor beta1 (TGF-β1) (PeproTech) was added to enhance chondrogenesis. Media were changed every other day, and aggregates were collected at 3 or 4 weeks for analysis.
The DNA content of the aggregates was determined using the Hoechst Dye 33258 method.32 Samples were digested overnight at 65°C in 100 μg/mL proteinase K (Sigma). Pellet digests were taken through three freeze–thaw cycles, and digest aliquots were diluted with 100 ng/mL Hoechst Dye 33258 (Sigma) in 10 mM Tris (pH 7.4), 1 mM Na2 ethylenediaminetetraacetic acid, 100 mM NaCl. The fluorescence intensity was measured with a DQ300 Fluorometer (Hoefer Scientific Instruments, San Francisco, CA), and DNA concentration was determined from a standard curve of calf thymus DNA (Sigma). Proteinase K digests were also analyzed for glycosaminoglycan (GAG) content using the dimethylmethylene blue (Sigma) dye binding assay.33 Aliquots of digest, diluted as necessary, were combined with dimethylmethylene blue solution, and sample absorbances were measured at 595 nm. GAG concentrations were interpolated from a standard curve of shark chondroitin sulphate (Sigma), and results were normalized by DNA content.
Before histology, aggregates were fixed for 30 min in 4% paraformaldehyde, encapsulated in 0.5% agarose gels, embedded in paraffin, and sectioned at 5 μm thickness. Sections were mounted onto glass slides, de-paraffinized with three xylene washes (5 min each), and re-hydrated in graded alcohol solutions. For detection of matrix proteoglycan, representative sections were stained with 1.0% (w/v) Toluidine blue (Sigma), pH 3.0 for 30 min. Slides were rinsed in deionized water, dehydrated in graded alcohols, rinsed three times in xylene, and cover slipped with Cytoseal™ XYL mounting medium (Richard-Allan Scientific, Kalamazoo, MI).
For immunohistochemistry, aggregate sections were digested with 0.1 U/mL chondroitinase ABC (Sigma) in PBS with 1% bovine serum albumin (Sigma) for 1 h at 37°C. Endogenous peroxidases were quenched in 0.3% hydrogen peroxide solution for 30 min. After blocking with normal horse serum for 1 h, slides were incubated overnight (2°C–8°C) with polyclonal rabbit anti-human collagen type II antibody (Santa Cruz) in blocking buffer. Antigens were observed using a biotinylated horse anti-rabbit secondary antibody and VectaStain® Elite ABC reagent (Vector Laboratories, Burlingame, CA). Slides were rinsed, counterstained with hematoxylin, dehydrated, and cover slipped.
Reverse transcriptase (RT)-polymerase chain reaction (PCR) analyses were used to evaluate expression of cartilage-specific genes. Total RNA was isolated from six aggregates, harvested after 21 days. To facilitate lysis of the cells in aggregate culture, they were first frozen in liquid nitrogen and ground to a powder with a custom-made pellet pestle. Total RNA was subsequently extracted using 1 mL of Trizol reagent with an additional purification step using RNeasy separation columns (RNeasy kit; Qiagen, Hilden, Germany) according to the manufacturer's instructions. For cDNA synthesis, 1 μg of total RNA from each group was reverse transcribed using random hexamer primers and MoMLV-H reverse transcriptase (Promega), according to the manufacturer's instructions. Equal amounts of each cDNA synthesized (100 ng) were used as templates for PCR amplification in a 50 μL reaction volume using Taq DNA polymerase (Amersham, Braunschweig, Germany) and 50 pmol of specific primers for the following human messenger (m)RNAs: collagen type II alpha 1 (COL II), aggrecan core protein (AGC), cartilage oligomeric matrix protein (COMP), and elongation factor 1α as an internal control. Primer sequences are provided in Table 2. The RT-PCR products were electrophoretically separated on 1.5% agarose gels containing 0.1 μg/mL ethidium bromide.
Table 2.
Primer Sequences (5′—3′) for Reverse Transcriptase–Polymerase Chain Reaction Analysis
| Target | Sense sequence | Antisense sequence | Product (bp) |
|---|---|---|---|
| AGC | TGAGGAGGGCTGGAACAAGTACC | GGAGGTGGTAATTGCAGGGAACA | 392 |
| ALP | TGGAGCTTCAGAAGCTCAACACCA | ATCTCGTTGTCTGAGTACCAGTCC | 454 |
| Cbfa1 | ACAGATGATGACACTGCCACC | CATAGTAGAGATATGGAGTGCTGC | 324 |
| COL I | GGACACAATGGATTGCAAGG | TAACCACTGCTCCACTCTGG | 461 |
| COL II | TTTCCCAGGTCAAGATGGTC | CTTCAGCACCTGTCCACCA | 374 |
| COMP | CAGGACGACTTTGATGCAGA | AAGCTGGAGCTGTC TGGTA | 312 |
| EF-1α | AGGTGATTATCCTGAACCATCC | AAAGGTGGATAGTCTGAGAAGC | 234 |
| Fibronectin | TGGAACTTCTACCAGTGCGAC | TGTCTTCCCATCATCGTAACAC | 451 |
| LPL | GAGATTTCTCTGTATGGCACC | CTGCAAATGAGACACTTTCTC | 276 |
| OC | ATGAGAGCCCTCACACTCCTC | GCCGTAGAAGCGCCGATAGGC | 294 |
| PPARγ2 | GCTGTTATGGGTGAAACTCTG | ATAAGGTGGAGATGCAGGCTC | 351 |
| Tenomodulin | CCATGCTGGATGAGAGAGGT | CTCGTCCTCCTTGGTAGCAG | 123 |
| Vimentin | GACCGCTTCGCCAACTACATCGAC | GGTCATCGTGATGCTGAGAACTTCG | 1060 |
AGC, aggrecan core protein; Cbfa1, core binding factor α1; COL I, collagen type I α2; COL II, collagen type II α1; COMP, cartilage oligomeric protein; EF-1α, elongation factor-1α; LPL, lipoprotein lipase; OC, osteocalcin; PPARγ2, proliferator-activator receptor γ2.
Osteogenesis
Cells were seeded at a density of 1×105 cells/cm2 in four-well chamber slides and 25 cm2 flasks (Thermo Fischer Scientific Nunc). At confluence, osteogenesis was induced by supplementing the media with 100 nM dexamethasone, 50 μg/mL ascorbate, 10 mM β-glycerophosphate, and 25 ng/mL recombinant human bone morphogenetic protein 2 (BMP2) (R&D Systems); control cultures lacked osteogenic supplements. Media were changed every 3 days for 3 weeks. Cultures were stained histochemically for ALP, using a commercial kit according to the manufacturer's protocol (Sigma), and for matrix mineralization using Alizarin red as described previously.17 Additionally, markers of osteogenesis were analyzed by RT-PCR, using the protocol described above. Expression of the following human mRNAs was examined: ALP, collagen type I alpha 2 (COL I), osteocalcin, and core binding factor alpha 1 (Runx2). Primer sequences are provided in Table 2.
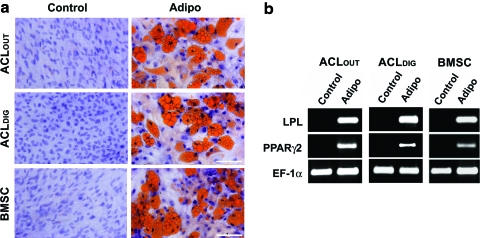
Adipogenesis
Cells were grown to 50%–70% confluence in four-well chamber slides, or 25 cm2 flasks (Thermo Fischer Scientific Nunc) in DMEM supplemented with 10% FBS. Adipogenesis was induced by supplementing the media with 1 μM dexamethasone, 1 μg/mL insulin, 0.5 mM 3-isobutyl-1-methylxanthine, and 100 μM indomethacin. Control cultures without adipogenic supplements were also maintained. After 3 weeks, cultures were examined for evidence of adipogenesis by fixing in 10% formalin and staining with freshly prepared Oil red-O solution for the detection of lipid droplets as previously reported.17 As additional markers of adipogenesis, RT-PCR, using the protocol described above, was used to detect expression of the following human genes: lipoprotein lipase and proliferator-activator receptor γ2. Primer sequences are provided in Table 2.
Ligamentogenesis
Ligamentogenic induction of cell cultures was performed using an in vitro assay that we described previously.34 Briefly, ACLOUT, ACLDIG, and BMSCs were seeded at 3.6×106 cells/175 cm2 flask and transduced at 100 multiplicities of infection of Ad.BMP12, in 5 mL of the serum-free medium. Marker gene (green fluorescent protein [GFP]) or untransduced cells were used as comparative controls. At 24 h after transduction, the cells were recovered and placed in collagen hydrogel constructs, with 3×105 cells being suspended in 100 μL gel neutralization solution, followed by the addition of 100 μL collagen type I stock solution (Arthro Kinetics AG, Esslingen, Germany). After polymerization, the constructs were cultured in 48-well plates with 500 μL complete culture medium with medium changes every 2–3 days throughout the 21-day culture period. All constructs were evaluated histologically (H&E, Azan, and M/G), immunohistochemically (collagen type III [COL III], elastin, vimentin, and fibronectin), and by RT-PCR (COL I, tenomodulin, fibronectin, and vimentin) after 21 days as described above. Primer sequences are provided in Table 2.
Statistical analysis
The numerical data from the ATP and GAG analyses were expressed as mean values±standard deviation. Each experiment was performed at least in triplicate per group and patient and repeated on at least 3 and up to 10 individual ACL/marrow preparations from different patients (n=3–10), as indicated in the respective experiments. Stainings for ALP, Alizarin red, and Oil red-O were evaluated histomorphometrically employing four high power fields of four representative slides per group and donor using the Zeiss Axio Vision Software Rel. 4.6.3 (Zeiss, Ulm, Germany). All numerical data were subjected to variance analysis (one- or two-factor analysis of variance) and statistical significance was determined by Student's t-test, with <0.05 considered significant.
Results
Cell morphology and proliferation of ACL outgrowth cells
Throughout this study, cells that migrated from the ACL during explant culture (ACLOUT) were compared to ACL cells released by collagenase digest (ACLDIG) and BMSCs. It was not possible to obtain ACL and bone marrow from the same individuals, so the samples are not patient- or age-matched. To prevent contamination of ligament cells by synoviocytes, the synovial sheaths were meticulously dissected from sections of human ACL before use.
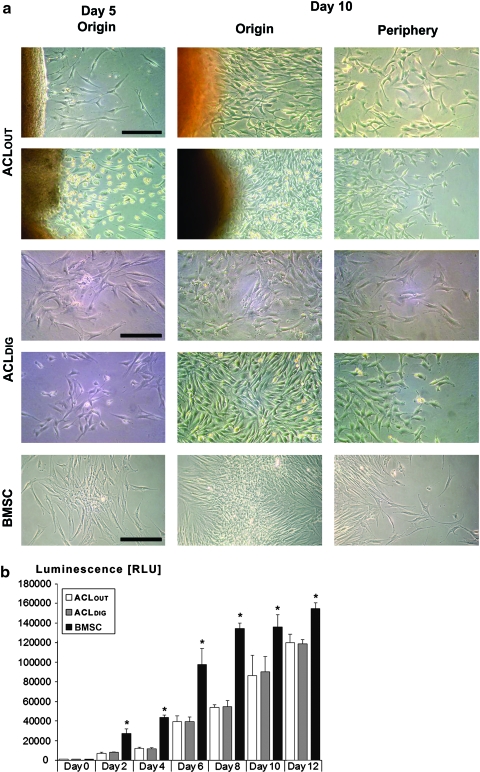
After human ACL samples were recovered, ligaments were dissected into explants (∼3 mm2) and placed into organ culture in 12-well plates. After a lag of ∼5 days, fibroblastic cells emigrated from the cut ends of the ACL. Although the rate of outgrowth and cell morphology varied between samples (Fig. 1a), all cultures formed confluent monolayers in ∼3 weeks.
FIG. 1.
Morphology and cell proliferation of anterior cruciate ligament outgrowth cells (ACLOUT). (a) Phase-contrast photomicrographs of adult human ACLOUT, collagenase-digested ACLDIG cultures, and human bone marrow stromal cells (BMSCs) after 5 and 12 days of culture. Fibroblastic cells migrated from ACL fragments after the first few days of culture (left). Rapidly increasing cell numbers were seen after 12 days, with monolayers reaching confluence after ∼3 weeks (not shown). Two different representative explants are shown for comparison (first and second row). After ACLDIG were placed in culture, where they formed distinct colonies within 5 days. ACLDIG colonies expanded substantially in size by day 12 of culture, and origins and periphery from colonies derived from of two different preparations are shown (third and fourth row). By 5 days after plating bone marrow, BMSCs (bottom row) had formed colonies. These expanded substantially in size by 12 days of culture, and origin and periphery from a representative colony are shown. Scale bar: 100 μm. (b) Comparative cell proliferation rates by means of luminescent measurement of ATP amounts present in all three cell types over time showed increased cell proliferation rates of the BMSCs at all time points compared to the ACLDIG and ACLOUT cultures, which showed no significant differences. Analyses of five different patients are included and n=10 different measurements per cell type, time point, and per patient were performed, with a p-value of 0.5 being considered significant between groups (Asterisks). Color images available online at www.liebertonline.com/tea
Cells recovered from collagenase-digested ACL (ACLDIG) were seeded directly into monolayer culture, where their growth pattern and morphology appeared almost indistinguishable from ACLOUT and BMSCs, which are shown for comparison (Fig. 1a).
We next compared cell proliferation rates in all three cell types, which were estimated by quantitation of ATP using the Cell Titer-Glo luminescent assay and revealed increased cell proliferation rates of the BMSCs at all time points measured compared to the ACLDIG and ACLOUT, which showed no significant differences (Fig. 1b).
Surface immunophenotype of ACL outgrowth cells
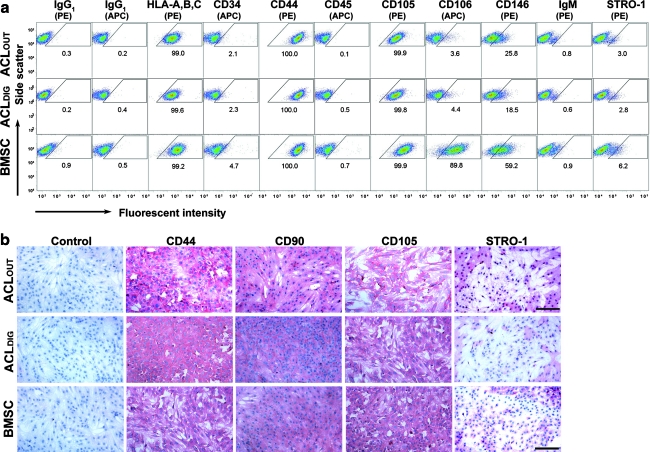
FACS was used to measure the presence of key surface markers that are displayed in Figure 2a and Table 1. ACLOUT and ACLDIG populations were both strongly positive for CD29, CD44, CD49c, CD73, CD90, CD97, CD105, CD146, CD166, HLA A, B, C, and ALP, weakly positive for CD14, but negative for CD11c, CD31, CD34, CD40, CD45, CD53, CD74, CD106 and CD133, CD144, CD163, and STRO-1 (Table 1). The ACLOUT populations appeared to contain a higher percentage of cells that were CD146 positive compared to the ACLDIG, although this observation did not reach levels of significance (p=0.33; Fig. 2a).
FIG. 2.
Comparative cell surface phenotying of ACLOUT. (a) Flow cytometric analyses of passage 2 cells revealed that ACLOUT represented nonhematopoietic populations with a very similar surface marker expression profile to that of ACLDIG and BMSCs. In particular, ACLDIG were almost indistinguishable in terms of their surface marker expression compared to the ACLOUT population, with differences in cluster of differentiation (CD)146+ being evident (not significant). However, smaller fractions of both ACL populations stained positively for CD106 and CD146 compared to BMSCs. PE, phycoerythrin; APC, allophycocyanin. Percentage of positive cells for each epitope is given within the corresponding panel. Cell preparations of five individuals for each cell type have been analyzed and representative images are shown. A more comprehensive surface profile comparison is presented in Table 1. (b) Immunocytochemical staining for CD44, CD90, CD105, and STRO-1 in ACLOUT, ACLDIG, and BMSC monolayer cultures revealed strong immunoreactivity (red staining) for the markers CD44, CD90, and CD105. Staining for STRO-1 was also positive, but less widespread as shown in the respective panels, whereas no immunoreactivity was found in the controls. Representative images from cell preparations of 10 donors for each cell type are shown. Scale bar=100 μm. Color images available online at www.liebertonline.com/tea
BMSCs displayed a similar, although not identical, pattern (Table 1); in particular, they stained more strongly for CD106 and CD146, and were weakly positive for CD11c and STRO-1. However, they stained less strongly for CD97 (Table 1) compared to the ACL cell populations. FACS data for selected surface markers are shown in Figure 2a.
The expression of certain markers was confirmed by immunocytochemical staining of ACLOUT, ACLDIG, and BMSCs (Fig. 2b). In agreement with the FACS data, these cells showed strong immunoreactivity toward CD44, CD90, and CD105 (Fig. 2b). Staining for STRO-1 was stronger than expected from the FACS data (Fig. 2a). This may reflect the properties of the anti-STRO-1 immunoglobulin M antibody used in these experiments, or the fact that cells used for FACS had gone through more passages.35
Chondrogenic differentiation
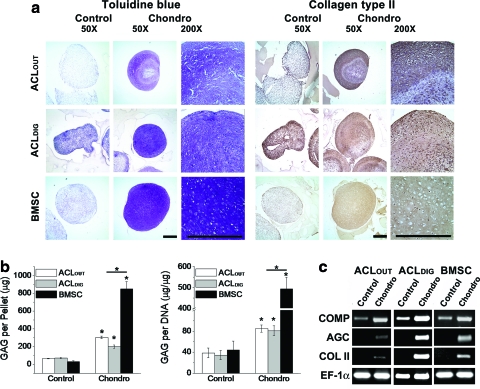
When placed into pellet culture, cells derived from both ACL sources and bone marrow underwent chondrogenic differentiation in the presence, but not absence, of TGF-β1 as judged by histology and immunohistochemistry (Fig. 3a), the accumulation of GAG (Fig. 3b), and the expression of transcripts encoding COMP, AGC, and COL II (Fig. 3c). Nevertheless, there were subtle differences between the chondrogenic behavior of ACLOUT, ACLDIG, and BMSCs. Pellets formed by the latter were larger and stained more uniformly for proteoglycans with toluidine blue and for cartilage-specific anti-type II collagen antibodies (Fig. 3a). Cartilaginous pellets formed by ACLOUT, and ACLDIG in contrast, had a core that stained only weakly with these reagents (Fig. 3a). Chondrogenic pellets from all cell types produced significantly more GAG compared to nonchondrogenic controls (Fig. 3b). However, chondrogenic pellets formed from ACLOUT and ACLDIG contained significantly less GAG than those formed from BMSCs, both in total and when normalized for DNA content (Fig. 3b). Consistent with this, chondrogenic pellets from all cell types produced significantly more mRNA transcripts encoding COMP and induced expression of AGC and COL II compared to the respective nonchondrogenic controls (Fig. 3c).
FIG. 3.
Chondrogenic differentiation by ACLOUT versus ACLDIG and BMSC cultures. After 4 weeks in the chondrogenic medium, aggregates derived from ACLOUT, ACLDIG, and BMSCs were analyzed for proteoglycan and type II collagen content as well as for expression of mRNAs associated with chondrogenesis. (a) Paraffin-embedded aggregate sections were stained with Toluidine blue or incubated with anti-human type II collagen antibody (collagen type II). Scale bars=500 μm at both magnifications. Unlike controls, aggregates treated with transforming growth factor beta1 (TGF-β1) (Chondro) from ACLOUT and ACLDIG populations produced an extracellular matrix rich in sulfated proteoglycans and type II collagen similar to the BMSC cultures, although the effect was more homogenous within BMSC aggregates. In proteoglycan- and collagen-rich regions of aggregates, cells were embedded within lacunae-like structures. At least 10 donors for each cell type have been analyzed and representative images are shown. (b) Cells undergoing chondrogenesis produced greater amounts of glycosaminoglycan (GAG) per pellet and per cell compared to controls. This was significantly more pronounced for cells derived from bone marrow. Graphs shown represent results obtained from each of five individual donors, and test have been performed in triplicate for each group and cell type. Asterisks indicate significant differences compared to controls. (c) Aggregate cultures of all three cell types collected at 3 weeks expressed the cartilage-specific genes encoding collagen type II (COL II) and aggrecan core (AGC) protein in response to TGF-β1 treatment, in contrast to controls lacking TGF-β1. Control aggregates expressed cartilage oligomeric matrix protein (COMP) mRNA at low levels, but expression was markedly increased in the presence of TGF-β1. The expression of elongation factor (EF)-1α was included as an internal control for RNA loading. Results are presented using representative patient populations from at least five independent experiments. Color images available online at www.liebertonline.com/tea
Osteogenic and adipogenic differentiation
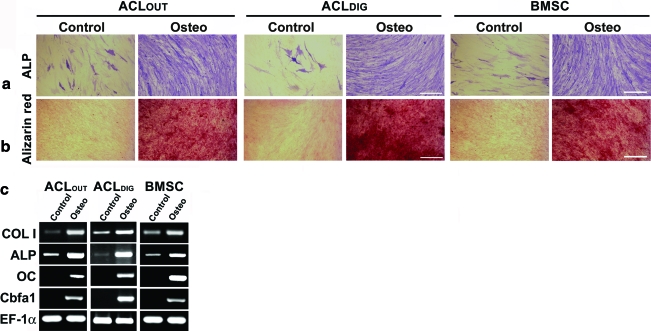
Monolayer cultures of ACLOUT, ACLDIG, and BMSC responded equivalently to the osteogenic medium in terms of staining for ALP by cytochemistry (Fig. 4a) and for mineralization with alizarin red (Fig. 4b). Histomorphometric analyses revealed significantly increased areas of positive staining for ALP (Fig. 4a) and Alizarin red (Fig. 4b) in the respective osteogenic cultures of all three cell types compared to controls. Among the osteogenic cultures the BMSCs showed the largest mean areas of positive staining for ALP (64.7%) and Alizarin red (70.2%), followed by the ACLDIG (56.8%/55.1%) and ACLOUT (52.9%/51.2%) cultures, respectively. However, significant differences between the cell types could not be resolved (p>0.07/p>0.09 for all comparisons). This corresponds to the induction of transcripts encoding osteocalcin and core binding factor alpha 1/Runx2, and the increased expression of ALP and the α-1 chain of COL I in all osteogenic cultures compared to controls (Fig. 4c).
FIG. 4.
Osteogenic differentiation by ACLOUT versus ACLDIG and BMSC cultures. At least 10 donors for each cell type have been analyzed and representative images are shown. Monolayer cultures were maintained without (Control) and with osteogenic (Osteo) supplements for 3 weeks, and then analyzed by (a, b) cytochemistry and (c) RT-polymerase chain reaction (PCR). (a) Cultures treated with osteogenic supplements contained increased numbers of alkaline phosphatase (ALP)-positive cells (original magnification 200×; bar=100 μm), and (b) produced a mineralized extracellular matrix as shown by intense staining for Alizarin red (original magnification 100×; bar=200 μm). Control cultures contained few ALP+ cells and did not produce a mineralized matrix. Corresponding histomorphometry revealed significant differences of the osteogenic cultures compared to controls for all cell types, but differences between cell types could not be resolved. (c) RT-PCR revealed that ACLOUT as well as ACLDIG and BMSC cultures expressed the osteoblast-related genes osteocalcin (OC), and core binding factor α1 (Cbfa1), in response to osteogenic stimuli, and increased expression of ALP and type I collagen (COL I) compared to controls. Expression of EF-1α was included as an internal control for RNA loading. Color images available online at www.liebertonline.com/tea
Monolayer cultures of ACLOUT, ACLDIG, and BMSCs responded equivalently to adipogenic conditions, as assessed by staining with Oil red-O (Fig. 5a). Again, histomorphometry showed significantly increased areas of positive staining for Oil red-O in the respective adipogenic cultures of all three cell types compared to controls (Fig. 5a). Again the adipogenic BMSCs showed the largest mean areas of positive staining for Oli red-O (39.6%), followed by the ACLOUT (33.8%) and the ACLDIG (30.6%) cultures; however, differences were not significant. Equivalently, adipogenic culture conditions induced production of transcripts encoding lipoprotein lipase and proliferator-activator receptor γ2 compared to controls within all groups (Fig. 5b).
FIG. 5.
Adipogenic differentiation of ACLOUT versus ACLDIG and BMSC cultures. Monolayer cultures were maintained without (Control) and with adipogenic supplements (Adipo) for 3 weeks, and analyzed for adipogenesis by cytochemistry and RT-PCR. (a) Cultures treated with adipogenic supplements contained cells accumulating large amounts of lipid droplets as shown by intense staining for Oil red-O, in contrast to the respective control cultures (original magnification 200×; scale bar=100 μm), which was significant for all cell types in histomorphometric evaluations (not shown). Differences between cell types were not significant. (b) RT-PCR analysis for expression of lineage-specific genes revealed that ACLOUT, as well as ACLDIG and BMSCs expressed the adipocyte-related lipoprotein lipase (LPL) and proliferator-activator receptor γ2 (PPARγ2) in response to adipogenic medium. Expression of EF-1α was included as a constitutive control for RNA loading. Color images available online at www.liebertonline.com/tea
This trilineage differentiation capacity of ACLDIG and ACLOUT was also confirmed, when intact ACLs from osteoarthritic donors were used and were also confirmed on single colony level for each donor (data not shown).
Ligamentogenesis
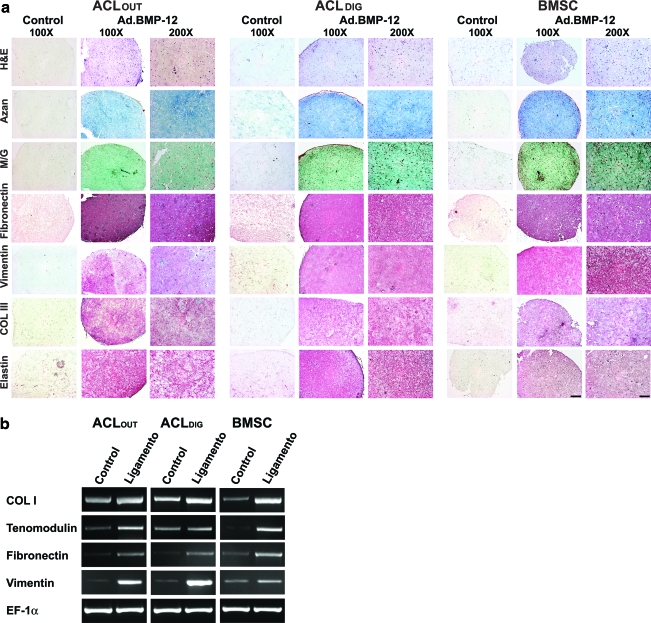
To show that ACLOUT have the capacity for self-renewal, ligamentogenesis was induced using an in vitro assay, which was employed previously to reveal the ligamentogenic capacities of ACLDIG and BMSC populations.34
After 21 days, staining with H&E showed a homogenous cell distribution of ACLOUT, ACLDIG, and BMSC fibroblasts within the hydrogel constructs (Fig. 6a). Moreover, the BMP12-modified constructs revealed that the cells were embedded in a dense collagenous matrix compared to GFP-modified controls where less matrix formation was seen, as evidenced by matrix staining with Azan and M/G (Fig. 6a).
FIG. 6.
Ligamentogenic differentiation of ACLOUT versus ACLDIG and BMSC cultures. As self-renewal is an important stem cell criterion, we employed ACLOUT, ACLDIG, and BMSC populations in an in vitro assay of ligamentogenesis using bone morphogenetic protein 12 (BMP12) encoding adenovirus vectors and three-dimensional culture in collagen hydrogels for differentiation induction compared to controls, which were maintained after marker gene transduction (green fluorescent protein, GFP) in a similar fashion. (a) At 21 days, hematoxylin and eosin (H&E) stainings revealed homogenous cell distribution of fibroblasts for all cell types within the hydrogel constructs (first row), and a strong accumulation of a collagenous matrix after modifications with BMP12 compared to GFP-modified controls where less matrix formation was seen, as evidenced by matrix staining with Azan (second row) and Masson/Goldner (M/G; third row). Immunohistochemical analyses for the ligament matrix proteins fibronectin (fourth row), vimentin (fifth row), collagen type III (Col III; sixth row), and elastin (seventh row) revealed more intense stainings in the Ad.BMP12 transduced hydrogel constructs of all cell types compared to corresponding controls, where no red staining of the matrix was detectable. Differences between cell types (ACLOUT vs. ACLDIG vs. BMSC) could not be detected. Scale bars: 100×bar=200 μm; 200×bar=100 μm. (b) RT-PCR analysis for expression of ligament-specific genes revealed that ACLOUT, as well as ACLDIG and BMSCs, increased expression of the factors COL I, tenomodulin, fibronectin, and vitronectin compared to the respective GFP controls, where only weak expression levels could be detected. Expression of EF-1α was included as a constitutive control for RNA loading. Color images available online at www.liebertonline.com/tea
Immunohistochemistry for the ligament matrix proteins fibronectin, vimentin, COL III, and elastin revealed more intense stainings in the Ad.BMP12 transduced hydrogel constructs compared to corresponding controls with ACLOUT, ACLDIG, and BMSC fibroblasts, where no red staining of the matrix was detectable (Fig. 6a). For each immunostaining, controls were performed without primary antibody, which were negative in all cases (not shown).
This is in correspondence with increased expression of mRNAs encoding COL I, tenomodulin, fibronectin, and vimentin in all ligamentogenic cultures compared to marker gene controls (Fig. 6b).
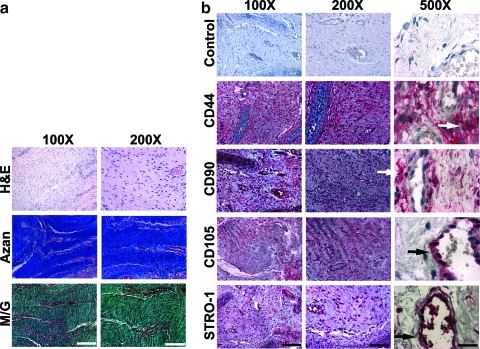
Immunohistochemical location of MSCs within the ACL
Longitudinal sections of ACLs stained with H&E, Azan, or M/G confirmed the typical pattern of undulating collagenous fibers along which align extended fibroblastic cells (Fig. 7a). Vasculature was represented by small, rare blood vessels.
FIG. 7.
Histological and immunohistochemical staining of ACL sections. Pieces of at least n=10 torn and intact ACLs were analyzed and representative images are shown. (a) H&E staining revealed ACL cells within the tissue matrix, as well as the vessel walls. Azan and M/G staining was used to illustrate the density and organization of the collagen fibers that form the tissue matrix within which the ACL cells are embedded. (b) Immunolocalization of antigens CD44, CD90, CD105, and STRO-1 revealed positive cells within the fascicles of the ACL, as well as the endothelium of the vasculature. Examples of such cells are denoted by white (fascicles) and black (endothelium) arrows, respectively, in 500×panels. This indicates that ACL-derived mesenchymal stem cells are likely to originate from both the bulk fascicle tissue as well as the small vessel walls of the ACL. Scale bars: 100×bar=200 μm; 200×bar=100 μm; 500×bar=40 μm. Color images available online at www.liebertonline.com/tea
Surprisingly, most of the fibroblastic cells, as well as cells associated with the blood vessels, stained positively for CD44, CD90, and CD105. Staining for STRO-1 was also positive, but less widespread (Fig. 7b). Examples of positively stained ACL fibroblasts within the matrix were labeled using white arrows, and near vasculature using black arrows, as shown in the 500×panels (Fig. 7b). We did not observe marked differences between torn and intact osteoarthritic ACLs, and representative sections of a 56-year-old male OA patient are shown (Fig. 7).
Thus, multipotent ACL cells appear to be common residents within the ligament, both as components of the ligament itself and associated with blood vessels. This indicates that they apparently do not arise from a separate, small, discrete pool of progenitors that are selected and amplified by the explant culture system.
Discussion
These data demonstrate unequivocally a population of multipotent mesenchymal progenitor cells within the human ACL. The recent identification of such progenitor cells by Cheng et al.25 that were isolated by collagenase digestion of anterior and posterior cruciate ligaments supports this conclusion. The present study, however, focuses on the repair relevant outgrowth ACL cell populations (ACLOUT) and offers a unique perspective on ACL progenitors by examining their origins within the tissue and by comparing them directly to ACL populations released by collagenase digestion (ACLDIG) and BMSCs from human bone marrow. Both ACL populations are very similar to human BMSCs, but differ in certain details. All cell types examined show the ability of colony formation (Fig. 1a) and revealed almost similar osteogenic (Fig. 4) and adipogenic (Fig. 5) capacities. However, the ACL populations show less cell proliferation (Fig. 1b) as well as chondrogenic capacity (Fig. 3), and do not express CD106 or CD146 as highly or STRO-1 as consistently, but they express higher levels of CD97 (Fig. 2 and Table 1). CD146 in particular was less expressed in the ACLOUT compared to the ACLDIG, although levels of significance were not reached (Fig. 2a). Expression of STRO-1 by MSCs declines with cell passage,35 which may explain the marginal staining for this epitope on those cells examined by FACS (Table 1). This would be consistent with the considerable staining for STRO-1 noted by immunocytochemistry, and immunohistochemistry, where less subculture is involved (Fig. 2). However, this study is limited to the examination of only a finite number of characteristic cell surface markers, and analyses of, for example, whole transcriptomes might be a suitable approach, to further resolve differences between the cell types.
Although cells with the general properties of MSCs have been recovered from a number of connective tissues, there are small phenotypic variations depending on the tissue source.14,16–24,26,36–39 The MSCs that are released from ACL tissues fit this pattern, but differ in subtle ways from the BMSCs used as a reference population. However, the three cell types could not be age- and patient-matched, so these variables could also account for the differences.
Progenitor cells were isolated from ligaments recovered 4–16 weeks after injury, at the time of surgical reconstruction. Given the evidence that MSCs migrate to sites of injury,40 there is the possibility that they are not normally present in the ACL. Indeed, a recent article by Morito et al.37 suggests that MSCs enter the synovial fluid after rupture of the ACL. The surface immunophenotype of the synovial fluid cells is similar to that reported here for ACL cells, except for a much higher expression of STRO-1.29 Despite the presence of these cells in the synovial fluid, our immunohistochemical findings argue against them being the source of the MSCs within the ACL. Cells positive for CD44, CD90, CD105, and STRO-1 were noted deep within the body of the ACL, closely aligned with the collagen fibers, as well as surrounding small blood vessels. The latter may be pericytes, long suspected of being a source of mesenchymal progenitors within vascularized tissues.39 The former, however, are morphologically indistinguishable from what are normally considered to be ligament fibroblasts, lying adjacent to the collagen bundles of this tissue. Further, we confirmed our results by using ACL outgrowth and digest populations from intact ACLs, derived from OA patients undergoing knee arthroplasty surgery, and noted an almost identical surface antigen expression and trilineage differentiation potential (not shown), compared to the data from cells recovered from torn ACLs (Figs. 1–5), supporting the concept of a tissue-inherent MSC population and confirming findings in the tendon.24
The capacity of self-renewal is also an important criterion of a stem cell that is met by the ACLOUT population as evidenced by an in vitro assay using BMP12 adenovirus and three-dimensional culture in a collagen hydrogel for ligamentogenic induction (Fig. 6). Using a similar approach, we were recently able to delineate the ligamentogenic potential of the ACLDIG and BMSC populations,34 which were used as comparative controls here (not shown). Our study, however, is limited to its in vitro nature, and only in vivo experiments can provide the necessary evidence for final validation of ACL regenerative approaches using inherent stem cells. The strong regenerative response of the BMP12 adenovirus, a strong ligamentogenic inductor of stem cells,41 in a rat model of Achilles tendon healing supports this concept.42
The results of our study differ in some ways to the data of Cheng and colleagues,43 who recently compared the cell proliferation and differentiation capacities of BMSCs to that of collagenase-digested ACL cells. In contrast to their study, where ACL cells revealed higher proliferation rates compared to BMSC,43 we found the BMSCs to elicit higher rates of cell proliferation compared to both ACLOUT and ACLDIG populations at all time points (Fig. 1b). Further, we found the BMSC to exhibit a higher chondrogenic potential and similar osteogenic potential compared to both ACL cells types (Figs. 3–5), whereas, in contrast, Cheng and colleagues found the BMSCs to have higher osteogenic capacity and similar chondrogenic capacity compared to ACL digest cells.43 We attribute the discrepancies in both studies to the different protocols that were used for cell isolation, maintenance, and differentiation as well as in differences in growth factor and FBS supplementation used to the respective cultures. However, the data in both studies correspond in that the adipogenic and ligamentogenic potential in all cell types examined were at comparable levels,43 undermining the relevance of ligament inherent stem cell populations and their implications over a possible use of this technology toward biological ACL repair. The fact that ACL outgrowth cells, the primary cell source that mediates any direct biological repair of the ACL,44 share these features further adds to this conclusion.
Putative stem/progenitor cells have recently been identified in similar locations of the tendon, suggesting that this may be a general phenomenon within fibrous tissues and several extracellular matrix components are important organizers of the stem cell niche.24 Not all fibroblasts within the ACL stained positively for MSC surface epitopes (Fig. 7), suggesting the presence of a mixed population of MSCs and ligament fibroblasts within the tissue. The heterogeneity of the cell population may be reflected in the nature of the cartilaginous pellets formed during chondrogenesis assays, where the inner core was much less cartilaginous than the outer component (Fig. 3a).
While the information gained in this study provided insight into the cellular characteristics of ACL outgrowth cell populations, the study is limited to its in vitro nature, and further in vivo work is required to determine its relevance to ligament repair including evaluation for structural and mechanical properties of the repair tissue.
The presence of MSCs within the ACL opens new opportunities for biological repair of this ligament. As noted above, biological repair has been largely ignored because of the belief that the ACL lacks regenerative capacity. Our data suggest, on the contrary, that the ACL houses a rich population of progenitors that are rapidly mobilized in response to injury. These cells migrate from the injured ligament, and can colonize a suitable, adjacent scaffold to synthesize repair tissue.9,10 Recent work by Cheng and colleagues revealed a vivid response of collagenase released ACL progenitors to several growth factors with respect to cell proliferation and differentiation.45 In animal models, the repair of tendons and ligaments can be enhanced by various morphogens and growth factors,38 such as BMP12,46 TGF-β,47 growth and differentiation factor (GDF)5,48 and platelet derived growth factor (PDGF).49 Gene transfer is an attractive technology for delivering these factors, especially if the vectors are associated with the matrix into which the ACL outgrowth cells migrate. Under these conditions, selective gene transfer to the ACL outgrowth cells occurs very efficiently in situ.50,51 This suggests one way in which these newly identified cells can be manipulated to improve healing of the ACL.
Acknowledgments
We are grateful to Nadja Karl, Viola Monz, Martina Regensburger, and Christa Amrehn for their excellent technical assistance. We thank Mr. Thomas Peter (Fraunhofer IGB, Stuttgart, Germany) for his expertise and help with the FACS analyses. Financial support was given by grants from BayFor (FORZEBRA TP1/WP1 to F.J., TP2/WP5 to A.F.S. and U.N.), BMBF (Grant No. 0313386E to U.N.), and NIH (RO1 AR052809 from NIH/NIAMS to C.H.E.; R01 AR054099 to M.M.M.; F32 EB005566 to R.M.P.).
Disclosure Statement
No competing financial interests exist.
References
- 1.The Burden of Musculoskeletal Diseases in the United States. Rosemont, IL: American Academy of Orthopaedic Surgeons; 2008. [Google Scholar]
- 2.Feagin J.A., Jr. Curl W.W. Isolated tear of the anterior cruciate ligament: 5-year follow-up study. Am J Sports Med. 1976;4:95. doi: 10.1177/036354657600400301. [DOI] [PubMed] [Google Scholar]
- 3.Lohmander L.S. Englund P.M. Dahl L.L. Roos E.M. The long-term consequence of anterior cruciate ligament and meniscus injuries: osteoarthritis. Am J Sports Med. 2007;35:1756. doi: 10.1177/0363546507307396. [DOI] [PubMed] [Google Scholar]
- 4.Ge Z. Yang F. Goh J.C. Ramakrishna S. Lee E.H. Biomaterials and scaffolds for ligament tissue engineering. J Biomed Mater Res A. 2006;77:639. doi: 10.1002/jbm.a.30578. [DOI] [PubMed] [Google Scholar]
- 5.West R.V. Harner C.D. Graft selection in anterior cruciate ligament reconstruction. J Am Acad Orthop Surg. 2005;13:197. doi: 10.5435/00124635-200505000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Murray M.M. Spector M. The migration of cells from the ruptured human anterior cruciate ligament into collagen-glycosaminoglycan regeneration templates in vitro. Biomaterials. 2001;22:2393. doi: 10.1016/s0142-9612(00)00426-9. [DOI] [PubMed] [Google Scholar]
- 7.Murray M.M. Bennett R. Zhang X. Spector M. Cell outgrowth from the human ACL in vitro: regional variation and response to TGF-beta1. J Orthop Res. 2002;20:875. doi: 10.1016/S0736-0266(01)00156-5. [DOI] [PubMed] [Google Scholar]
- 8.Murray M.M. Palmer M. Abreu E. Spindler K.P. Zurakowski D. Fleming B.C. Platelet-rich plasma alone is not sufficient to enhance suture repair of the ACL in skeletally immature animals: an in vivo study. J Orthop Res. 2009;27:639. doi: 10.1002/jor.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray M.M. Spindler K.P. Abreu E. Muller J.A. Nedder A. Kelly M. Frino J. Zurakowski D. Valenza M. Snyder B.D. Connolly S.A. Collagen-platelet rich plasma hydrogel enhances primary repair of the porcine anterior cruciate ligament. J Orthop Res. 2007;25:81. doi: 10.1002/jor.20282. [DOI] [PubMed] [Google Scholar]
- 10.Murray M.M. Spindler K.P. Devin C. Snyder B.S. Muller J. Takahashi M. Ballard P. Nanney L.B. Zurakowski D. Use of a collagen-platelet rich plasma scaffold to stimulate healing of a central defect in the canine ACL. J Orthop Res. 2006;24:820. doi: 10.1002/jor.20073. [DOI] [PubMed] [Google Scholar]
- 11.Pittenger M.F. Mackay A.M. Beck S.C. Jaiswal R.K. Douglas R. Mosca J.D. Moorman M.A. Simonetti D.W. Craig S. Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 12.Caplan A.I. Bruder S.P. Mesenchymal stem cells: building blocks for molecular medicine in the 21st century. Trends Mol Med. 2001;7:259. doi: 10.1016/s1471-4914(01)02016-0. [DOI] [PubMed] [Google Scholar]
- 13.Friedenstein A.J. Chailakhjan R.K. Lalykina K.S. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 14.Zuk P.A. Zhu M. Mizuno H. Huang J. Futrell J.W. Katz A.J. Benhaim P. Lorenz H.P. Hedrick M.H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 15.Jankowski R.J. Haluszczak C. Trucco M. Huard J. Flow cytometric characterization of myogenic cell populations obtained via the preplate technique: potential for rapid isolation of muscle-derived stem cells. Hum Gene Ther. 2001;12:619. doi: 10.1089/104303401300057306. [DOI] [PubMed] [Google Scholar]
- 16.Shi C. Zhu Y. Su Y. Cheng T. Stem cells and their applications in skin-cell therapy. Trends Biotechnol. 2006;24:48. doi: 10.1016/j.tibtech.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Noth U. Osyczka A.M. Tuli R. Hickok N.J. Danielson K.G. Tuan R.S. Multilineage mesenchymal differentiation potential of human trabecular bone-derived cells. J Orthop Res. 2002;20:1060. doi: 10.1016/S0736-0266(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 18.De Bari C. Dell'Accio F. Vanlauwe J. Eyckmans J. Khan I.M. Archer C.W. Jones E.A. McGonagle D. Mitsiadis T.A. Pitzalis C. Luyten F.P. Mesenchymal multipotency of adult human periosteal cells demonstrated by single-cell lineage analysis. Arthritis Rheum. 2006;54:1209. doi: 10.1002/art.21753. [DOI] [PubMed] [Google Scholar]
- 19.Sakaguchi Y. Sekiya I. Yagishita K. Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52:2521. doi: 10.1002/art.21212. [DOI] [PubMed] [Google Scholar]
- 20.Mauck R.L. Martinez-Diaz G.J. Yuan X. Tuan R.S. Regional multilineage differentiation potential of meniscal fibrochondrocytes: implications for meniscus repair. Anat Rec (Hoboken) 2007;290:48. doi: 10.1002/ar.20419. [DOI] [PubMed] [Google Scholar]
- 21.Dowthwaite G.P. Bishop J.C. Redman S.N. Khan I.M. Rooney P. Evans D.J. Haughton L. Bayram Z. Boyer S. Thomson B. Wolfe M.S. Archer C.W. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117:889. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 22.Hiraoka K. Grogan S. Olee T. Lotz M. Mesenchymal progenitor cells in adult human articular cartilage. Biorheology. 2006;43:447. [PubMed] [Google Scholar]
- 23.Risbud M.V. Guttapalli A. Tsai T.T. Lee J.Y. Danielson K.G. Vaccaro A.R. Albert T.J. Gazit Z. Gazit D. Shapiro I.M. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine. 2007;32:2537. doi: 10.1097/BRS.0b013e318158dea6. [DOI] [PubMed] [Google Scholar]
- 24.Bi Y. Ehirchiou D. Kilts T.M. Inkson C.A. Embree M.C. Sonoyama W. Li L. Leet A.I. Seo B.M. Zhang L. Shi S. Young M.F. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- 25.Cheng M.T. Yang H.W. Chen T.H. Lee O.K. Isolation and characterization of multipotent stem cells from human cruciate ligaments. Cell Prolif. 2009;42:448. doi: 10.1111/j.1365-2184.2009.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pountos I. Corscadden D. Emery P. Giannoudis P.V. Mesenchymal stem cell tissue engineering: techniques for isolation, expansion and application. Injury. 2007;38(Suppl 4):S23. doi: 10.1016/s0020-1383(08)70006-8. [DOI] [PubMed] [Google Scholar]
- 27.Evans C.H. Palmer G.D. Pascher A. Porter R. Kwong F.N. Gouze E. Gouze J.N. Liu F. Steinert A. Betz O. Betz V. Vrahas M. Ghivizzani S.C. Facilitated endogenous repair: making tissue engineering simple, practical, and economical. Tissue Eng. 2007;13:1987. doi: 10.1089/ten.2006.0302. [DOI] [PubMed] [Google Scholar]
- 28.Bonadio J. Smiley E. Patil P. Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5:733. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 29.Kolf C.M. Cho E. Tuan R.S. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solchaga L.A. Penick K. Porter J.D. Goldberg V.M. Caplan A.I. Welter J.F. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2005;203:398. doi: 10.1002/jcp.20238. [DOI] [PubMed] [Google Scholar]
- 31.Penick K.J. Solchaga L.A. Welter J.F. High-throughput aggregate culture system to assess the chondrogenic potential of mesenchymal stem cells. Biotechniques. 2005;39:687. doi: 10.2144/000112009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S.C. Szybalski W. Amplification of cloned DNA as tandem multimers using BspMI-generated asymmetric cohesive ends. Gene. 1988;71:1. doi: 10.1016/0378-1119(88)90071-6. [DOI] [PubMed] [Google Scholar]
- 33.Farndale R.W. Buttle D.J. Barrett A.J. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 34.Haddad-Weber M. Prager P. Kunz M. Seefried L. Jakob F. Murray M.M. Evans C.H. Noth U. Steinert A.F. BMP12 and BMP13 gene transfer induce ligamentogenic differentiation in mesenchymal progenitor and anterior cruciate ligament cells. Cytotherapy. 2010;12:505. doi: 10.3109/14653241003709652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gronthos S. Zannettino A.C. Hay S.J. Shi S. Graves S.E. Kortesidis A. Simmons P.J. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci. 2003;116:1827. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- 36.Lucas P.A. Calcutt A.F. Southerland S.S. Alan Wilson J. Harvey R.L. Warejcka D. Young H.E. A population of cells resident within embryonic and newborn rat skeletal muscle is capable of differentiating into multiple mesodermal phenotypes. Wound Repair Regen. 1995;3:449. doi: 10.1046/j.1524-475X.1995.30409.x. [DOI] [PubMed] [Google Scholar]
- 37.Morito T. Muneta T. Hara K. Ju Y.J. Mochizuki T. Makino H. Umezawa A. Sekiya I. Synovial fluid-derived mesenchymal stem cells increase after intra-articular ligament injury in humans. Rheumatology (Oxford) 2008;47:1137. doi: 10.1093/rheumatology/ken114. [DOI] [PubMed] [Google Scholar]
- 38.Evans C.H. Cytokines and the role they play in the healing of ligaments and tendons. Sports Med. 1999;28:71. doi: 10.2165/00007256-199928020-00001. [DOI] [PubMed] [Google Scholar]
- 39.Diaz-Flores L. Gutierrez R. Varela H. Rancel N. Valladares F. Microvascular pericytes: a review of their morphological and functional characteristics. Histol Histopathol. 1991;6:269. [PubMed] [Google Scholar]
- 40.Devine M.J. Mierisch C.M. Jang E. Anderson P.C. Balian G. Transplanted bone marrow cells localize to fracture callus in a mouse model. J Orthop Res. 2002;20:1232. doi: 10.1016/S0736-0266(02)00051-7. [DOI] [PubMed] [Google Scholar]
- 41.Wolfman N.M. Hattersley G. Cox K. Celeste A.J. Nelson R. Yamaji N. Dube J.L. DiBlasio-Smith E. Nove J. Song J.J. Wozney J.M. Rosen V. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-beta gene family. J Clin Invest. 1997;100:321. doi: 10.1172/JCI119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majewski M. Betz O. Ochsner P.E. Liu F. Porter R.M. Evans C.H. Ex vivo adenoviral transfer of bone morphogenetic protein 12 (BMP-12) cDNA improves Achilles tendon healing in a rat model. Gene Ther. 2008;15:1139. doi: 10.1038/gt.2008.48. [DOI] [PubMed] [Google Scholar]
- 43.Cheng M.T. Liu C.L. Chen T.H. Lee O.K. Comparison of potentials between stem cells isolated from human anterior cruciate ligament and bone marrow for ligament tissue engineering. Tissue Eng Part A. 2010;16:2237. doi: 10.1089/ten.TEA.2009.0664. [DOI] [PubMed] [Google Scholar]
- 44.Murray M.M. Current status and potential of primary ACL repair. Clin Sports Med. 2009;28:51. doi: 10.1016/j.csm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng M.T. Yang H.W. Chen T.H. Lee O.K. Modulation of proliferation and differentiation of human anterior cruciate ligament-derived stem cells by different growth factors. Tissue Eng Part A. 2009;15:3979. doi: 10.1089/ten.TEA.2009.0172. [DOI] [PubMed] [Google Scholar]
- 46.Lou J. Tu Y. Burns M. Silva M.J. Manske P. BMP12 gene transfer augmentation of lacerated tendon repair. J Orthop Res. 2001;19:1199. doi: 10.1016/S0736-0266(01)00042-0. [DOI] [PubMed] [Google Scholar]
- 47.Kashiwagi K. Mochizuki Y. Yasunaga Y. Ishida O. Deie M. Ochi M. Effects of transforming growth factor-beta 1 on the early stages of healing of the Achilles tendon in a rat model. Scand J Plast Reconstr Surg Hand Surg. 2004;38:193. doi: 10.1080/02844310410029110. [DOI] [PubMed] [Google Scholar]
- 48.Rickert M. Wang H. Wieloch P. Lorenz H. Steck E. Sabo D. Richter W. Adenovirus-mediated gene transfer of growth and differentiation factor-5 into tenocytes and the healing rat Achilles tendon. Connect Tissue Res. 2005;46:175. doi: 10.1080/03008200500237120. [DOI] [PubMed] [Google Scholar]
- 49.Wang X.T. Liu P.Y. Tang J.B. Tendon healing in vitro: genetic modification of tenocytes with exogenous PDGF gene and promotion of collagen gene expression. J Hand Surg [Am] 2004;29:884. doi: 10.1016/j.jhsa.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Steinert A.F. Weber M. Kunz M. Palmer G.D. Noth U. Evans C.H. Murray M.M. In situ IGF-1 gene delivery to cells emerging from the injured anterior cruciate ligament. Biomaterials. 2008;29:904. doi: 10.1016/j.biomaterials.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 51.Pascher A. Steinert A.F. Palmer G.D. Betz O. Gouze J.N. Gouze E. Pilapil C. Ghivizzani S.C. Evans C.H. Murray M.M. Enhanced repair of the anterior cruciate ligament by in situ gene transfer: evaluation in an in vitro model. Mol Ther. 2004;10:327. doi: 10.1016/j.ymthe.2004.03.012. [DOI] [PubMed] [Google Scholar]