Although recent advances in non-viral gene delivery vectors have greatly improved delivery and expression, effective tumor targeting of these vectors still poses a significant challenge for systemic cancer treatment. Tumor cells have been know to upregulate many cell surface receptors such as growth factor and adhesion molecule (integrin) receptors, which have been of interest for targeting [1]. Thus, the general approach to target tumors systemically is to introduce ligands that bind the upregulated cell surface receptors to the surface of non-viral vectors. These ligands include small molecules (e.g. folate and galactose), proteins (e.g. transferrin and antibodies) [2] as well as Arg-Gly-Asp (RGD) peptides [3, 4]. Direct conjugation of the αvß3 ligand, RGD, to DNA/poly(ethylene imine) (PEI) polyplexes has been show to increases the transfection efficiency in vitro and in vivo [3-5]. However, these approaches have not been able to differentiate targeting between tumors that express high levels of αvß3 over tumors that express medium levels of αvß3 (or over cell types that express low amounts of αvß3). The ability to preferentially target non-viral vectors towards tumors that highly express αvß3 integrin receptors would increase the applicability of non-viral vectors in cancer treatment by targeting metastatic tumors and the tumor vasculature.
We hypothesized that the introduction of clustered RGD ligands on the surface of non-viral vectors would increase targetability towards cells with high αvß3 integrin receptors. We recently reported that by introducing clustered RGD ligands on the surface of DNA/PEI polyplexes, in vitro transgene expression was enhanced, with the enhancement being larger for cells that contained higher levels of αvß3 integrin receptors[6]. The enhancement was hypothesized to be due to the increased avidity of the clustered RGDs over unclustered RGDs. However, enhanced targeting could not be tested in vitro. Previous reports have shown that the introduction of clustered RGD ligands to small molecular drugs [7] or labeling agents [8-10] increases targeting to tumor in vivo and demonstrated increased effects over monovalent binding [11-14].
Herein, we demonstrate the use of a clustered integrin binding approach to enhance the targeting of gene delivery vectors toward tumors that highly express αvß3 integrin receptors. Most cell types express low or medium levels of integrin receptors, while many metastatic cancers and the tumor vasculature over express these receptors. The ability to target high integrin expressing cells is beneficial since therapeutics would ideally be targeted to only these cells. In our approach, gold nanoparticles were modified with the integrin binding peptide sequence RGD to form RGD nanoclusters as previously described [6] that were subsequently used to modify the surface of DNA/PEI polyplexes (abbreviated as DNA/PEI-Au) (Figure 1). Thus, the resulting RGD nanocluster modified polyplexes target cells through RGD peptide clusters rather than individual RGD peptides. A mouse xenograph model was used in which subcutaneous tumors were formed in SCID mice above the left and right shoulders by injection of HeLa (medium αvß3) and U87MG (high αvß3) cells. Flow cytometry and western blotting show the difference in expression level of αvß3 integrin expression of these two cell lines (Supporting Figure S1). Studies examining integrin expression of in vitro cell lines have found only slight differences when they are later cultured in vivo[15]. A high and medium integrin expressing cell line was selected rather than a low expression or knockout of αvß3 to be able to study how targeting by clustered ligands is affected by the number of receptors at the cell surface.
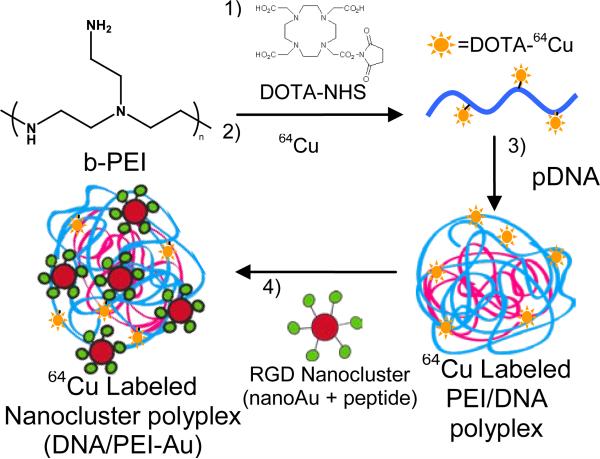
Figure 1.
64Cu labeled RGD nanocluster modified DNA/PEI polyplexes (DNA/PEI-Au). DOTA conjugated PEI is formed by reaction with NHS-DOTA (1). Radiolabeling of PEI is achieved by chelating 64Cu chloride to DOTA conjugated PEI (2). 64Cu labeled PEI is mixed with plasmid DNA allowing for polyplex formation (3). RGD nanoclusters are conjugated to the 64Cu labeled DNA/PEI polyplexes via UV activated crosslinker [6](4).
Positron emission tomography (PET) was used to visualize and quantify the in vivo biodistribution of PEI conjugated with 64Cu and RGD nanoclusters used to form the modified polyplexes. Prior to polyplex formation, DOTA-NHS was conjugated to branched PEI to function as a chelator for the positron emitting metal ion. The metal ion, 64Cu chloride, was mixed with the DOTA modified PEI prior to polyplex formation and RGD nanoparticle conjugation. 64Cu labeled DNA/PEI polyplexes with either none, low (0.1 pmol/μg DNA), or high (0.2 pmol/μg DNA) amounts of RGD nanoclusters (DNA/PEI-Au) were injected into the tail vein of anesthetized SCID mice. Micro-PET/CT imaging of the mice was performed and dynamic scans were taken over a period of 1 hr after injection and static scans after 20 hrs after injection. Extensive modeling of 64Cu-DOTA kinetics has recently been studied and the extended time point of 20 hrs is commonly used in PET studies involving 64Cu-DOTA[16, 17].
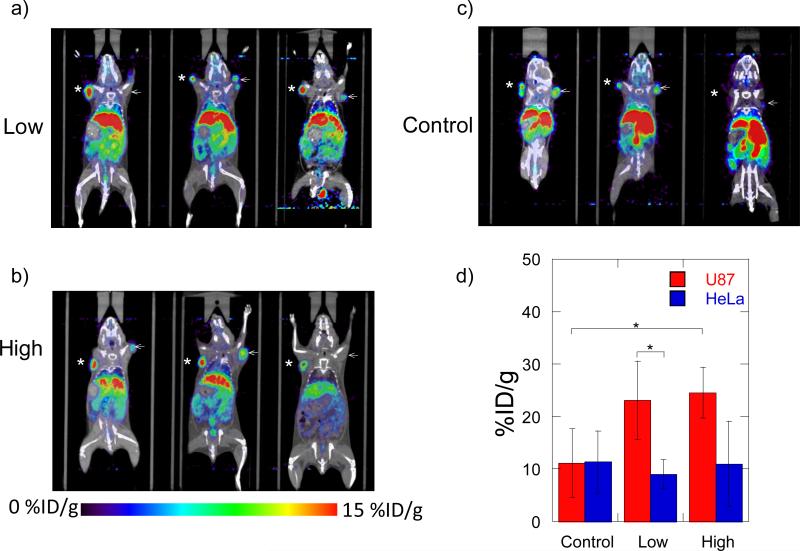
The Micro-PET/CT studies show enhanced targeting to the tumor with high αvß3 expression (U87MG) cells. The differences in accumulation in the tumors for all 3 conditions can be observed in the static scans after 20 hrs (Figure 2). In both cases, all conditions containing the RGD nanocluster modified polyplexes (high and low) have increased accumulation in the high αvß3 expression (U87MG) cells compared to the medium αvß3 expression (HeLa) cells (Figure 2a, b, d). The accumulation in the U87MG tumor varies from a 40 to 470% increase (3.8-22% of ID/g) compared to the HeLa tumor. The controls which consist of the polyplexes without the nanoclusters also show a varying amount of accumulation (4-15% of ID/g) (Figure 2a) although the accumulations in the controls show no statistical significance between the two tumors (Figure 2d). The nonspecific accumulation of the polyplexes in the tumors is expected due to the enhanced permeability and retention effect (EPR) of the tumor vasculature [18, 19]. The similar polyplex accumulations in both U87MG and HeLa tumors suggest a similar tumor vascularity.
Figure 2.
MicroPET/CT static biodistribution of RGD nanocluster modified (high and low) and unmodified polyplexes in individual SCID mice with U87 and HeLa tumors 20 hrs post injection. a) Images with low RGD nanocluster modified polyplexes b) Images with high RGD nanocluster modified polyplexes c) Images with unmodified polyplexes. d) Comparison of accumulations in U87 and HeLa tumors. (*=U87, ← = HeLa). The numbers correspond to the average of all animals used (n = 3). (*P<0.05, using a two tailed t-test)
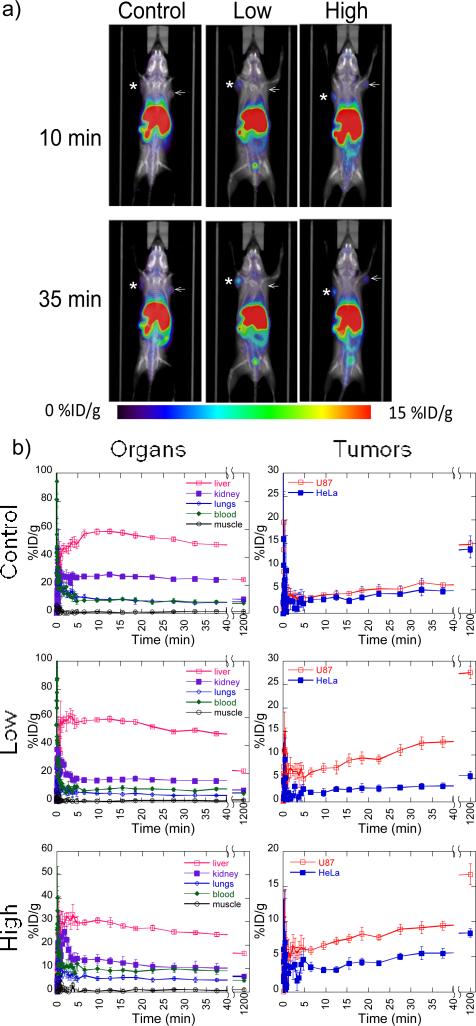
Dynamic biodistribution scans reveal that the accumulation of the RGD nanocluster modified polyplexes in the U87MG tumor increases gradually over time while accumulations in the HeLa tumors remain roughly the same within 40 min (Figure 3). A large increase in accumulation of the modified polyplexes is observed between 40 min and 20 hrs with the differences in the U87MG and HeLa tumor increasing ~2-fold for both low and high RGD nanocluster conditions (Figure 3b). The biodistribution of polyplexes in the examined organs show similar trends for all 3 conditions. A low accumulation is seen (<10% ID/g) in the lungs, blood, and muscle and high accumulation in the liver (30-60% ID/g) and kidney (15-30% ID/g) is seen within 40 min (Figure 3b). Many systemically delivered nanoparticles are known to accumulate in the reticuloendothelial system or are cleared through renal uptake depending on the size and properties of the particles [20, 21]. The biodistribution of the polyplexes in this study show similar high accumulations in the liver and kidneys in studies using unshielded PEI polyplexes in vivo[22]. Although rapid clearance through the “first pass” organs (lung, liver, spleen) is common for systemically delivered particles, others have reduced the accumulation and increased circulation time by PEGylation of the PEI polyplexes[20, 21]. At 20 hrs, the levels drop significantly in the liver (~20% ID/g) to the accumulation we seen in the tumors and kidney (<10% ID/g) to the other organs (Figure 3b), which indicates that a large amount of the particles have been cleared from the system [23]. Since 64Cu is not the optimal metal ion for binding with DOTA, the high accumulation in the liver could also be due to free 64Cu. However, 64Cu-DOTA binding has been shown to be stable under physiological condition in which 64Cu labeled antibodies only lose 0.3% per day in serum stability studies[24]. Since testing of gene expression from the polyplexes was not conducted for the tumors, we are unable to determine if the accumulation in the tumors was due to intact polyplexes or free PEI due to decomplexation after opsinization. Although successful gene delivery to the tumors was not shown, the specific accumulation to high αvß3expression cells of the modified vector backbone shows the potential targeting applications for other non-viral gene delivery nanoparticles by clustered integrin ligands.
Figure 3.
MicroPET/CT dynamic biodistribution of RGD nanocluster modified (high and low) and unmodified polyplexes in SCID mice with U87MG and HeLa tumors. a) MicroPET/CT images of RGD nanocluster modified and unmodified (control) DNA/PEI polyplexes at 10 min and 35 min. b) Graphs of accumulation in organs (left) and tumors (right) for RGD nanocluster modified and unmodified (control) DNA/PEI polyplexes. (*=U87MG, ← = HeLa)
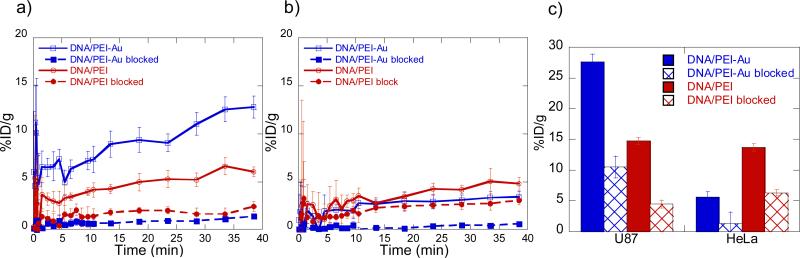
The increases in accumulation of the RGD modified polyplexes in the high αvß3 expressing tumor over time coupled with the decrease in other organs suggests a preferential targeting due to the RGD nanoclusters. To ensure that the enhanced tumor targeting to the high αvß3 expressing tumor is due to the RGD nanoclusters binding to the integrin receptors, competitive binding using free RGD was conducted. Prior to injection of the modified polyplexes, 150 μg of RGD peptide was injected through tail vein injection. In addition, 100 μg of peptide was injected along with the modified polyplexes to further block binding. Dynamic biodistribution scans of the competitive binding after 40 min show that blocking with free RGD peptide greatly reduces (<2% ID/g) accumulation of both modified and unmodified polyplexes in the U87MG tum o r (Figure 4a). Accumulations in the HeLa tumors remain low (<5% ID/g) as expected although the competition with free RGD shows to decreases the overall accumulation (Figure 4b). Comparing the accumulations after 20 hrs (Figure 4c); we can see that both unblocked and RGD blocked for unmodified polyplexes are similar for both U87MG and HeLa tumors but is greatly reduced as seen in the dynamic scans. For the modified polyplexes, accumulations in the U87MG tumor are higher for both unblocked and RGD blocked compared to the HeLa tumor although accumulations are also greatly reduced for the RGD blocked case. It is shown that the blocking of the modified polyplexes in the U87MG tumor decreases the accumulation to below what is seen for the unmodified polyplexes when not blocked. This data suggests that free RGD peptide causes the blocking of the modified polyplexes to the high αvß3 expressing tumor but also causes the blocking of both modified and unmodified PEI/DNA polyplexes. This is consistent with reports that adhesion receptors play a role in mediating binding and internalization of cationic non-viral vectors and it has been shown that blocking integrin binding can reduce lipoplex binding up to 50% [25]. Nevertheless, the blocking observed for the RGD clusters was much higher than for untargeted particles demonstrating that the enhanced targeting occurs through RGD receptors. Additional studies were conducted using RGD directly conjugated to PEI as an unclustered comparison instead of the RGD nanoclusters prior to forming PEI/DNA polyplexes. Using the same charge ratios and RGD concentrations, we encountered toxicity issues as seen in by others for DNA/PEI polyplexes[26] and were unable to continue the experiments to compare the two studies. In conclusion, although this method could not conclude that intact polyplexes were localized to the tumors since the DNA was not labeled, clustered integrin binding in association with non-viral vectors showed improved targeting of the vector towards tumors with high levels of αvß3 integrin expression over tumors with low αvß3 integrin expression. The increase in targeting was observed after 40 minutes and 20 hours and was observed with PET. We believe that the use of clustered ligands will result in more specific targeting of other receptors. Further, the approach presented here for ligand clustering could be applied easily to other peptide ligands and be modified for larger ligands. These findings can provide a basis for future research in the use of ligand clustering for improved targeting of other non-viral vectors and nanoparticles for drug delivery.
Figure 4.
MicroPET/CT accumulation data of RGD nanocluster modified (DNA/PEI-Au) and unmodifiedcontrol (DNA/PEI)polyplexes with and without blocking using free RGD peptide. Graphs of accumulation over 40 min in a) U87 and b) HeLa tumors. c) Graphs of accumulation after 20 hrs in the tumors.
Experimental Section
DOTA-PEI conjugation
All solutions used were treated with 1.2 mg/mL Chelex-100 (Biorad, Hercules, CA) and filtered using 0.22μm filters prior to use. A solution of branched PEI (25 kDa, Sigma, St. Louis, MO) in phosphate buffer (pH 7.4) was reacted with a 10 fold molar excess of 1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetraacetic acid mono(N-hydroxysuccinimide ester) (DOTA-NHS, Macrocyclics, Dallas, TX) dissolved in DMSO. The solution was reacted with shaking for 1 hr at room temperature. Unreacted DOTA-NHS was removed by dialysis using a 8k MWCO dialysis tubing (Fisher, Pittsburgh, PA) in water. The solution was then lyophilized and stored at -20°C.
64Cu chelation with DOTA-PEI
All solutions used were treated with 1.2 mg/mL Chelex-100 (Biorad, Hercules, CA) and filtered using 0.22μm filters prior to use. 64Cu (10-15 mCi) was diluted with 0.1M ammonium acetate (pH 5.5) to a final volume of 100 μL. Lyophilized DOTA-PEI was resuspended to 10 mg/mL in 0.1M ammonium acetate (pH 5.5). DOTA-PEI solution was mixed with 64Cu solution in a 800 fold molar excess and incubated at 60°C for 1 hr. 64Cu labeled PEI was filtered through Centricon YM3 filters at 14,000g for 15 min and washed with milliQ water. Labelling efficiency was measured before and after labelling by measuring the radioactivity in the filter, the filtrate and the retentate and found to be >90% efficient.
RGD nanocluster modified polyplexes (DNA/PEI-Au)
RGD nanoclusters were synthesized as previously reported[6]. Briefly, the peptides CCVVVT-COOH (Cap) and Ac-CCVVVTGRGDSP-SSK-COOH (Cap-RGD) were used to modify the surface of citrate stabilized gold nanoparticles. The resulting RGD nanoclusters were immobilized to the surface of the DNA/PEI polyplex via a UV-activated heterobifunctional crosslinker (NHS-ASA) bound to the Cap-RGD peptide (Cap-RGD-ASA). DNA/PEI polyplexes were formed by mixing equal volumes of plasmid DNA with 64Cu-PEI to get an N/P (nitrogen to phosphate ratio) of 10 in 5% glucose solution in water. PEI was added to the DNA solution, vortexed for 10 seconds, and incubated at room temperature for 15 minutes. Specified concentrations of RGD nanoclusters (16 (low) or 32 (high) x1010 particles/μg DNA) were added to the DNA/PEI polyplexes and exposed to ambient light for 15 minutes.
Micro-PET/CT imaging
SCID mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal manipulations were performed with sterile technique and were approved by the University of California at Los Angeles Animal Research Committee. A mouse xenograph model following the B16 mouse melanoma model was used in which subcutaneous tumors were formed in SCID mice above the left and right shoulders by injection of 1×106 HeLa (medium αvß3) and U87MG (high αvß3) cells in matrigel. Tumors were allowed to grow to ~1 cm3 over 2 weeks. Imaging was performed with a micro-PET FOCUS 220 PET scanner (Siemens, Malvern, PA) and a MicroCAT II CT scanner (Siemens). Fifteen minutes before imaging, mice were anesthetized by using 1.5–2% isoflurane in a heated (30°C) induction chamber, then transferred to a heated isolation/imaging chamber. 64Cu labeled DNA/PEI-Au polyplexes (50 μg DNA in 175 μL) in 5% glucose with either none, high, or low amounts of RGD nanoclusters were injected into the tail vein into anesthetized SCID mice while the anesthetized animals were positioned on the scanner bed. Dynamic micro-PET scans of the mice were taken over a period of 40 min. Immediately after the micro-PET scan, mice were moved to the micro-CT in the same isolation/imaging chamber and underwent a 7-min micro-CT scan, using routine image acquisition parameters. The micro-CT scan was used for anatomical localization of the tissue concentrations of the 64Cu over time by micro-PET. Static micro-PET scans were acquired on the following day (20hrs after injection) with another micro-CT scan for anatomical co-registration. PET images were reconstructed by filtered back projection. To determine temporal changes of tracer concentration in various tissues, ellipsoid regions of interest were placed in the region that exhibited the highest 64Cu activity as determined by visual inspection. To ensure accurate anatomical positioning, regions of interest were placed on fused micro-PET/CT images generated by the AMIDE software. To minimize partial volume effects, care was taken not to include the anatomical borders of the organs. Considering the size of the studied organs and tumors and the spatial resolution of the PET scanner, partial volume effects are not expected to have a major impact on the results of quantitative analysis. Activity concentrations are expressed as percent of the decay-corrected injected activity per cm3 of tissue (can be approximated as percentage ID/g), using the AMIDE software, and these values are normalized to an elliptic cylinder region of interest drawn over the entire mouse.
Supplementary Material
Supplementary Information
Methods:
Integrin quantification with flow cytometry and western blotting:U87MG and HeLa surface αvβ3 integrin expression was analyzed by flow cytometry and Western blot. Cells were harvested and resuspended in FACS buffer (Dulbecco's phosphate-buffered saline with 2% BSA) and stained with the anti-integrin αvβ3 monoclonal antibody (MAB1976, Chemicon International, Temecula, CA). Samples were read on a FACScan Analytic Flow Cytometer (Becton Dickinson). Cell lysates were assessed using αv Antibody (CD51) (611012 BD Biosciences, San Jose, CA) and Integrin β3 Antibody (4702, Cell Signaling Technology, Beverly, MA). Blotting was performed according to the manufacturer's instructions and imaged with the LI-COR Odyssey Imager using IR dye conjugated secondary antibodies.
Figure S1: Integrin characterization of tumor cell lines used for in vivo assay. a) FACS of αvβ3 integrin expression in U87 and HeLa cells b) Protein quantification using western blotting for integrins (αv and β3) and actin.
Figure S2: Dynamic biodistribution of DNA/PEI or DNA/PEI-Au polyplexes in SCID mice with U87 and HeLa tumors. PET/CT images (left) and signals in organs/tumors (right). a) DNA/PEI (control), b) DNA/PEI-Au (low), c) DNA/PEI-Au (high), (*=U87, ← = HeLa, Z in mm)
S3: Static 3D reconstruction of MicoPET/CT biodistribution for RGD nanocluster modified DNA/PEI (low)
S4: Static 3D reconstruction of MicoPET/CT biodistribution for unmodified (control) DNA/PEI
S5: Dynamic 3D reconstruction of MicoPET/CT biodistribution for RGD nanocluster modified DNA/PEI (low)
Acknowledgements
Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
Footnotes
Supporting information for this article is available on the WWW under http://www.chemmedchem.org or from the author.
References
- 1.Hofmeister V, Schrama D, Becker J. Cancer Immunol. Immunother. 2008;57(1):1–17. doi: 10.1007/s00262-007-0365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis ME. Curr. Opin. Biotechnol. 2002;13(2):128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 3.Erbacher P, Remy JS, Behr JP. Gene Ther. 1999;6(1):138–145. doi: 10.1038/sj.gt.3300783. [DOI] [PubMed] [Google Scholar]
- 4.Kunath K, Merdan T, Hegener O, Haberlein H, Kissel T. J. Gene Med. 2003;5(7):588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- 5.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Nucleic Acids Res. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng QKT, Sutton MK, Soonsawad P, Xing L, Cheng H, Segura T. Mol. Ther. 2009;17(5):828–836. doi: 10.1038/mt.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiffelers RM, Koning GA, ten Hagen TLM, Fens MHAM, Schraa AJ, Janssen APCA, Kok RJ, Molema G, Storm G. J. Controlled Release. 2003;91(1-2):115–122. doi: 10.1016/s0168-3659(03)00240-2. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy JR, Weissleder R. Adv Drug Delivery Rev. 2008;60(11):1241–1251. doi: 10.1016/j.addr.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almutairi A, Rossin R, Shokeen M, Hagooly A, Ananth A, Capoccia B, Guillaudeu S, Abendschein D, Anderson CJ, Welch MJ, Fréchet JMJ. Proceedings of the National Academy of Sciences. 2009;106(3):685–690. doi: 10.1073/pnas.0811757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Zhang X, Xiong Z, Cheng Z, Fisher DR, Liu S, Gambhir SS, Chen X. J. Nucl. Med. 2005;46(10):1707–1718. [PubMed] [Google Scholar]
- 11.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. J. Med. Chem. 2006;49(20):6087–6093. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 12.Dijkgraaf I, Kruijtzer J, Liu S, Soede A, Oyen W, Corstens F, Liskamp R, Boerman O. Eur. J. Nucl. Med. Mol. Imaging. 2007;34(2):267–273. doi: 10.1007/s00259-006-0180-9. [DOI] [PubMed] [Google Scholar]
- 13.Kiessling LL, Gestwicki JE, Strong LE. Angew. Chem. 2006;45(15):2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kok RJ, Schraa AJ, Bos EJ, Moorlag HE, Asgeirsdottir SA, Everts M, Meijer DKF, Molema G. Bioconjug. Chem. 2002;13(1):128–135. doi: 10.1021/bc015561+. [DOI] [PubMed] [Google Scholar]
- 15.Mette S, Pilewski J, Buck C, Albelda S. Am. J. Respir. Cell Mol. Biol. 1993;8(5):562. doi: 10.1165/ajrcmb/8.5.562. [DOI] [PubMed] [Google Scholar]
- 16.Ferl GZ, Dumont RA, Hildebrandt IJ, Armijo A, Haubner R, Reischl G, Su H, Weber WA, Huang S-C. J. Nucl. Med. 2009;50(2):250–258. doi: 10.2967/jnumed.108.054049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z.-b., Cai W, Cao Q, Chen K, Wu Z, He L, Chen X. J. Nucl. Med. 2007;48(7):1162–1171. doi: 10.2967/jnumed.107.039859. [DOI] [PubMed] [Google Scholar]
- 18.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J. Controlled Release. 2000;65(1-2):271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 19.Ogris M, Wagner E. Drug Discov. Today. 2002;7(8):479–485. doi: 10.1016/s1359-6446(02)02243-2. [DOI] [PubMed] [Google Scholar]
- 20.Soo Choi H, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Nat Biotech. 2007;25(10):1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brigger I, Dubernet C, Couvreur P. Adv Drug Delivery Rev. 2002;54(5):631–651. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 22.Merdan T, Kunath K, Petersen H, Bakowsky U, Voigt KH, Kopecek J, Kissel T. Bioconjug. Chem. 2005;16(4):785–792. doi: 10.1021/bc049743q. [DOI] [PubMed] [Google Scholar]
- 23.Walker GF, Fella C, Pelisek J, Fahrmeir J, Boeckle S, Ogris M, Wagner E. Mol. Ther. 2005;11(3):418–425. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Meares CF. Bioconjug. Chem. 1993;4(4):275–283. doi: 10.1021/bc00022a005. [DOI] [PubMed] [Google Scholar]
- 25.Zuhorn IS, Kalicharan D, Robillard GT, Hoekstra D. Mol. Ther. 2007;15(5):946–953. doi: 10.1038/mt.sj.6300139. [DOI] [PubMed] [Google Scholar]
- 26.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. Gene Ther. 1999;6(4):595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Methods:
Integrin quantification with flow cytometry and western blotting:U87MG and HeLa surface αvβ3 integrin expression was analyzed by flow cytometry and Western blot. Cells were harvested and resuspended in FACS buffer (Dulbecco's phosphate-buffered saline with 2% BSA) and stained with the anti-integrin αvβ3 monoclonal antibody (MAB1976, Chemicon International, Temecula, CA). Samples were read on a FACScan Analytic Flow Cytometer (Becton Dickinson). Cell lysates were assessed using αv Antibody (CD51) (611012 BD Biosciences, San Jose, CA) and Integrin β3 Antibody (4702, Cell Signaling Technology, Beverly, MA). Blotting was performed according to the manufacturer's instructions and imaged with the LI-COR Odyssey Imager using IR dye conjugated secondary antibodies.
Figure S1: Integrin characterization of tumor cell lines used for in vivo assay. a) FACS of αvβ3 integrin expression in U87 and HeLa cells b) Protein quantification using western blotting for integrins (αv and β3) and actin.
Figure S2: Dynamic biodistribution of DNA/PEI or DNA/PEI-Au polyplexes in SCID mice with U87 and HeLa tumors. PET/CT images (left) and signals in organs/tumors (right). a) DNA/PEI (control), b) DNA/PEI-Au (low), c) DNA/PEI-Au (high), (*=U87, ← = HeLa, Z in mm)
S3: Static 3D reconstruction of MicoPET/CT biodistribution for RGD nanocluster modified DNA/PEI (low)
S4: Static 3D reconstruction of MicoPET/CT biodistribution for unmodified (control) DNA/PEI
S5: Dynamic 3D reconstruction of MicoPET/CT biodistribution for RGD nanocluster modified DNA/PEI (low)