Abstract
Exact calculations of model posterior probabilities or related quantities are often infeasible due to the analytical intractability of predictive densities. Here new approximations to obtain predictive densities are proposed and contrasted with those based on the Laplace method. Our theory and a numerical study indicate that the proposed methods are easy to implement, computationally efficient, and accurate over a wide range of hyperparameters. In the context of GLMs, we show that they can be employed to facilitate the posterior computation under three general classes of informative priors on regression coefficients. A real example is provided to demonstrate the feasibility and usefulness of the proposed methods in a fully Bayes variable selection procedure.
Keywords: Laplace Approximation, GLM, Normal Prior, Power Prior, Conjugate Prior, Asymptotic Normality, Logistic Regression
1 Introduction
Bayesian applications in a number of problems need to be able to evaluate marginal probability distributions of the data, often called predictive densities, or their ratios known as Bayes factors, for a set of competing models, which are often analytically intractable. Calculations of such quantities have been addressed by several authors, including sampling or Monte Carlo methods (e.g., Gelfand and Smith 1990, Verdinelli and Wasserman 1995, Han and Carlin 2001) and analytic approximations based on the Laplace method (e.g., Tierney and Kadane 1986, Tierney et al. 1989, Gelfand and Dey 1994, Raftery 1996, Boone et al. 2005). The first part of this paper presents new analytical methods to approximate the predictive densities, where we also discuss and compare their theoretical properties with those of Laplace approximations.
The proposed methods are then applied to Bayesian analysis of Generalized Linear Models (GLMs). Under a hierarchical Bayes setup for model uncertainty, we show that our proposed methods, when approximating predictive densities, are applicable to three general classes of informative priors, namely the normal prior (Dellaportas and Smith 1993, Meyer and Laud 2002), the conjugate prior (Diaconis and Ylvisaker 1979, Meyer and Laud 2002, Chen et al. 2008), and the power prior (Zellner 1988, Chen et al. 2000), for regression coefficients. The proposed approximations can also greatly facilitate the posterior computation. Raftery (1996) presented asymptotic analytics to approximate Bayes factors and accounted for model uncertainty in GLMs. He assumed independent normal priors on regression coefficients, which is a special case of our approach. Wang and George (2007) proposed several closed-form Bayesian selection criteria for GLMs, using normal priors with empirical covariance structures on regression coefficients. An integrated Laplace method was used to achieve the analytical tractability, which is a special case of our proposed methods; the classes of the conjugate and power priors were not discussed there. Chen et al. (1999) and Ibrahim et al. (2000) concentrated on variable selection for logistic and Poisson regression models, respectively. Their computation of model posteriors was purely sample based without using any analytic approximations. What distinguishes our work from these papers are the generality of our approach and the novelty of our analytical approximations.
The remainder of the paper is organized as follows. In Section 2 we introduce new analytic methods for approximating predictive densities. Section 3 describes their applications in GLMs. Section 4 presents two examples, one providing a simulation evaluation and comparison of various approximations, and the other using a real dataset with 19 predictors and a binary response variable to illustrate the approximations in a Fully Bayes variable selection procedure. Section 5 concludes the paper with discussions.
2 Methods for Approximating Predictive Densities
We consider the problem of approximating a predictive density of the form
| (1) |
where β is a parameter vector whose domain Ω is Rm, Ln(β) is a likelihood function based on n observations, and π (β) is a prior density on β. In this paper we always assume that I exists and is finite.
2.1 The Case of Normal Priors
We begin with the case where π(β) is a normal prior density with a mean vector β0 and a covariance matrix λΣ0, λ > 0. The matrix Σ0 reflects relative uncertainties of all parameters in β and the correlations among them. It can be specified a priori or empirically. After Σ0 is chosen, the hyperparameter λ quantifies the strength of prior information relative to the data. When λ = 0, π reduces to a point mass at β0. When λ gets large, π approaches a flat prior for β.
Theorem 2.1
Suppose π(β) ~ N(β0, λΣ0), and {ln = logLn(β) : n = 1, 2, …} is a Laplace-regular sequence of log-likelihood functions having strict local maxima {(β̂n : n = 1, 2, …} and positive definite matrices { }. Let
| (2) |
where I is a n × n identity matrix. Then Ĩ = I(1 + O(n−1)).
Laplace regularity has been discussed in depth in Kass et al. (1990). Briefly speaking, it requires that ln(β) is six-time continuously differentiable, all partial derivatives (up to the sixth order) are bounded in the neighborhood of β̂n, the determinants of the Hessians are bounded and positive, and ln has a dominant peak at β̂n. Note that the result in Theorem 2.1 is an analytical assessment for given sequences of well-behaved (i.e. Laplace-regular) data. In Kass et al. (1990), general conditions are discussed that guarantee the well-behaved data will occur with probability one. In particular, since those conditions hold for the exponential family, the results of Theorem 2.1 hold for GLMs almost surely.
A full proof of the theorem is given in Appendix A. Below is a brief description of the idea. When {ln : n = 1, 2, …} is Laplace regular, we know from Lemma 2 in Kass et al. (1990) that, ∃ c > 0 and δ > 0, such that for all sufficient large n,
| (3) |
where Bδ (β̂n) denotes an open ball centered at β̂n with a radius δ. This reduces our consideration of I to the integral of Ln(β) π(β) over a neighborhood Bδ (β̂n) because (3) ensures that I only depends on the behavior of Ln near its maximum when n is large. Thus Ĩ can be obtained by first approximating ln(β) with a second-order Taylor series expansion at β̂n, say l̂n(β), over the region Bδ (β̂n), and then inserting it in (1) and integrating out β from the approximate integrand exp{l̂(β)} π (β). As a result, Ĩ is derived by taking advantage of the normality assumption for π (β).
2.2 Comparison with the Laplace’s Method
Under the conditions of Theorem 2.1, the well-known Laplace’s method is legitimate for the integral I in (1). The resulting approximations are not unique, depending on how one defines the Laplace-regular sequence {ln}.
Denning ln = log Ln yields the standard-form Laplace approximation.
| (4) |
In practice, this approximation is easy to compute since it only requires the MLE of β and the observed Fisher information, which are reported by nearly all statistical software.
Next we consider the fully exponential form of Laplace’s method (Tierney and Kadane, 1986; Tierney et al., 1989) by defining ln = log Ln + log π. Suppose {log Ln + logπ : n = 1, 2, …} have strict local maxima {β̃n : n = 1, 2, …} and positive definite matrices { }. Then the approximation is given by
| (5) |
Usually ĨLF is quite accurate, but it is computationally intensive because it requires the derivation of β̃n (i.e., the posterior mode) and the inverse Hessian of log posterior evaluated at β̃n, which are not directly available to users in most statistical software, and so have to be computed via numerical algorithms.
In addition, Raftery (1996) proposed a method for approximating Bayes factors based on the fully exponential form of Laplace’s method, followed by another approximation via Newton’s method to reduce the computational cost. For example, he approximated β̃n from β̂n using one step of Newton’s method. In our context, applying his idea yields the approximation
| (6) |
which is less accurate but is easier to compute than ĨLF.
Note that the errors of Ĩ, ĨL, ĨLF and ĨR are all O(n−1). Another similarity of the four is that for large λ with fixed Σ0, Ĩ ≈ ĨL ≈ ĨLF ≈ ĨR. This can be seen from the fact that as λ gets large, any pairwise ratio converges to 1, e.g., limλ→+∞ Ĩ/ĨL = 1.
Since computing Ĩ is as easy as computing ĨL and ĨR, we first contrast Ĩ with ĨL and ĨR. First of all, when Ln(β) is proportional to ψ, the normal density function, (2) gives the exact value, i.e. Ĩ = I. However, it is easy to verify that ĨL ≠ I and ĨR ≠ I in the normal case. Secondly, for small λ with fixed Σ0, approximations by Ĩ may differ substantially from those by ĨL and ĨR. The limit of I, as λ approaches zero, is limλ→0 I = Ln(β0) since β is fixed at β0 when λ = 0. For Ĩ,
| (7) |
Thus,
| (8) |
whenever β̂n → β0 as n gets large, which occurs with probability one under mild regularity conditions for many commonly used models including GLMs. But this limiting equality does not hold for either ĨL or ĨR, since
and limλ→0 ĨR = +∞. Neither of them converges to the true value. As a result, Ĩ is more accurate than ĨL and ĨR for small λ. This improvement in accuracy becomes important in situations where the predictive density needs to be evaluated for a range of values of λ including small ones, like a sensitivity analysis on λ or a Fully Bayes approach that entails choosing a prior on λ over (0, +∞).
Now we proceed to compare Ĩ with ĨLF. Note that ĨLF has excellent performance at small λ because limλ→0 ĨLF = limλ→0 I holds for every n. As will be shown in our simulation, Ĩ can achieve nearly the same performance as ĨLF, but requires much less computing time. In addition, in a fully Bayes (FB) method that treats λ as random, there does not exist a conjugate prior of λ based on ĨLF. In contrast, when Σ0 is chosen to be Σn, Ĩ can yield great analytical tractability when further integrating λ out. In this case, equation (2) becomes
which suggests a conjugate prior of λ in the form of 1/(λ + 1) ~ Truncated Gamma(a, b). This would simplify the FB computation since a closed form of the posterior can be derived.
Finally, Table 1 summarizes the discussions about comparing the four analytical approximations to the integral I.
Table 1.
Comparison of different analytical approximations to the integral I. Note that the limiting properties when λ → 0 indicate their performance at small λ. And ψ denotes the normal pdf.
| Approximations to I | Ĩ | ĨL | ĨLF | ĨR |
|---|---|---|---|---|
| Methods | Theory 2.1 | standard Laplace | fully-exponential Laplace | fully-exponential Laplace + Newton |
| Normal case: Ln ∝ ψ | Ĩ = I | ĨL ≠ I | ĨLF = I | ĨR ≠ I |
| Convergence when λ → 0? | Yes for GLMs with large n | No | Yes | No |
| Existence of conjugate priors on λ? | Yes | Yes | No | No |
| Ease of computation? | Yes | Yes | No | Yes |
2.3 The Case of General Priors
Here we discuss methods for approximating the predictive density in (1) with a general form of the prior π (β). A straightforward extension of Theorem 2.1 leads to the following result.
Corollary 2.1
Suppose {ln = log Ln(β) : n= 1, 2, …} satisfy the same conditions as in Theorem 2.1; π (β) is a four-time continuously differentiate prior density on β with a mode β0; and Σ0 = {− λ [log π (β0)]″}−1 is positive definite, λ > 0. Let
| (9) |
where Ĩ is given in (2), ĨL is given in (4) and . Then Ĩ* = I(1 + O (n−1)).
Proof
Let πN(β) denote the pdf of N(β0, λ Σ0). Note that we can write
| (10) |
Applying Theorem 2.1 to the first integral at the right-hand side of (10) and applying the standard-form Laplace approximation to the second integral yields (9) immediately.
One can treat Ĩ* as an improved Laplace approximation to I under the condition that π (β) can be approximated by πN(β), which is often satisfied when the prior is constructed from the likelihood or posterior of β based on historical or imaginary data with valid asymptotic normality. This is because in (9), is indeed the standard-form Laplace approximation to I; the remaining term Ĩ − ĨL provides a correction factor, using the difference between the Laplace’s method and Theorem 2.1 based on the normality of πN(β). This correction is useful when λ is small or when the prior sample size n0 (i.e., the size of historical data) is not significantly smaller than n. Under this case, the standard-form Laplace approximation may not work well as Ln does not dominate π; Ĩ* can achieve better performance because π − πN is dominated by Ln when π is well approximated by πN.
Like Ĩ for normal priors, an advantage of Ĩ* over a Laplace approximation to I is its nice limiting property. For example, when λ approaches 0, π (β) reduces to a point mass at its mode β0 so that limλ→0 I = Ln(β0); in this case, the Laplace approximation to the second integral in (10) is zero because π (β) = πN(β). This leads to Ĩ* = Ĩ and the equality limn→+∞ limλ→0 Ĩ* = limλ→0 I follows directly from (8) whenever β̂n → β0. We also note that if β̂n is within a small neighborhood of β0, then . This occurs sometimes in practice, for example, when the information contained in the current data agrees well with that in the historical data where the prior of β is from. In this case, I can be approximated by Ĩ only.
3 Application to Bayesian Inference in GLMs
3.1 Background
Suppose are independent observations from an exponential family distribution
| (11) |
indexed by unknown canonical parameters and a dispersion parameter φ. The functions and , assumed to be known, jointly determine the type of the distribution. The n × n matrix W is diagonal with its ith diagonal element being wi, a known weight for the ith observation. And J is the n × 1 vector of all 1’s.
In the GLM setup θ may depend on p observed covariates X1,…, Xp. Let γ = 1,…, 2P index all subsets of the covariates, qγ be the size of the γth subset, and Xγ be a n × (qγ + 1) covariate matrix with 1’s in the first column and the γth subset of Xj’s in the remaining columns. Further, denote the entire data by  = (Y, X, W), and the subset data for model γ by
= (Y, X, W), and the subset data for model γ by  = (Y, Xγ, W). Under model γ, g(E(Y)) = Xγ/βγ, where g is a known link function that by definition is monotonic and differentiable, and (βγ is a(qγ + 1)×1 vector of regression coefficients. Thus, the likelihood function of model γ is given by
= (Y, Xγ, W). Under model γ, g(E(Y)) = Xγ/βγ, where g is a known link function that by definition is monotonic and differentiable, and (βγ is a(qγ + 1)×1 vector of regression coefficients. Thus, the likelihood function of model γ is given by
| (12) |
Here we denote θ(Xγβγ) explicitly for θ because θ = b′−1 ◦ g−1 (Xγβγ) under model γ, where ◦ denotes function composition.
Bayesian inferences of GLMs, when model uncertainty exists, often require the calculation of the model posterior, which reply heavily on the evaluation of the predictive density:
| (13) |
In (13), π(βγ|γ, λ) is the prior on βγ, where λ is a hyperparameter. Note that the dispersion parameter φ is assumed to be known throughout this paper. This indeed occurs in Poisson, Binomial and Negative Binomial GLMs, in which φ is 1. For other members of the exponential family, φ is usually unknown and might be replaced by an estimate (McCullagh and Nelder 1989). Alternatively one can assess a prior distribution on φ so that it can be integrated out from the model posterior (Raftery 1996 and references therein). In either case, it will not affect the evaluation of the predictive density.
We consider three classes of informative priors on βγ in the literature, namely the normal, conjugate and power prior denoted by πN, πC and πP, respectively. Let λN, λC and λP denote the hyperparameters introduced by these priors. The normal prior (Dellaportas and Smith 1993, Meyer and Laud 2002) is
| (14) |
where mγ is the prior mean and Uγ is a multiple of the prior covariance matrix of βγ. The conjugate prior (Diaconis and Ylvisaker 1979, Meyer and Laud 2002, Chen et al. 2008) can be regarded as the likelihood of parameters (βγ, λC φ) with data :
| (15) |
where
and μ0 is the prior guess for Y. The power prior (Zellner (1988), Chen et al. 2000) is based on historical data 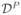 = (Y0, X0, W0) containing the same response and covariates as the current study:
= (Y0, X0, W0) containing the same response and covariates as the current study:
| (16) |
The three classes of priors are closely related via their large sample properties. The conjugate prior πC is asymptotically normal as n → ∞:
| (17) |
where is the MLE of βγ using μ0 rather than Y as the response vector; and is minus the inverse of , the Hessian matrix of evaluated at . This result can be obtained from Theorem 2.1 in Chen (1985) under some mild normality conditions. Similarly, the power prior πP is asymptotically normal as the historical sample size n0 → ∞:
| (18) |
where is the MLE of βγ based on the historical data , and is minus the inverse of , the Hessian matrix of evaluated at .
As revealed by (14), (17) and (18), λN, λC and λP essentially play the same important role, each as a multiplier to the prior covariance matrix, weighing the impact of the prior information relative to the current data. Thus, our proposed methods can provide a unified way to compute the predictive densities (13) under the three classes of priors.
3.2 Approximate Predictive Densities
In what follows we will use λ to denote any of λN, λC and λP.
A direct application of Theorem 2.1 yields an approximation for p(Y|γ, λ) based on the normal prior (14), namely
| (19) |
where β̂γ is the MLE of βγ using the current data  , and V̂γ = −H−1(β̂γ;
, and V̂γ = −H−1(β̂γ;  ). When Y is normally distributed so that the model is the familiar linear model, this approximation is exact, i.e. p̃(Y|γ, λ) = p(Y|γ, λ). This makes the results for linear models consistent with those for GLMs, as expected under the same Bayesian framework. However, it is not true if using the standard Laplace method or Raftery’s method. Another attractive feature of (19) is that, as discussed in Section 2.2, if Uγ is set to V̂γ, it can provide great analytical tractability in a Fully Bayes approach that entails integrating λ out. As shown by Wang and George (2007), in the context of model selection, approximation (19) leads to selection criteria that have computational simplicity and adaptive performance.
). When Y is normally distributed so that the model is the familiar linear model, this approximation is exact, i.e. p̃(Y|γ, λ) = p(Y|γ, λ). This makes the results for linear models consistent with those for GLMs, as expected under the same Bayesian framework. However, it is not true if using the standard Laplace method or Raftery’s method. Another attractive feature of (19) is that, as discussed in Section 2.2, if Uγ is set to V̂γ, it can provide great analytical tractability in a Fully Bayes approach that entails integrating λ out. As shown by Wang and George (2007), in the context of model selection, approximation (19) leads to selection criteria that have computational simplicity and adaptive performance.
Corollary 2.1 can be employed to approximate p(Y|γ, λ) in (13) with the conjugate and the power prior of βγ. For notation simplicity, denote 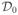 the prior data, which is
the prior data, which is  for the conjugate prior and is
for the conjugate prior and is 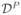 for the power prior; denote β̂0γ the MLE of βγ based on
for the power prior; denote β̂0γ the MLE of βγ based on  , the prior data for model γ; denote V̂0γ minus the inverse of H(βγ;
, the prior data for model γ; denote V̂0γ minus the inverse of H(βγ;  ) evaluated at β̂0γ. First note that the normalizing constant ∫ L(βγ, λφ;
) evaluated at β̂0γ. First note that the normalizing constant ∫ L(βγ, λφ;  )dβγ in π(βγ|γ, λ) varies from model to model, and thus cannot be ignored when model uncertainty is under consideration. We calculate the constant from the Laplace method, namely
)dβγ in π(βγ|γ, λ) varies from model to model, and thus cannot be ignored when model uncertainty is under consideration. We calculate the constant from the Laplace method, namely
| (20) |
Then Corollary 2.1 yields
| (21) |
where β̂γ and V̂γ are defined in (19). Due to the asymptotic normality in (17) and (18), (21) is in general better than the standard-form Laplace approximation, as discussed in Section 2.3. When β̂γ is close to β̂0γ, the part following the “+” sign in the second line of (21) is about zero, so p(Y|γ, λ) can be approximated only by the first term, which simplifies the calculation in some degree.
One can easily calculate (19) and (21) using statistical packages for GLMs, in which MLEs and observed Fisher information matrices are often standard outputs. For example, V̂γ can be calculated from the estimated covariance matrix divided by φ̂γ that is fitted to estimate φ, based on the current data  ; and V̂0γ can be calculated in the same way but based on the prior data
; and V̂0γ can be calculated in the same way but based on the prior data  for the conjugate or power prior of βγ.
for the conjugate or power prior of βγ.
4 Examples
4.1 Evaluation in a Simple Case
We first study the performance of the method proposed in Theorem 2.1 on a very simple Poisson linear model with a canonical link function. Two datasets, with n = 10 and 100 respectively, are simulated by generating (x1i, x2i) from N(0,I) and yi from independent Poisson with mean μi, where log μi = 1 + x1i, −0.5 x2i for i = 1,…, n. Note β = (1,1, −0.5)T, and φ = 1, b(θi) = μi = exp(θi) and c(yi, φ) =− log(yi!) in (12). For comparison, we adopt various methods to calculate the marginal likelihoods of the simulated data, p(Y|λ) = ∫ p (Y|β) π (β| λ)dβ, based on normal priors on β in the form of N(mγ, λUγ). For each dataset, we consider eight different λ values and two prior means, mγ = (β¯0, 0, 0)T and mγ = (1, 1, −0.5)T, where β¯0 is chosen to be the MLE of β0 under the null model as in Chipman et al. (2003). Further, to see the impact of prior covariances, we consider two prior covariance matrices, Uγ = I and Uγ = V̂ γ. The methods include Prop. (the method proposed in Theorem 2.1), SL (standard-form Laplace), FEL (fully-exponential Laplace), R (fully-exponential Laplace + Newton, Raftery 1996), and IS (Importance Sampling). The formulas for these methods are available in Appendix B. Since the true value of the integral is unknown, we keep generating blocks of 20, 000 sample units until the second decimal place of the log marginal likelihood value, estimated by IS cumulatively using all the generated blocks, stabilizes for 5 consecutive blocks. Since the absolute value of the log marginal likelihood varies from about 15 to 470, the variation in the third digit has a negligible impact on the relative error (i.e., the estimated value minus the true value, then divided by the true value). So the results from IS can be treated as surrogates of true values. Here we report the relative error in the log scale, |(log Ĩ − log I)/log I|, instead of the absolute error |Ĩ − I|. Thus, as n gets large, the relative error in log is not expected to decrease.
The absolute values of relative errors compared against estimates of IS are reported in Table 2. The proposed method is much better than SL and R for small λ. When λ is large, all methods work equally well. When λ is small, SL and R are sensitive to the prior variances Uγ as their relative errors for Uγ = I are much smaller than those for Uγ = V̂ γ: by contrast, the proposed method appears to be robust to the choice of Uγ. Furthermore, all methods except for FEL are sensitive to m since their relative errors for m = β are smaller than m = (β¯0, 0, 0)T, but the proposed one appears to be less sensitive than the other two.
Table 2.
Absolute Values of Relative Errors in Approximate Log Marginal Likelihoods of Poisson Model
| λ | n = 10, mγ = (β¯0, 0, 0), Uγ = I | n = 10, mγ = (β¯0, 0, 0), Uγ = V̂γ | ||||||
|---|---|---|---|---|---|---|---|---|
| SL | R | Prop. | FEL | SL | R | Prop. | FEL | |
| 1.0E-10 | 3.E+08 | 3.E+08 | 0.031 | 0.000 | 4.E+09 | 4.E+09 | 0.031 | 0.000 |
| 0.0001 | 297.593 | 297.731 | 0.030 | 0.000 | 3524.682 | 3524.964 | 0.031 | 0.000 |
| 0.001 | 29.328 | 29.335 | 0.031 | 0.001 | 351.894 | 352.032 | 0.030 | 0.000 |
| 0.01 | 2.615 | 2.572 | 0.034 | 0.000 | 34.782 | 34.780 | 0.028 | 0.000 |
| 0.1 | 0.110 | 0.119 | 0.017 | 0.000 | 3.178 | 3.058 | 0.015 | 0.000 |
| 1 | 0.009 | 0.003 | 0.003 | 0.001 | 0.157 | 0.107 | 0.002 | 0.000 |
| 10 | 0.000 | 0.001 | 0.001 | 0.001 | 0.004 | 0.001 | 0.000 | 0.001 |
| 100 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.001 | 0.001 | 0.001 |
| λ | n = 10, mγ = (1, 1, −0.5), Uγ = I | n = 10, mγ = (1, 1, −0.5), Uγ = V̂γ | ||||||
| SL | R | Prop. | FEL | SL | R | Prop. | FEL | |
| 1.0E-10 | 1.E+07 | 1.E+07 | 0.001 | 0.000 | 1.E+08 | 1.E+08 | 0.001 | 0.000 |
| 0.0001 | 13.503 | 14.180 | 0.001 | 0.000 | 104.586 | 105.494 | 0.001 | 0.000 |
| 0.001 | 0.951 | 1.398 | 0.000 | 0.000 | 9.851 | 10.530 | 0.001 | 0.000 |
| 0.01 | 0.108 | 0.123 | 0.001 | 0.000 | 0.584 | 1.034 | 0.001 | 0.000 |
| 0.1 | 0.066 | 0.006 | 0.002 | 0.000 | 0.143 | 0.088 | 0.002 | 0.000 |
| 1 | 0.010 | 0.000 | 0.000 | 0.000 | 0.063 | 0.005 | 0.003 | 0.000 |
| 10 | 0.000 | 0.001 | 0.001 | 0.001 | 0.008 | 0.000 | 0.000 | 0.001 |
| 100 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.001 | 0.001 | 0.001 |
| λ | n = 100, mγ = (β¯0, 0, 0), Uγ = I | n = 100, mγ = (β¯0, 0, 0), Uγ = V̂γ | ||||||
| SL | R | Prop. | FEL | SL | R | Prop. | FEL | |
| 1.0E-10 | 2.E+07 | 2.E+07 | 0.077 | 0.000 | 6.E+09 | 6.E+09 | 0.077 | 0.000 |
| 0.0001 | 15.703 | 15.083 | 0.046 | 0.004 | 6309.329 | 6308.576 | 0.077 | 0.000 |
| 0.001 | 1.262 | 1.231 | 0.108 | 0.089 | 630.802 | 630.042 | 0.077 | 0.000 |
| 0.01 | 0.078 | 0.047 | 0.012 | 0.007 | 62.937 | 62.175 | 0.078 | 0.000 |
| 0.1 | 0.001 | 0.000 | 0.000 | 0.000 | 6.149 | 5.444 | 0.080 | 0.002 |
| 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.324 | 0.255 | 0.061 | 0.088 |
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.011 | 0.000 | 0.001 | 0.000 |
| 100 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| λ | n = 100, mγ = (1, 1, −0.5), Uγ = I | n = 100, mγ = (1, 1, −0.5), Uγ = V̂γ | ||||||
| SL | R | Prop. | FEL | SL | R | Prop. | FEL | |
| 1.0E-10 | 6.E+05 | 6.E+05 | 0.001 | 0.000 | 1.E+08 | 1.E+08 | 0.001 | 0.000 |
| 0.0001 | 0.602 | 0.613 | 0.001 | 0.000 | 124.862 | 124.918 | 0.001 | 0.000 |
| 0.001 | 0.045 | 0.045 | 0.000 | 0.000 | 12.430 | 12.469 | 0.001 | 0.000 |
| 0.01 | 0.001 | 0.001 | 0.000 | 0.000 | 1.202 | 1.224 | 0.001 | 0.000 |
| 0.1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.096 | 0.103 | 0.000 | 0.000 |
| 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.003 | 0.000 | 0.000 |
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 |
| 100 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
It is helpful to discuss the methods from a computational perspective. SL, R and the proposed method only require the MLE of β, the estimated covariance matrix and likelihood, so they are easy to program using standard statistical software. FEL requires more programming efforts because one needs to calculate the posterior mode of β via an optimization algorithm and the corresponding posterior covariance matrix involving detailed formulas. IS, as a sampling-based method, is easy to code but requires fine tune for fast convergence. Turning to the CPU time, based on calculations for the dataset of size 100 and a fixed λ on a workstation with a 1.8 GHz Xeon processor and 1 GB of RAM, it takes 33 ms for SL, R and the proposed method, 68 ms for FEL, and for IS to get an estimate with an error within ±0.05, the time varies from 270 ms to 51 seconds for different λ. Overall, it appears that the proposed method is promising because of greater accuracy, less human efforts and faster computing speed.
4.2 Intensive Care Unit Data
We provide an example that incorporates our approximation methods in an application of Bayesian variable selection. We consider a dataset in Hosmer and Lemeshow (1989) with 200 subjects from a study on survival of patients following admission to an adult intensive care unit (ICU). The response STA is the vital status of a patient at the time of hospital discharge (0=Lived, 1=Died). There are 19 predictor variables in the dataset: (1) Age; (2) Sex; (3) Race (White/Black/Other); (4) Service at ICU admission (SER, Medical/Surgical); (5) Cancer Part of Present Problem (CAN, No/Yes); (6) History of Chronic Renal Failure (CRN, No/Yes); (7) Infection Probable at ICU Admission (INF, No/Yes); (8) CPR prior to ICU Admission (CPR, No/Yes); (9) Systolic Blood Pressure at ICU Admission (SYS); (10) Heart Rate at ICU Admission (HRA); (11) Previous Admission to an ICU within 6 Months (PRE, No/Yes); (12) Type of Admission (TYP, Elective/Emergency); (13) Long Bone, Multiple, Neck, Single Area, or Hip Fracture (FRA, No/Yes); (14) PO2 from Initial Blood Gases (PO2, > 60/≤ 60); (15) PH from Initial Blood Gases (PH, ≥ 7.25/< 7.25); (16) PCO2 from Initial Blood Gases (PCO, ≤ 45/> 45); (17) Bicarbonate from Initial Blood Gases (BIC, ≥ 18/< 18); (18) Creatinine from Initial Blood Gases (CRE, ≤ 60/> 60); (19) Level of Consciousness at ICU Admission (LOC, No Coma/Deep Stupor/Coma). The aim is to select models with highest posterior probabilities out of 219 or 524288 possible logistic regression models to predict the probability of survival and study the risk factors associated with ICU mortality. Note for logistic regression, φ = 1, b(θi) = log(1 + eθi) and c(yi, φ) = 0 in (12).
Here, a fully Bayes method (FB) is used for illustration. Often, when a single value of λ is difficult to specify, like in this case, there is a general agreement that fully Bayes methods are well suited in summarizing posterior uncertainties, which entail integrating λ out:
| (22) |
where π(γ) and π(λ) denote the prior on γ and λ, respectively. Except for certain restricted examples, the resulting model posterior p(γ|Y) (up to a normalizing constant) is analytically intractable. Our approximation methods would allow MCMC algorithms to only sample over a two-dimension space of γ and λ. We consider conjugate priors (15) on regression coefficients and an inverse Gamma hyperprior IG(a, b) on the hyperparameter λ. For illustrative purposes, the prior mean μ0 was obtained from a prior prediction using the logistic regression model reported in Lemeshow et al. (1988),
| (23) |
We explored four sets of (a, b) values with prior means of λ at 2, 5, 10, and 50, respectively. We found that the posterior of λ was sensitive to the choice of (a, b). Finally we chose (a, b) = (2.25, 62.5) because this hyperprior has a heavier right tail than the others, indicating the prior guess (23) is not to have much impact compared to the data. For prior model probabilities π(γ), to reflect no real prior information, we assigned equal probability to each possible model. To efficiently search high posterior models, the evolutionary Monte Carlo algorithm (Liang and Wong 2000) was applied in this example. Four chains were simulated in parallel with a temperature ladder {1,2,3,4} and each running 50,000 iterations. Within each iteration, for any proposed γ and λ, when calculating the transition and exchange probabilities, p(Y|λ, γ)π(λ|a, b) was approximated by p̃(Y|λ, γ)π(λ|a, b) using our methods, which greatly simplified the MCMC because there was no need to sample from the space of regression coefficients βγ.
Table 3 reports the top 50 models from the MCMC search. The posterior probability of model γ was estimated from the frequency of γ in the chain divided by 40,000 (the first 10,000 iterations were discarded for the burn-in process). For comparison, we also give AIC and BIC values and ranks for each of the models: AICγ = log L(β̂γ;  ) − qγ and BICγ = log L(β̂γ;
) − qγ and BICγ = log L(β̂γ;  ) − qγ log n/2. We can see from Table 3 that the “best” model selected by the FB procedure contains 6 covariates: RACE, SER, CAN, TYP, FRA, and LOC, denoted γFB; the “best” model given by AIC contains 10 covariates: AGE, RACE, SER, CAN, PRE, TYP, FRA, PH, PCO and LOC, denoted γA; and the “best” model given by BIC contains only three covariates: CAN, TYP, and LOC, denoted γB. It is well known that AIC tends to favor large models while BIC tends to favor small models, so it is interesting to see that γB ⊂ γFB ⊂ γA here. Also, γFB is a model that ranks high in both AIC and BIC (i.e., the model with a small rank sum). Another interesting feature for this dataset is, none of the top 50 models selected by AIC agrees with the top 50 models selected by BIC; for example, γA ranks 8098 in BIC, γB ranks 2588 in AIC, but both of them are in the top 50 list of the FB procedure.
) − qγ log n/2. We can see from Table 3 that the “best” model selected by the FB procedure contains 6 covariates: RACE, SER, CAN, TYP, FRA, and LOC, denoted γFB; the “best” model given by AIC contains 10 covariates: AGE, RACE, SER, CAN, PRE, TYP, FRA, PH, PCO and LOC, denoted γA; and the “best” model given by BIC contains only three covariates: CAN, TYP, and LOC, denoted γB. It is well known that AIC tends to favor large models while BIC tends to favor small models, so it is interesting to see that γB ⊂ γFB ⊂ γA here. Also, γFB is a model that ranks high in both AIC and BIC (i.e., the model with a small rank sum). Another interesting feature for this dataset is, none of the top 50 models selected by AIC agrees with the top 50 models selected by BIC; for example, γA ranks 8098 in BIC, γB ranks 2588 in AIC, but both of them are in the top 50 list of the FB procedure.
Table 3.
Top 50 Models From MCMC for the ICU Data. Note that the estimated posterior probabilities π̂(γ|Y) were multiplied by 1,000.
| Model | No. Variables | Est. Posterior | AIC (rank) | BIC (rank) |
|---|---|---|---|---|
| 3,4,5,12,13,19 | 6 | 4.70 | −51.20 (11) | −63.75 (61) |
| 2,3,4,5,12,13,19 | 7 | 4.67 | −51.72 (44) | −65.66 (593) |
| 3,4,5,12,19 | 5 | 3.95 | −52.28 (173) | −63.43 (41) |
| 3,4,5,11,12,13,19 | 7 | 3.87 | −51.12 (8) | −65.06 (312) |
| 2,3,4,5,11,12,13,19 | 8 | 3.15 | −51.74 (45) | −67.07 (2326) |
| 3,4,5,6,12,13,19 | 7 | 2.70 | −51.83 (58) | −65.76 (657) |
| 1,3,4,5,8,11,12,13,16,19 | 10 | 2.55 | −51.64 (33) | −69.76 (16867) |
| 3,4,5,6,11,12,13,15,16,19 | 10 | 2.50 | −51.79 (51) | −69.91 (18400) |
| 1,3,4,5,11,12,13,19 | 8 | 2.45 | −51.28 (12) | −66.61 (1501) |
| 1,3,4,5,12,13,16,19 | 8 | 2.35 | −51.14 (9) | −66.47 (1318) |
| 2,3,4,5,11,12,13,18,19 | 9 | 2.20 | −52.41 (228) | −69.13 (11251) |
| 3,5,12,19 | 4 | 2.15 | −53.44 (1435) | −63.19 (30) |
| 3,4,5,12,13,15,19 | 7 | 2.07 | −51.96 (87) | −65.89 (752) |
| 3,4,5,6,10,12,13,19 | 8 | 2.00 | −52.78 (470) | −68.11 (5364) |
| 3,4,5,8,11,12,13,16,19 | 9 | 2.00 | −52.00 (98) | −68.72 (8428) |
| 3,4,5,12,13,16,19 | 7 | 1.97 | −51.60 (31) | −65.54 (530) |
| 2,3,4,5,6,12,19 | 7 | 1.87 | −53.16 (916) | −67.10 (2382) |
| 3,4,5,8,11,12,19 | 7 | 1.80 | −52.87 (562) | −66.81 (1786) |
| 3,4,5,11,12,13,14,19 | 8 | 1.80 | −51.91 (78) | −67.25 (2696) |
| 5,8,12,15,16,19 | 6 | 1.77 | −53.42 (1399) | −64.57 (174) |
| 3,4,5,11,12,19 | 6 | 1.77 | −52.57 (301) | −65.11 (325) |
| 2,3,4,5,6,8,11,12,13,19 | 10 | 1.75 | −52.99 (701) | −71.10 (36334) |
| 3,4,5,10,11,12,13,19 | 8 | 1.72 | −52.10 (115) | −67.43 (3114) |
| 5,12,19 | 3 | 1.67 | −53.81 (2588) | −60.78 (1) |
| 3,5,10,12,19 | 5 | 1.67 | −54.17 (4250) | −65.32 (413) |
| 1,2,3,4,5,12,13,15,16,19 | 10 | 1.65 | −51.39 (19) | −69.51 (14521) |
| 1,3,4,5,9,11,12,13,16,18,19 | 11 | 1.65 | −52.79 (478) | −72.30 (64620) |
| 3,4,5,12,15,19 | 6 | 1.57 | −52.86 (556) | −65.41 (450) |
| 1,2,3,4,5,12,13,16,19 | 9 | 1.57 | −51.69 (37) | −68.41 (6714) |
| 2,3,4,5,11,12,13,15,16,19 | 10 | 1.57 | −51.38 (17) | −69.50 (14404) |
| 3,4,5,7,8,10,12,13,19 | 9 | 1.55 | −53.56 (1730) | −70.28 (23084) |
| 1,3,4,5,12,13,15,16,19 | 9 | 1.50 | −51.08 (7) | −67.81 (4238) |
| 3,4,5,11,12,13,16,19 | 8 | 1.47 | −51.30 (13) | −66.63 (1526) |
| 3,4,5,8,12,19 | 6 | 1.45 | −52.78 (468) | −65.32 (415) |
| 3,4,5,8,12,13,19 | 7 | 1.42 | −51.96 (89) | −65.90 (761) |
| 3,4,5,11,12,15,19 | 7 | 1.32 | −53.03 (742) | −66.96 (2065) |
| 3,4,5,9,11,12,19 | 7 | 1.30 | −52.97 (679) | −66.90 (1948) |
| 1,3,4,5,11,12,13,15,16,19 | 10 | 1.30 | −50.55 (1) | −68.67 (8098) |
| 2,3,4,5,12,13,18,19 | 8 | 1.27 | −52.51 (265) | −67.84 (4350) |
| 2,3,4,5,12,19 | 6 | 1.25 | −52.86 (549) | −65.40 (446) |
| 5,12,15,19 | 4 | 1.22 | −53.84 (2723) | −62.20 (7) |
| 3,4,5,7,12,13,19 | 7 | 1.22 | −51.70 (40) | −65.63 (579) |
| 1,3,4,5,11,12,13,17,19 | 9 | 1.22 | −52.27 (168) | −68.99 (10156) |
| 3,4,5,9,11,12,13,15,16,19 | 10 | 1.22 | −51.57 (28) | −69.69 (16167) |
| 4,5,12,19 | 4 | 1.20 | −53.02 (727) | −61.38 (2) |
| 3,4,5,7,11,12,13,19 | 8 | 1.20 | −51.66 (35) | −66.99 (2126) |
| 3,4,5,8,11,12,13,16,17,19 | 10 | 1.20 | −52.96 (669) | −71.08 (35795) |
| 1,4,5,12,19 | 5 | 1.17 | −53.11 (837) | −62.87 (14) |
| 3,5,8,12,19 | 5 | 1.17 | −53.47 (1519) | −64.62 (188) |
| 3,4,5,6,9,12,19 | 7 | 1.17 | −52.86 (554) | −66.80 (1776) |
It is worth noting that the “best” model γFB represents only 0.47% of the total posterior probability, indicating a fair amount of model uncertainty in the ICU data. This reveals that in this example, model averaging may be more appropriate than simply selecting a “best” model. More importantly, our fully Bayes method gives the top 50 models covering more than 95% posterior probability, so that model averaging can be done over these 50 models (not all the 524,288 possible logistic regression models), with weights already provided.
In conclusion, our approximation methods, when applied to the fully Bayes variable selection, seem to work reasonably well.
5 Discussion
In this paper, we have proposed new methods to approximate predictive distributions that are easy to implement and computationally efficient, and discussed their performance both in theory and through simulation. In simulation studies we have shown that the relative error associated with the proposed methods is clearly superior to the Laplace-based methods when λ is small, say λ < 1. Whether the better performance of Ĩ at small λ has practical importance is worth discussing.
As we know, a large λ might be often used in practice to allow for a diffuse and vague prior distribution on (β). However, there exist situations when a wide range of λ, including small values, is of interest, like in a sensitivity analysis about λ or a Fully Bayes approach that entails choosing a prior on λ over the entire support (0,+∞). Using our proposed methods can help to avoid mathematical pitfalls that may be extremely hard to detect for practitioners, with little cost, as opposed to other existing analytical approximation methods.
Further, small λ can be chosen through empirical Bayes methods (George and Foster 2000) in data analysis. This can be seen from the formula given in George and Foster (2000) to estimate λ from data (in their denotation c is equivalent to λ):
| (24) |
where (·)+ is the positive part of the function, γ represents the γth regression model in the model space, qγ is the number of variables in model γ; and SSγ is the regression sum of squares of model γ, measuring the part of the variability of Yis which can be explained by the regression line. Clearly, if the signal-to-noise ratio, measured by SSγ/(σ2qγ), is too low (say close to 1, or even small than 1), then λ would be chosen to be close to 0 or even 0 based on (24).
Finally, we would like to mention that in Bayesian applications of GLM, when model uncertainty can not be ignored, MCMC are commonly used to sample from a joint space of models and parameters. For large-sized problems, the computation of high-dimensional integrations via MCMC can be intensive or slow in convergence sometimes. As mentioned in Han and Carlin (2001), “…the ability of joint model and parameter search methods to sample effectively over such large spaces is very much in doubt” and they often require substantial time and effort (both human and computer). Thus, it would be useful for practitioners to try our proposed methods when MCMC-based algorithms do not work well.
Acknowledgments
This work was supported in part by NSF grant DMS-0906545 and NIDA grant 1R21DA027592.
A Proof of Theorem 2.1
This proof is similar in part to that of Theorem 1 in Kass et al. (1990). Without loss of generality, we consider the case m = 1 for simplicity. The higher-dimensional case involves straightforward modifications. Let hn(β) ≡ −ln(β)/n so that the integrand Ln(β)π(β) of (1) can be written as exp[−nhn(β)]π(β), and let u ≡ n1/2(β − β̂n) so that for a fixed u, (β − β̂n)k is of O(n−k/2). Now expanding nhn(β) about β̂n and e−x about zero to the terms of order smaller than O(1) yields
where Rn(u) is of order O(n−1) uniformly on Bδ(β̂n) defined in (3). Then
where
and
Let’s look at E1 first. Note by changing the variable from β to u, we have
| (25) |
where δ(n) = n1/2 δ. Since the integration region Bδ(n)(0) is expanding at the rate O(n1/2) as n → +∞, replacing this region by the whole real line incurs an error of exponentially decreasing order for the integral in (25), and yields (after some algebra)
| (26) |
in which Ĩ is given in (2). For E2, expanding π(β) about β̂n and changing the variable from β to u, we have
| (27) |
where Sn(u) is in the order O(n−1) uniformly on Bδ(β̂n). Using the same reasoning as for E1, we replace Bδ(n)(0) by the real line in (27) and note that the third central moment of a normal distribution vanishes, so
| (28) |
holds as long as the ith derivative of hn(i ≤ 4) is uniformly bounded, which is satisfied automatically from the Laplace regularity of ln. Combining (26), (28) and (3) yields Ĩ = I(1 + O(n−1)), which completes the proof.
B Formulas For Calculations in Section 4.1
For a GLM described in (12), consider the marginal density of the data p(Y|γ, λ) based on a normal prior π(βγ) in the form of N(mγ, λφUγ), λ > 0. Below are the formulas we derive for approximating p(Y|γ, λ) using the fully-exponential Laplace and the important sampling methods. The other methods involve trivial applications of formulas (2), (4) and (6).
-
Full-exponential Laplacewhere β̃γ is the posterior mode of βγ, , Dγ is diagonal with its ith diagonal element equal to dγi, and θ̃γi = b′−1 ◦ g−1(Xγiβ̃γ),
-
Importance Sampling:
where , t = 1, …, M, are independent samples, each with probability r generated from π(βγ), i.e., N(mγ, λφUγ) and with probability 1 − r generated from N(β̂γ, φV̂γ); and h(βγ) is the density of the mixture h(βγ) = rπ (βγ) + (1 − r) f(βγ), where f(βγ) is the pdf of N(β̂γ, φV̂γ). In our experiment, for λ = 1E − 20, we set r equal to 0.8; for λ = 0.001, we set r = 0.1 and for any larger λ, set r = 0.5. M is chosen to be large enough so that the estimate p̃IS(Y|γ, λ, φ) stabilizes within a small region.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Min Chen, Email: Min.Chen@UTSouthwestern.edu, Department of Clinical Sciences, University of Texas Southwestern Medical Center, Dallas, TX 75390.
Xinlei Wang, Email: swang@mail.smu.edu, Department of Statistical Science, Southern Methodist University, 3225 Daniel Avenue, P O Box 750332, Dallas, Texas 75275-0332.
References
- Boone EL, Ye K, Smith EP. Assessment of two approximation methods for computing posterior model probabilities. Computational Statistics & Data Analysis. 2005;48(2):221–234. [Google Scholar]
- Chen CF. On asymptotic normality of limiting density functions with Bayesian implications. Journal of the Royal Statistical Society, Series B. 1985;47(3):540–546. [Google Scholar]
- Chen M-H, Huang L, Ibrahim JG, Kim S. Bayesian variable selection and computation for generalized linear models with conjugate priors. Bayesian Anal. 2008;3(3):585–614. doi: 10.1214/08-BA323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Ibrahim JG, Shao QM. Understanding the Metropolis-Hastings algorithm. Journal of Statistical Planning and Inference. 2000;84:121–137. [Google Scholar]
- Chen MH, Ibrahim JG, Yiannoutsos C. Prior elicitation, variable selection and Bayesian computation for logistic regression models. Journal of the Royal Statistical Society, Series B. 1999;61:223–242. [Google Scholar]
- Chipman HA, George EI, McCulloch RE. Bayesian treed generalized linear models. In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics. Vol. 7. Clarendon Press; Oxford: 2003. pp. 85–104. [Google Scholar]
- Dellaportas P, Smith AFM. Bayesian inference for generalised linear and proportional hazards models via Gibbs sampling. Applied Statistics. 1993;42:443–459. [Google Scholar]
- Diaconis P, Ylvisaker D. Conjugate priors for exponential families. The Annals of Statistics. 1979;7:269–281. [Google Scholar]
- Gelfand AE, Dey D. Bayesian model choice: Asymptotics and exact calculations. Journal of the Royal Statistical Society, Series B. 1994;56(3):501–514. [Google Scholar]
- Gelfand AE, Smith AFM. Sampling-based approaches to calculating marginal densities. Journal of the American Statistical Association. 1990;85(410):398–409. [Google Scholar]
- George EI, Foster DP. Calibration and empirical Bayes variable selection. Biometrika. 2000;87:731–747. [Google Scholar]
- Han C, Carlin BP. Markov chain Monte Carlo methods for computing Bayes factors: A comparative review. Journal of the American Statistical Association. 2001;96(455):1122–1132. [Google Scholar]
- Hosmer DW, Lemeshow S. Applied Logistic Regression. John Wiley —& Sons; 1989. [Google Scholar]
- Ibrahim JG, Chen MH, Ryan LM. Bayesian variable selection for time series count data. Statistica Sinica. 2000;10(3):971–987. [Google Scholar]
- Kass RE, Tierney L, Kadane JB. The validity of posterior expansions based on Laplace’s method. In: Geisser S, Hodges JS, Press SJ, Zellner A, editors. Bayesian and Likelihood Methods in Statistics and Econometrics. Elsevier Science; Amsterdam: 1990. pp. 473–483. [Google Scholar]
- Lemeshow S, Teres D, Avrunin JS, Pastides H. Predicting the outcome of intensive care unit patients. Journal of American Statistical Association. 1988;83(402):348–356. [Google Scholar]
- Liang F, Wong WH. Evolutionary Monte Carlo: Applications to cp model sampling and change point problem. Statistica Sinica. 2000;10:317–342. [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. 2 Chapman and Hall; London: 1989. [Google Scholar]
- Meyer MC, Laud PW. Predictive variable selection in generalized linear models. Journal of the American Statistical Association. 2002;97(459):859–871. [Google Scholar]
- Raftery A. Approximate Bayes factors and accounting for model uncertainty in generalised linear models. Biometrika. 1996;83:251–266. [Google Scholar]
- Tierney L, Kadane JB. Accurate approximation for posterior moments and marginal densities. Journal of the American Statistical Association. 1986;81:82–86. [Google Scholar]
- Tierney L, Kass RE, Kadane JB. Approximate marginal densities of nonlinear functions. Biometrika. 1989;76:425–437. [Google Scholar]
- Verdinelli I, Wasserman L. Computing Bayes factors using a generalization of the Savage-Dickey density ratio. Journal of the American Statistical Association. 1995;90(430):614–618. [Google Scholar]
- Wang X, George EI. Adaptive Bayesian criteria in variable selection for generalized linear models. Statistica Sinica. 2007;17:667–690. [Google Scholar]
- Zellner A. Optimal information processing and bayes’s theorem. The American Statistician. 1988;42(4):278–280. [Google Scholar]


