Summary
Silent information regulator 2 (Sir2) orthologs are an evolutionarily conserved family of NAD-dependent protein deacetylases that regulate aging and longevity in model organisms. The mammalian Sir2 ortholog Sirt1 regulates metabolic and stress responses through the deacetylation of many transcriptional regulatory factors. To elucidate the mechanism by which Sirt1 controls gene expression in response to nutrient availability, we devised a bioinformatic screen combining gene expression analysis with phylogenetic footprinting to identify transcription factors as new candidate partners of Sirt1. One candidate target was HNF-1α, a homeodomain transcription factor that regulates pancreatic β cell and hepatocyte functions and is commonly mutated in patients with maturity onset diabetes of the young (MODY). Interestingly, Sirt1 physically interacts with HNF-1α in vitro but does so in vivo only in nutrient-restricting conditions. This interaction requires 12–24 hr of nutrient restriction and is dependent on protein synthesis. Both nutrient restriction and Sirt1 suppress HNF-1α transcriptional activity and the expression of one of its target genes, C-reactive protein (Crp), in mouse primary hepatocytes. Pharmacological inhibition of Sirt1 blocks the suppression of Crp by nutrient restriction. Similarly, Crp expression is also suppressed in fasted and diet-restricted liver. Furthermore, Sirt1 and HNF-1α co-localize on two HNF-1α binding sites on the Crp promoter, leading to decreased acetylation of lysine 16 of histone H4 at these sites only in response to nutrient restriction. These findings reveal a novel nutrient-dependent interaction between Sirt1 and HNF-1α and provide important insight into the molecular mechanism by which Sirt1 mediates the anti-aging effects of diet restriction.
Keywords: Sirt1, bioinformatics, HNF-1α, primary hepatocytes, nutrient restriction, Crp
Introduction
Sir2 (silent information regulator 2) proteins or sirtuins are an evolutionarily conserved family of NAD-dependent protein deacetylases/ADP-ribosyltransferases that regulate critical biological processes in response to nutrient availability and impact the process of aging and longevity in a variety of experimental organisms (Finkel et al. 2009; Haigis & Sinclair 2010; Imai & Guarente 2010). In mammals, it has been well established that the Sir2 ortholog Sirt1 mediates metabolic and stress responses to acute and chronic energy limitation, such as fasting and diet restriction, by deacetylating many transcriptional regulatory factors and affecting gene expression (Finkel et al. 2009; Haigis & Sinclair 2010; Imai & Guarente 2010). Diet restriction (DR), the single, most reliable regimen known to retard aging and extend lifespan in a wide variety of organisms, has provided a unique dietary intervention to investigate the physiological relevance of Sirt1 and other mammalian sirtuins in the metabolic regulation of aging and longevity in mammals (Sinclair 2005; Bishop & Guarente 2007; Schwer & Verdin 2008; Imai 2009b). It has been speculated that sirtuins, Sirt1 in particular, play a critical role in mediating the beneficial effects of DR, but the molecular details are still poorly understood.
Sirt1 activity is regulated in response to acute and chronic nutritional limitation through multiple mechanisms. First, its enzymatic activity depends on the availability of its co-substrate, NAD (Imai & Guarente 2010). Nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme in mammalian NAD biosynthesis, promotes NAD biosynthesis by converting nicotinamide into nicotinamide mononucleotide (NMN), a key NAD intermediate, and enhances Sirt1 enzymatic activity (Revollo et al. 2004; Garten et al. 2009; Imai 2009a). The NAMPT-Sirt1 pathway also comprises a novel circadian rhythm-regulatory feedback loop with the core clock components, CLOCK and BMAL1 (Nakahata et al. 2009; Ramsey et al. 2009). Fasting and DR indeed increase the expression of NAMPT, raising NAD levels in tissues and organs (Yang et al. 2007, our unpublished obvervation; Nakagawa et al. 2009). Second, the expression of Sirt1 is regulated by nutrient availability through alterations in transcription (Nemoto et al. 2004), mRNA stability (Yamakuchi et al. 2008; Rane et al. 2009), and/or protein stability (Ford et al. 2008). It has been reported that fasting and DR increases Sirt1 protein levels in several tissues and organs (Al-Regaiey et al. 2004; Cohen et al. 2004; Nemoto et al. 2004; Rodgers et al. 2005). Third, Sirt1 activity is suppressed by an interaction with DBC1 (deleted in breast cancer-1) (Kim et al. 2008; Zhao et al. 2008). In mouse liver, Sirt1 activity increases by decreased Sirt1-DBC1 interaction in response to 24-hr fasting (Escande et al. 2010). Regulated by these different mechanisms, Sirt1 controls networks of gene expression by interacting with and deacetylating many transcriptional regulatory factors, particularly major metabolic transcription factors including PPARγ, PGC-1α, FOXO1, CRTC2 (also known as TORC2), LXRα, and STAT3 (Finkel et al. 2009; Haigis & Sinclair 2010; Imai & Guarente 2010). A growing number of tissue-specific Sirt1 target factors are still being identified, raising the questions of how many additional targets have yet to be identified and how important each of the known interactions is.
To identify new protein targets of Sirt1, we devised a bioinformatic screen combining gene expression analysis with phylogenetic footprinting and identified several transcription factors as candidate partners of Sirt1. One of the candidates identified by this screen was HNF-1α, a homeodomain transcription factor that interacts with a complex network of transcription factors to regulate gene expression in the liver, kidney, and pancreatic β cells (Pontoglio 2000). In humans, mutations in HNF-1α profoundly impair insulin secretion and are responsible for the most common type of monogenic diabetes, MODY3 (Shih & Stoffel 2002). HNF-1α-null mice show corresponding defects in liver and β cell functions and have hyperglycemia, hypercholesterolemia, and low levels of IGF-1 (Lee et al. 1998; Shih et al. 2001a; Shih et al. 2001b). We found that Sirt1 and HNF-1α directly bind to each other in vitro, but only co-immunoprecipitate in vivo under conditions of nutrient restriction. Sirt1 suppresses HNF-1α transcriptional activity and the expression of one of its target genes, C-reactive protein (Crp), in mouse primary hepatocytes and in fasted and diet-restricted liver. On the Crp proximal promoter, Sirt1 is recruited to the HNF-1α binding sites, resulting in Sirt1-mediated deacetylation of lysine 16 of histone H4 in response to nutrient restriction. These findings validate our bioinformatics approach, identify a new mode of regulation of Sirt1 in response to nutrient restriction, and indicate that HNF-1α may be an important mediator of Sirt1-dependent transcriptional regulation during fasting and DR.
Results
Bioinformatic identification of candidate transcription factor partners of Sirt1
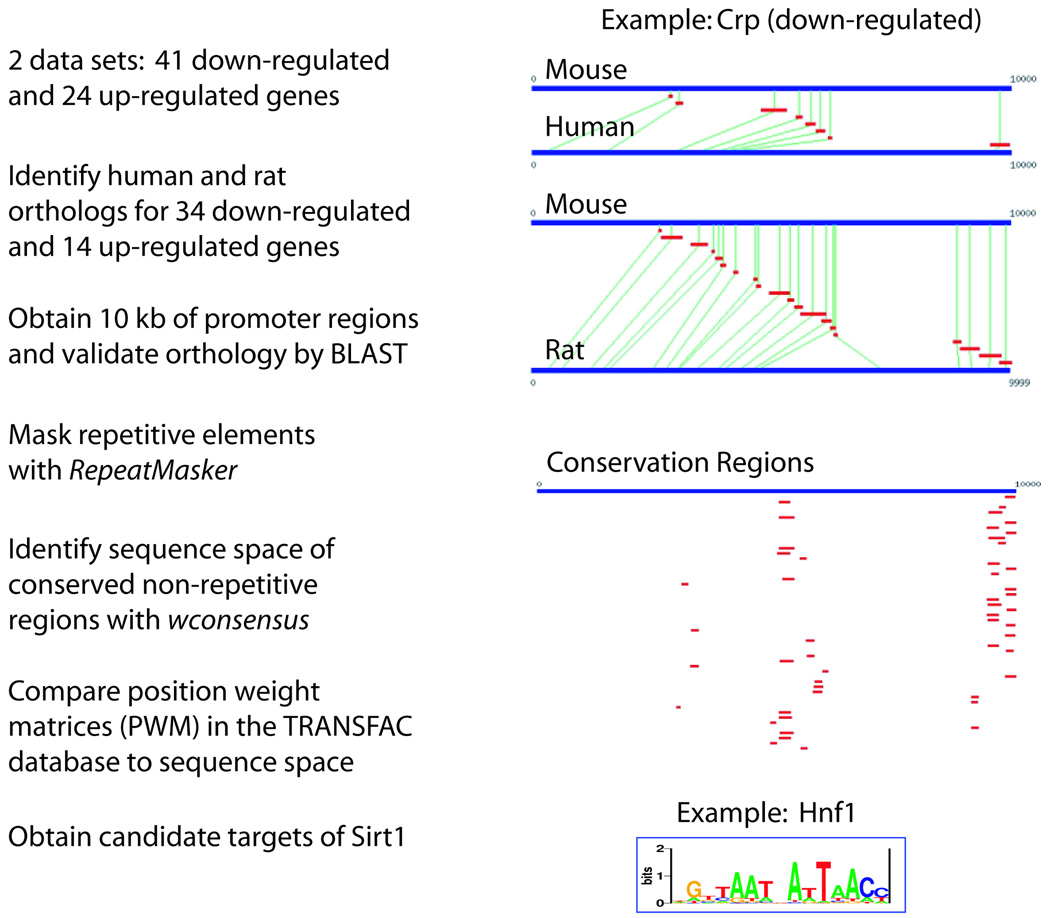
In a previous study, we analyzed gene expression profiles of mouse MIN6 pancreatic β cell lines that stably overexpress Sirt1 (Moynihan et al. 2005). Using these results, we attempted to identify transcription factor partners of Sirt1 through a bioinformatic analysis. The scheme of this analysis is illustrated in Fig. 1. We started with two datasets that we analyzed independently: 41 and 24 mouse genes down-regulated and up-regulated, respectively, by Sirt1 overexpression. We identified rat and human orthologs for 34 down-regulated and 14 up-regulated genes and collected up to 10 kb of promoter sequences and up to 5 kb of 5’ UTRs for each gene from each species. We masked repetitive elements using RepeatMasker (Chen 2004) and used wconsensus (Hertz & Stormo 1999) to identify putative regulatory regions in each promoter that are evolutionarily conserved. Using statistical algorithms previously described (Wang & Stormo 2003; Hu et al. 2004), we then assessed the enrichment of conserved transcription factor binding sites represented as position weight matrices (PWMs) in the TRANSFAC database. Binding sites of 13 transcription factors were found to be enriched in each group. The list of transcription factors’ PWMs, their sequence logos, and the statistical enrichment in the up- and down-regulated gene promoter regions are shown in Table 1. Surprisingly, 6 factors were common to both sets. The likelihood of this overlap occurring by chance is 1.3 × 10−7. This overlap not only validates the datasets as genes coordinately regulated by common transcription factors, but also indicates that Sirt1 affects the activity of individual transcription factors in both positive and negative ways. This phenomenon has previously been observed for the effect of Sirt1 on FOXO3 (Brunet et al. 2004). To verify that the overlapping factors were not identified by a bias in our algorithm, we examined the likelihood of binding of each factor in a similarly sized dataset of random gene promoters and found that the overlapping factors were not enriched (see Table 1).
Figure 1. Strategy to identify transcription factor partners of Sirt1 in mammalian cells.
Based on our previous microarray results from mouse MIN6 pancreatic β cell lines (Moynihan et al. 2005), genes affected by Sirt1 overexpression were grouped into up-regulated and down-regulated gene sets. After orthologous human and rat genes were identified, promoters in rats and humans were aligned with BLAST. An example is shown for the Crp promoter. Evolutionarily conserved motifs were identified with wconsensus after masking repetitive sequences with RepeatMasker. Comparison of the conserved regions to the TRANSFAC database was performed using multiple statistics previously described (Wang & Stormo 2003; Hu et al. 2004). A visual analog of the HNF-1 matrix is shown.
Table 1. Enrichment of transcription factor binding sites in the promoters of Sirt1 target genes.
The names of the transcription factor binding matrices enriched in promoters of the up-and down-regulated genes by Sirt1 are shown in the first column. For each matrix, a sequence logo is shown in the second column. The conventional name(s) of the transcription factors it represents are given in the third column. The score shown for each matrix is the sum of three separate statistics previously described (Wang & Stormo 2003; Hu et al. 2004), and shown only if the matrix is enriched in the respective dataset. The final column shows the scores for the top matrices when compared to a random set of genes, indicating that they were not enriched due to a bias in our statistics.
| Matrix | Logo | Factor(s) | Classification | Group | Enrichment in downregulated genes |
Enrichment in upregulated genes |
Enrichment in random set |
|---|---|---|---|---|---|---|---|
| POU1F1 | Pit-1 | homeodomain | both | 2.62 | 3.64 | 0.19 | |
| ZID | ZID | BTB/POZ | both | 2.43 | 2.82 | 0.11 | |
| NFE2 | NF-E2 | BRLZ | both | 1.82 | 3.01 | 0.29 | |
| OCT | OCT | homeodomain | both | 1.76 | 4.31 | −0.41 | |
| PIT1 | Pit-1 | homeodomain | both | 1.86 | 2.74 | 0.52 | |
| CDC5 | CDC5 | Myb, SANT | both | 1.92 | 2.89 | −0.53 | |
| XBP1 | XBP-1 | BRLZ | down | 3.02 | |||
| HNF1 | HNF-1 | homeodomain | down | 2.76 | |||
| AP1 | AP-1, Jun | BRLZ | down | 2.13 | |||
| HLF | HLF | BRLZ | down | 2.04 | |||
| ICSBP | ICSBP | IRF | down | 1.95 | |||
| CRX | CRX | homeodomain | down | 1.87 | |||
| BACH | BACH1/2 | BRLZ | down | 1.86 | |||
| PAX1 | PAX-1 | PAX | up | 5.20 | |||
| HNF6 | HNF-6a/b | homeodomain | up | 5.20 | |||
| HFH | HFH, FoxJ/I/F | Forkhead | up | 4.50 | |||
| STAT5B | STAT5B | STAT | up | 4.38 | |||
| S8 | S8, Prx2 | homeodomain | up | 3.99 | |||
| NKX2 | NKX2 | homeodomain | up | 3.96 | |||
| FOX | FOX | Forkhead | up | 2.61 |
Interestingly, most of the transcription factors identified in this analysis fall into one of two groups. The first group contains POU domain/homeodomain transcription factors that are important for tissue-specific endocrine regulation and/or cell-type specification (POU1F1/PIT1, OCT, HNF1, HNF6, CRX, and S8), and the second group contains bZIP (a.k.a. beta-rich leucine zipper (BRLZ)) family transcription factors that commonly regulate stress responses, such as anti-oxidant and unfolded protein responses (NFE2, XBP1, AP1, HLF, and BACH) (see Table 1). These findings suggest that Sirt1 regulates metabolism and stress responses, both of which have been implicated to control the process of aging in response to nutrient availability and environmental stress.
Sirt1 and HNF-1α physically interact in nutrient-restricting conditions
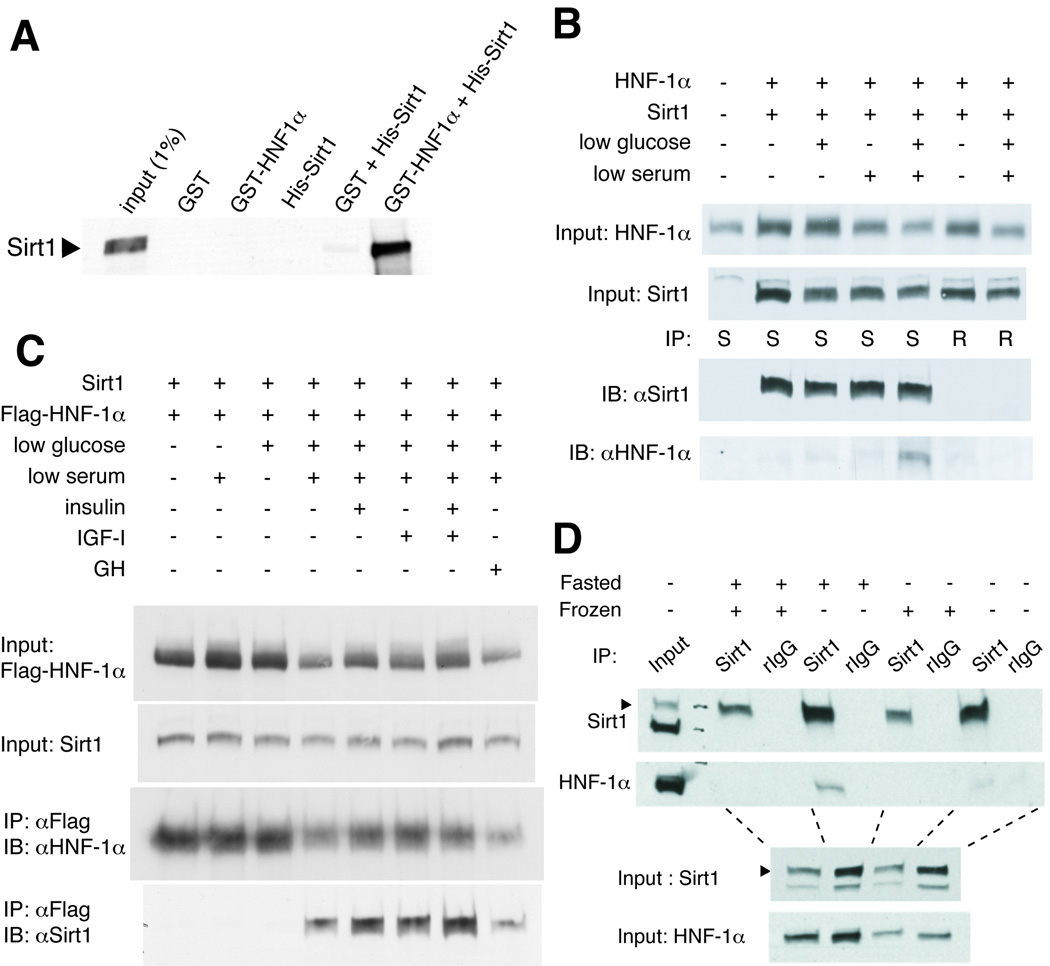
Among those factors that we identified, we were particularly interested in “HNF1” whose binding sites are enriched in the promoters of genes down-regulated by Sirt1. This PWM is common to HNF-1α and β, whose mutations cause maturity onset diabetes of the young (MODY) types 3 and 5, respectively (Shih & Stoffel 2002). Since HNF-1α is more highly expressed in adult tissues and has stronger transcriptional activity (Pontoglio 2000), we examined whether Sirt1 and HNF-1α could directly interact in vitro. We ran purified recombinant His-tagged Sirt1, GST, or GST-tagged HNF-1α alone or together through a glutathione-Sepharose column. While His-Sirt1 alone did not bind to the glutathione-Sepharose beads and bound only very weakly to GST alone, Sirt1 co-purified with GST-HNF-1α, indicating that the interaction of these proteins was specific and direct (Fig. 2A).
Figure 2. Interaction of Sirt1 and HNF-1α under nutrient restriction.
(A) Sirt1 and HNF-1α interact in vitro. His-Sirt1, GST, and GST-HNF-1α were purified from E. coli and combined as indicated. Proteins complexes were purified with glutathione-conjugated sepharose beads, eluted, and immunoblotted for Sirt1. (B) Complex formation of Sirt1 and HNF-1α in cells cultured in the media with low serum and low glucose. HepG2 cells were transfected with the indicated expression vectors and cultured for 24 hours in media with normal (10%) or low (0.5%) serum and/or normal (25 mM) or low (0.5 mM) glucose. Extracts were immunoprecipitated with anti-Sirt1 (S) or control rabbit IgG (R), and the eluted immune complexes were immunoblotted with the indicated antibodies. (C) The nutrient-dependent association of Sirt1 and HNF-1α is not blocked by insulin, IGF-1, or growth hormone. HepG2 cells were transfected with the indicated expression vectors and cultured for 24 hours with normal (10%) or low (0.5%) serum and/or normal (25 mM) or low (0.5 mM) glucose supplemented with insulin (5.73 ng/ml), IGF-1 (114 ng/ml), or growth hormone (GH, 500 ng/ml). Extracts were immunoprecipitated with anti-Flag antibody-conjugated agarose beads, and the immune complexes were immunoblotted for the indicated proteins. (D) Complex formation of Sirt1 and HNF-1α in fasted mouse liver. Mice were fed or fasted for 48 hours and sacrificed. Extracts of fresh or frozen liver samples were immunoprecipitated with anti-Sirt1 or control rabbit IgG (rIgG), and the immune complexes were immunoblotted for the indicated proteins.
Because HNF-1α is normally expressed in both pancreatic β cells and hepatocytes, we further characterized the interaction between Sirt1 and HNF-1α in HepG2 hepatocarcinoma cells and liver. We transfected HepG2 cells with plasmids encoding untagged HNF-1α and Sirt1 and examined whether Sirt1 and HNF-1α can be co-immunoprecipitated. Different from the in vitro results, HNF-1α did not associate with Sirt1 in extracts of cells cultured in regular DMEM media containing 25 mM glucose and 10% heat-inactivated FBS, even when high amounts of both proteins were overexpressed (Fig. 2B). Surprisingly, however, a robust interaction between these two proteins was detected in the extracts of cells cultured in media in which glucose and serum concentrations were reduced to 0.5 mM and 0.5%, respectively, for 24 hours (Fig. 2B). To confirm this interaction, we conducted the reciprocal experiment by using FLAG-tagged HNF-1α and Sirt1. Sirt1 associated with FLAG-HNF-1α weakly when the cells were cultured in media with either low glucose or low serum, and strongly when both glucose and serum were reduced together (Fig. 2C). Identical results were obtained from immunoprecipitations using Myc-tagged HNF-1α (Supplementary Figure 1). These findings indicate that the interaction between Sirt1 and HNF-1α is regulated by a combination of nutritional and hormonal cues. Notably, this interaction was not triggered by increased expression of either protein, as has been previously described between Sirt1 and other binding partners such as PGC-1α (Rodgers et al. 2005).
To confirm the physiological relevance of the interaction between these two proteins, we then co-immunoprecipitated Sirt1 and HNF-1α from whole liver extracts of fed and fasted mice. While no interaction was observed in liver extracts from ad libitum-fed mice, HNF-1α formed a complex with Sirt1 in liver extracts of mice fasted for 48 hours (Fig. 2D). During this experiment, we noticed that freezing liver samples appeared to destroy the Sirt1-HNF-1α complex (Fig. 2D), implying a labile nature of this complex. These results suggest that the interaction between Sirt1 and HNF-1α is also regulated by nutrient restriction in vivo.
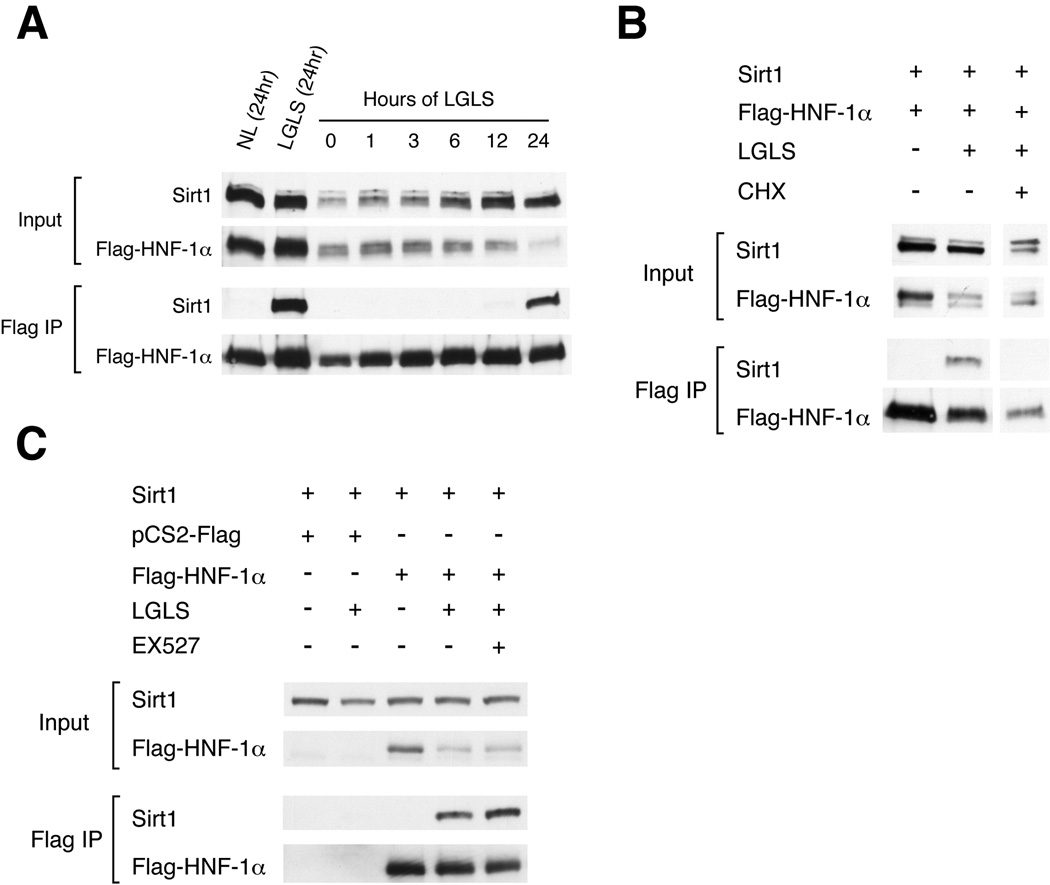
We then attempted to elucidate the mechanism that regulates formation of the Sirt1-HNF-1α complex. We first added back various hormones to the low glucose/low serum culture condition, and repeated the co-immunoprecipitations. None of the factors tested, including insulin, IGF-1, and growth hormone, disrupted the physical interaction of Sirt1 and HNF-1α (Fig. 2C), suggesting that in addition to the reduction of glucose, the reduction of an unidentified factor of the serum triggers the association of Sirt1 and HNF-1α. We also examined the dynamics of the association of Sirt1 with HNF-1α and found that the formation of the Sirt1-HNF-1α complex requires 12–24 hrs after reduction of glucose and serum (Fig. 3A). This extended time frame suggests that the complex formation or stabilization might be dependent on protein synthesis. Indeed, we found that the interaction was prevented by cycloheximide (Fig. 3B), indicating that formation or stabilization of the Sirt1-HNF-1α complex requires de novo synthesis of one or more additional proteins in response to nutrient restriction, although the identity of these proteins is currently unknown. The interaction was not suppressed by blocking Sirt1 enzymatic activity with EX527, a specific chemical inhibitor of Sirt1 (Peck et al. 2010) (Fig. 3C). Furthermore, we did not find alterations in NAD levels or NAD flux in primary hepatocytes cultured in the same conditions (data not shown), suggesting that the interaction was not driven by increased NAD biosynthesis or flux.
Figure 3. The time course of Sirt1/HNF-1α complex formation and its requirement of new protein synthesis.
(A) Time course for the complex formation of Sirt1 and HNF-1α. HepG2 cells were transfected with Sirt1 and Flag-tagged HNF-1α and cultured in media with normal or low glucose and serum (NL vs. LGLS). Extracts were collected at the indicated time points and immunoprecipitated with anti-Flag antibody-conjugated agarose beads, and the immune complexes were immunoblotted for the indicated proteins. (B) Cycloheximide disrupts Sirt1 and HNF-1α complex formation. HepG2 cells were transfected with Sirt1 and Flag-tagged HNF-1α and cultured for 24 hrs in normal or LGLS media with or without cycloheximide (CHX; 10µg/ml). Extracts were immunoprecipitated with anti-Flag antibody-conjugated agarose beads, and the immune complexes were immunoblotted for the indicated proteins. (C) The association of Sirt1 and HNF-1α in cells cultured in LGLS media is not blocked by a specific Sirt1 inhibitor, EX527. Co-immunoprecipitation was conducted as above, with the addition of 10µM EX527.
Sirt1 suppresses HNF-1α-mediated transcriptional activation of the Crp promoter
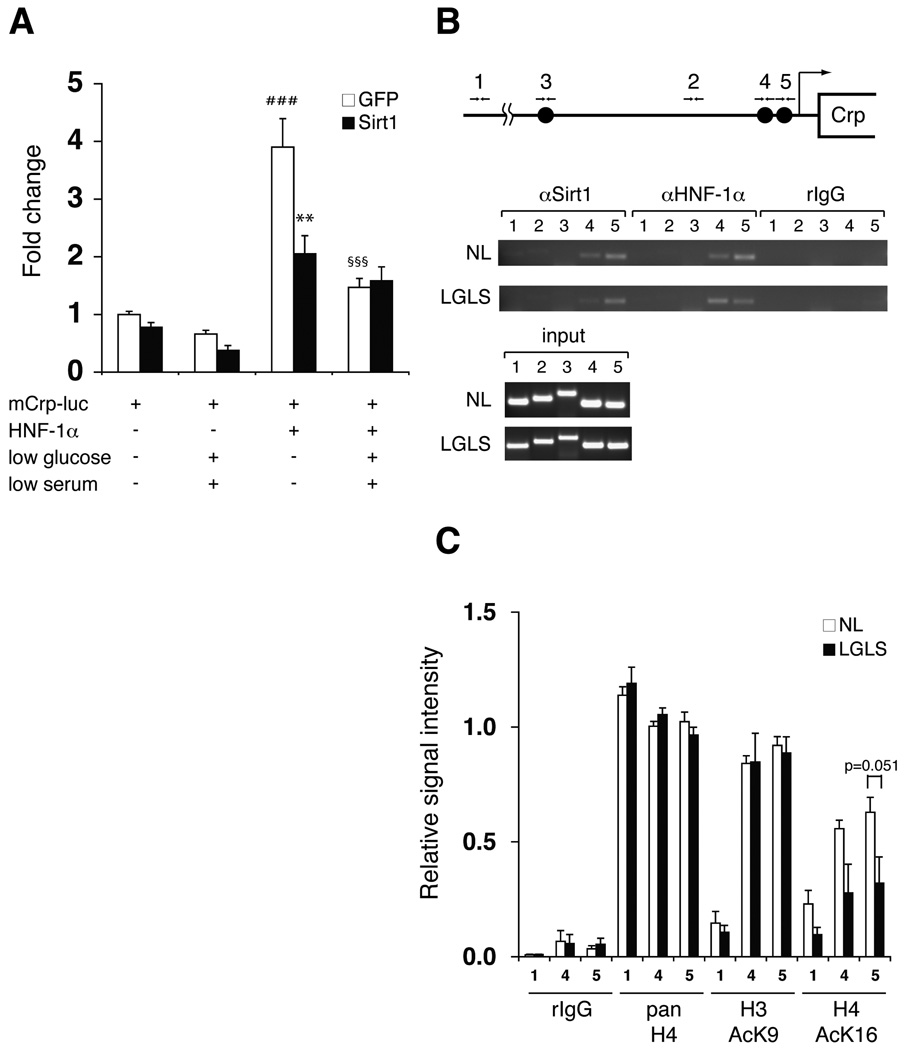
The enrichment of HNF-1 binding sites in the promoters of genes down-regulated by Sirt1 (Table 1) suggests that Sirt1 suppresses HNF-1α-mediated transcriptional activation. To test this hypothesis, we examined the effect of Sirt1 on HNF-1α-mediated transcriptional activation of the mouse C-reactive protein (Crp) gene, which showed the highest enrichment score for the HNF-1 PWM in our bioinformatics analysis (data not shown). We infected mouse primary hepatocytes with Sirt1- or GFP-expressing adenoviruses and then transfected these cells with HNF-1α-expressing or control vectors and a luciferase reporter driven by a ~1.8 kb fragment of the mouse Crp promoter containing three putative HNF-1α binding sites. Under regular culture conditions, HNF-1α robustly activated transcription from the Crp promoter about 4-fold (Fig. 4A). Co-overexpression of Sirt1 significantly suppressed HNF-1α-dependent activation of the promoter in primary hepatocytes even under normal culture conditions, while the reduction of glucose and serum in the media suppressed HNF-1α-dependent activation of the promoter to a similar extent. Interestingly, in this transient reporter assay, this nutrient restriction-dependent suppression was not enhanced further by the simultaneous overexpression of Sirt1, implying that Sirt1 amounts are not limiting in this condition. Taken together, consistent with our bioinformatic analysis, the results clearly show that Sirt1 suppressed HNF-1α-mediated transcriptional activation of its target gene, Crp, in primary hepatocytes.
Figure 4. Suppression of HNF-1α-mediated transcriptional activation of the Crp promoter by Sirt1.
(A) Nutrient restriction and Sirt1 suppress HNF-1α-mediated activation of the Crp promoter. Primary hepatocytes were infected with GFP- or Sirt1-expressing adenoviruses, transfected with pGL3-mCrp-1777 (mCrp-luc) and HNF-1α expression or control vectors, and cultured in normal or LGLS media as indicated for 48 hours prior to extract collection. Luciferase activity was normalized to protein concentration. Results are shown as mean values ± SEM (n=3, ###p<0.001, compared to GFP-expressing cells transfected with mCrp-luc under normal and LGLS media; **p<0.01 (p=0.001), §§§p<0.001, compared to GFP-expressing cells transfected with mCrp-luc and HNF-1α under normal media, by one-way ANOVA with the Tukey-Kramer post hoc test). (B) Chromatin immunoprecipitation (ChIP) for the Sirt1-HNF-1α complex on HNF-1α binding sites in the mouse Crp promoter. Chromatin-DNA adducts were extracted from mouse primary hepatoctyes cultured in normal media (NL) or media with low glucose and serum (LGLS). After immunoprecipitation with the indicated antibodies, DNA was purified and amplified by PCR with the following primer sets: primer sets 1 and 2 for control regions located ~5 kb and 965bp upstream of the transcription start site, respectively; primer set 3 for a region containing a distal HNF-1α binding site located 1.8-kb upstream; primer sets 4 and 5 for regions containing proximal HNF-1α binding sites locate 273 bp and 113 bp upstream, respectively. (C) Quantitation of ChIP for acetylated histones H3 and H4 under normal and nutrient-restricting conditions. Chromatin-DNA adducts were extracted from mouse primary hepatoctyes cultured in normal media (NL) or media with low glucose and low serum (LGLS) and immunoprecipitated with the indicated antibodies. After PCR amplification, band intensities were quantitated and normalized to the signal intensity of each input. Relative signal intensity is shown for primer sets 1, 4, and 5 as mean values ± SEM. The results were assessed by one-way ANOVA with the Tukey-Kramer post hoc test (n=4).
Sirt1 and HNF-1α reside together on proximal HNF-1α binding sites of the Crp promoter
To examine the mechanism of the suppression of HNF-1α-mediated transcriptional activation by Sirt1, we initially evaluated the acetylation status of HNF-1α. Neither acetyl-lysine antibodies nor mass spectrometric analysis could detect acetylated HNF-1α in regular and nutrient-restricting conditions with nicotinamide and trichostatin A (data not shown), suggesting that HNF-1α may not be regulated by acetylation/deacetylation.
Another possible mechanism is that Sirt1 is recruited to the HNF-1α binding sites through its interaction with HNF-1α and deacetylates surrounding histones. To address this possibility, we examined whether Sirt1 and HNF-1α reside together on the mouse Crp promoter using chromatin immunoprecipitation (ChIP) in mouse primary hepatocytes. We found both proteins resided on two proximal HNF-1α binding sites in the mouse Crp promoter under both regular and nutrient-restricting conditions, but not on a distal HNF-1α binding site about 1.8-kb upstream (Fig. 4B). We then conducted ChIP for the same Crp promoter regions with antibodies specific for acetylated histones. Interestingly, the acetylation levels of lysine 16 on histone H4, the evolutionarily conserved target for Sir2 orthologs (Imai et al. 2000), were reduced around the two proximal HNF-1α binding sites in response to nutrient restriction, whereas the acetylation levels of lysine 9 on histone H3 did not change (Fig. 4C). Given that formation or stabilization of the HNF-1α-Sirt1 complex requires de novo protein synthesis, these results suggest that the Sirt1-HNF-1α complex, which might interact with an unknown component induced by nutrient restriction, mediates the suppression of Crp expression by allowing Sirt1 to deacetylate lysine 16 on histone H4 near HNF-1α binding sites on the Crp promoter.
Sirt1 regulates the expression of endogenous HNF-1α target genes in primary hepatocytes
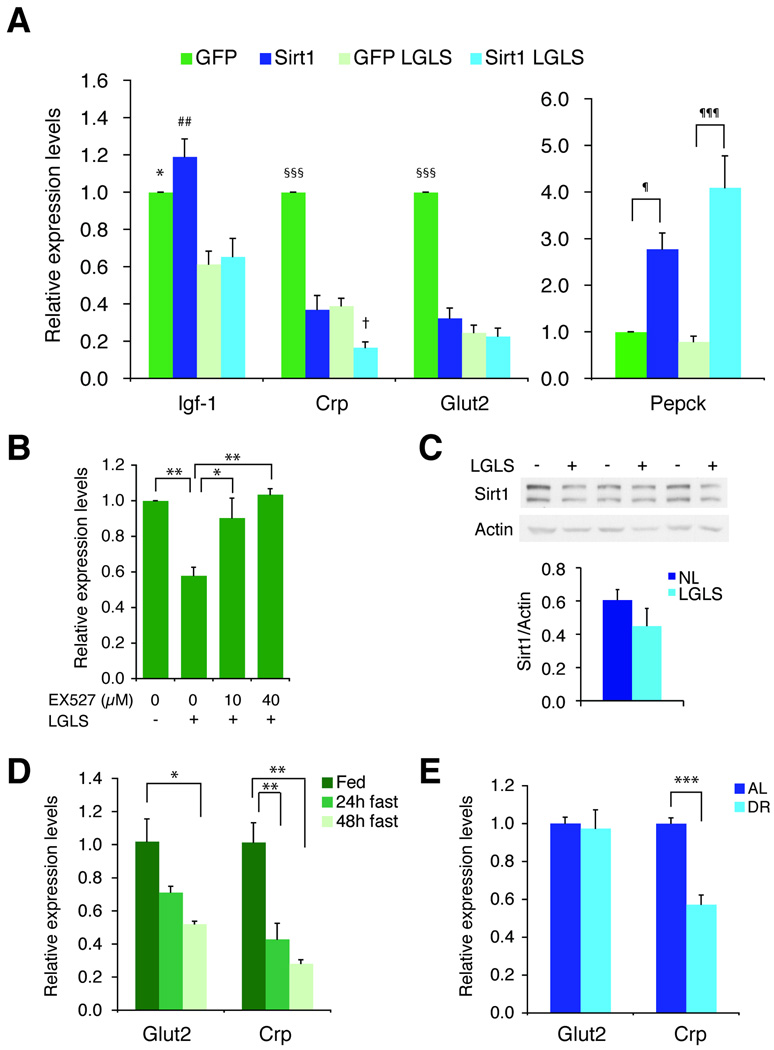
To investigate the functional effect of Sirt1 on endogenous HNF-1α target genes, we measured expression levels of known HNF-1α target genes (Lee et al. 1998; Pontoglio 2000; Shih et al. 2001a; Shih et al. 2001b) in primary mouse hepatocytes infected with GFP- or Sirt1-adenoviruses. The expression of endogenous Crp and Glut2 were suppressed by overexpression of Sirt1, as well as by nutrient restriction to a similar extent (Fig. 5A). Interestingly, an additive effect of Sirt1 plus nutrient restriction was observed for Crp expression, but not for Glut2 expression, in primary hepatocytes. Igf-1 levels were suppressed by nutrient restriction, but not by Sirt1. Contrarily, Pepck levels were up-regulated by Sirt1 as reported previously (Rodgers et al. 2005), but not by nutrient restriction.
Figure 5. The effects of Sirt1 and nutrient restriction on the expression of endogenous HNF-1α target genes in mouse primary hepatocytes.
(A) Sirt1 affects the expression of HNF-1α target genes in mouse primary hepatocytes. Hepatocytes were infected with GFP- or Sirt1-expressing adenoviruses and cultured for 48 hrs in normal or low glucose/low serum (LGLS) media. Gene expression was determined by quantitative RT-PCR and normalized to GAPDH and the respective control treatment. Igf1, insulin-like growth factor 1; Crp, C-reactive protein; Glut2, glucose transporter 2; Pepck, phosphoenolpyruvate carboxykinase 1. Results are shown as mean values ± SD (n=3 for Igf-1, Crp, and Glut2 and n=6 for Pepck, *p≤0.05, ##p<0.01, compared to GFP- or Sirt1-expressing cells under LGLS media [GFP LGLS and Sirt1 LGLS, respectively]; §§§p<0.001, compared to three other conditions [Sirt1, GFP LGLS, and Sirt1 LGLS] for each gene; †p≤0.05, compared to Sirt1 and GFP LGLS; ¶p<0.05, ¶¶¶p<0.001 for the indicated comparisons, by one-way ANOVA with the Tukey-Kramer post hoc test). (B) EX527 blocks suppression of Crp by nutrient restriction. Primary hepatocytes were infected with GFP-expressing adenovirus and cultured for 48 hrs in normal or low glucose/low serum (LGLS) media with the indicated concentration of EX527. Gene expression for Crp was determined by quantitative RT-PCR and normalized to Gapdh and normal culture conditions. Results are shown as mean values ± SEM (n=3, *p<0.05, **p<0.01 for the indicated comparisons by one-way ANOVA with the Tukey-Kramer post hoc test.) (C) Representative western blot of Sirt1 in primary hepatocytes grown under normal culture conditions (NL) or in low glucose and serum (LGLS) for 48 hrs. For quantitation, protein levels of Sirt1 were normalized to those of β-actin. Results are shown as mean ± SEM (n=3). (D) mRNA expression of Glut2 and Crp in fed and fasted mouse liver. The expression of Glut2 and Crp was measured by quantitative RT-PCR normalized to Gapdh. Results are shown as mean values ± SD (n=3, *p<0.05, **p<0.01 by one-way ANOVA with the Tukey-Kramer post hoc test). (E) mRNA expression of Glut2 and Crp in mice fed ad libitum (AL; n=5) or subjected to 60–70% calorie restriction (CR; n=4) for 5 months. Gene expression was normalized to Gapdh. Results are shown as mean ± SD (***, p ≤ 0.001 by unpaired Student’s t test).
To verify that the suppression of Crp by nutrient restriction was mediated by Sirt1, we treated primary mouse hepatocytes with EX527, a highly specific Sirt1 inhibitor (Peck et al. 2010). EX527 abolished the suppressive effect of nutrient restriction on Crp expression in a dose-dependent manner (Fig. 5B), suggesting that Sirt1 activity is required for the suppression of Crp. Because Sirt1 protein levels did not change in primary hepatocytes in response to nutrient restriction (Fig 5C), the observed increase in Sirt1 activity is likely mediated through the nutrient-sensitive formation of the Sirt1-HNF-1α complex.
Crp expression is reduced by fasting and DR
To further determine whether nutrient-sensitive regulation of HNF-1α target gene expression occurs in a physiologically relevant context, we examined whether the expression of Crp and Glut2 in the liver responded to fasting and DR. Consistent with our findings in mouse primary hepatocytes, Crp expression levels were significantly reduced in both fasting and DR (Figs. 5D and 5E). Glut2 expression was also significantly reduced in fasting, but not in DR. These results are also consistent with the observation that circulating Crp levels are reduced in diet-restricted human subjects (Fontana et al. 2004). Together, these results support our conclusion that a nutrient-sensitive interaction between Sirt1 and HNF-1α regulates transcription in response to energy-limiting conditions, such as fasting and DR, in mammals.
Discussion
In this study, we combined gene expression analysis with phylogenetic footprinting to identify transcription factors as candidate targets of Sirt1. We identified HNF-1α, a homeodomain transcription factor that regulates pancreatic β cell and hepatocyte functions (Pontoglio 2000) and is commonly mutated in patients with maturity onset diabetes of the young (MODY) (Shih & Stoffel 2002). Sirt1 physically interacts with HNF-1α in vitro, but the interaction in vivo is induced by nutrient restriction. This nutrient-sensitive interaction is not triggered by alterations in expression of Sirt1 or HNF-1α, but by the induction of an unidentified protein that is likely able to promote formation or stabilization of the Sirt1-HNF-1α complex in response to nutrient restriction. Consistent with the results from our bioinformatic analysis, Sirt1 inhibits HNF-1α-mediated transcriptional activation of the Crp promoter by deacetylating lysine 16 of histone H4 around the proximal HNF-1α binding sites in response to nutrient restriction. Sirt1 also suppresses Crp expression in mouse primary hepatocytes, and nutrient restriction further enhances this effect. Furthermore, pharmacological inhibition of Sirt1 demonstrates that the enzymatic activity of Sirt1 is required for suppression of Crp by nutrient restriction. This nutrient-sensitive suppression of Crp expression is also observed in fasted and diet-restricted liver. These findings reveal a new mode of regulation of Sirt1 function in response to nutrient restriction and demonstrate the importance of the complex of Sirt1 and HNF-1α in transcriptional regulation in response to energy-limiting conditions in mammals.
Significance of the bioinformatic approach
Because Sirt1 does not possess direct DNA binding capability, we hypothesized that gene expression changes caused by Sirt1 are mediated by sequence-specific transcription factors that interact with Sirt1. Approaches to identify regulatory DNA motifs have traditionally analyzed the conservation of a single gene promoter across multiple species, or the overrepresentation of motifs in multiple genes within a single species. By combining gene expression data with phylogenetic footprinting, we were able to use orthogonal information to identify a new protein-protein interaction, namely the Sirt1-HNF-1α interaction, indicating that such methods can be used to elucidate the function of other transcriptional co-regulators that do not directly bind DNA.
The candidate transcription factors identified in this bioinformatic analysis are worth further consideration. Most of those factors are categorized into either a group of POU domain/homeodomain transcription factors that regulate tissue-specific endocrine function or a group of bZIP/BRLZ family transcription factors that regulate stress and damage responses. Intriguingly, these two categories are consistent with two major features of Sirt1 function, metabolic regulation and stress response regulation (Imai & Guarente 2007; Finkel et al. 2009; Haigis & Sinclair 2010). Indeed, the enrichment of forkhead protein or AP-1 binding sites is consistent with the reported role of Sirt1 in regulating the transcriptional activity of FOXO family members (Brunet et al. 2004; Daitoku et al. 2004; Motta et al. 2004; van der Horst et al. 2004; Qiao & Shao 2006) and c-Jun, a component of AP-1 (Zhang et al. 2009; Zhang et al. 2010). It is conceivable that the POU domain/homeodomain and bZIP domain might be common structural motifs that interact with Sirt1 and mediate major aspects of Sirt1 function. Further investigation will be necessary to examine whether other transcription factors in Table 1 also interact with Sirt1 and mediate metabolic and/or stress response regulations in mammals.
Regulation of the interaction between Sirt1 and HNF-1α
We observed that the association of Sirt1 with HNF-1α in cultured cells is dramatically strengthened by reduction of glucose and serum in combination, but not individually, and that the association is also detected in vivo in the liver after 48-hr fast, indicating that the interaction is regulated by alterations in multiple signaling pathways. The time course of the complex formation and the inhibition by cycloheximide indicate that the complex formation likely requires an unidentified protein that is induced by nutrient restriction (Fig. 6). Although this protein has not yet been identified, our findings demonstrate a novel nutrient-sensitive mechanism that integrates nutritional and possibly hormonal signals into Sirt1-mediated transcriptional adaptation to energy-limiting conditions. To attempt to characterize such nutritional/hormonal signals, we tested several different possibilities, including hormones such as insulin, IGF-I, growth hormone, and NAD biosynthesis. Although we identified decreased glucose as one of the required signals, the identity of the other necessary signal(s) is currently unknown. It will be of great importance to uncover the molecular details of this new regulatory mechanism of Sirt1 function.
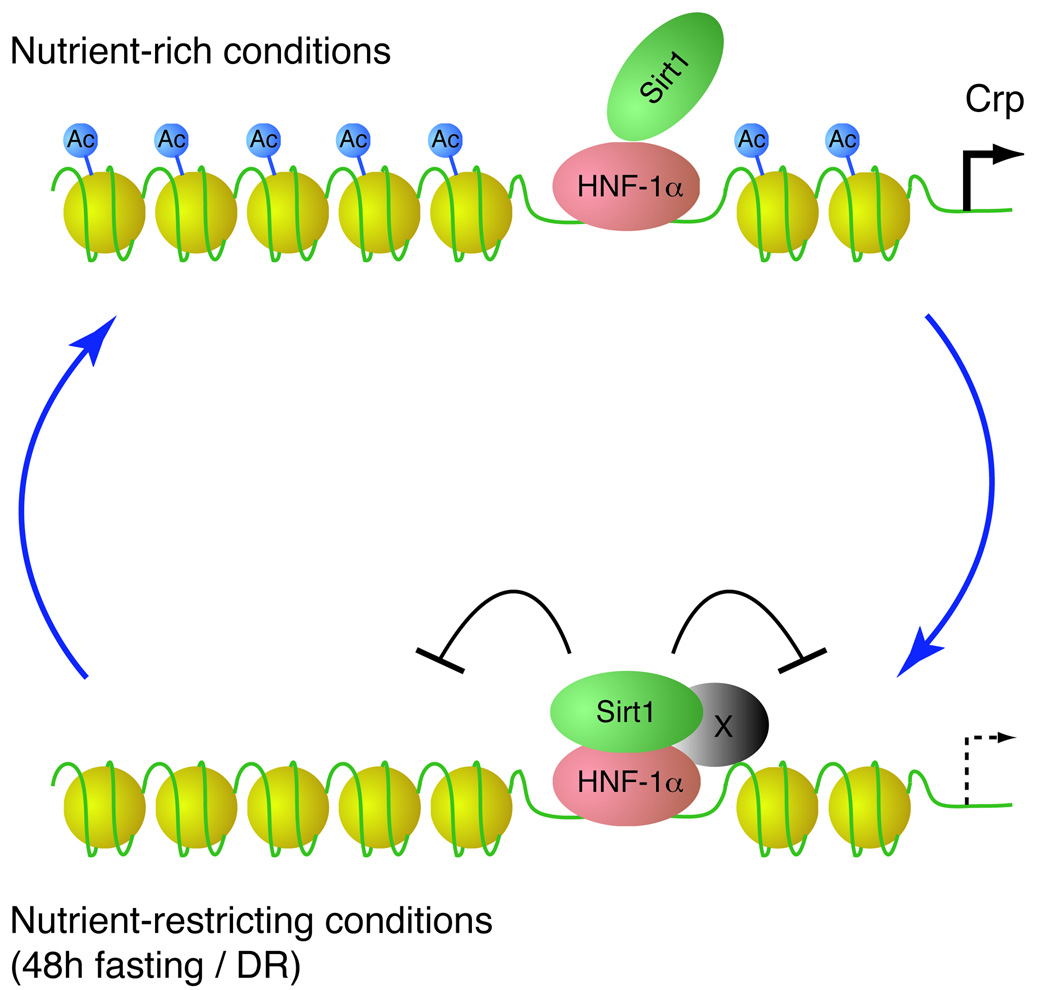
Figure 6. A model for the nutrient-sensitive regulation of gene expression by the Sirt1/HNF-1α complex.
Sirt1 weakly associates with HNF-1α on HNF-1α binding sites under nutrient-rich conditions. Under nutrient-restricting conditions, such as 48h fasting and diet restriction (DR), Sirt1 and HNF-1α become more tightly associated, possibly with an unidentified factor that is induced by nutrient restriction (represented by X). Sirt1 then deacetylates lysine 16 of histone H4, suppressing transcription of HNF-1α target genes, such as Crp.
The physiological importance of Sirt1 in nutrient-sensitive transcriptional regulation
The Sirt1-HNF-1α complex induced by nutrient restriction leads to the transcriptional suppression of the Crp gene through the deacetylation of lysine 16 on histone H4 (Fig. 6). Interestingly, blood CRP levels are known to decline in diet-restricted human subjects (Fontana et al. 2004). Conversely, the high CRP producer phenotype is associated with a shorter life expectancy in humans (Hurme et al. 2007), implying that the CRP gene might be a longevity gene in humans. Although production of Crp has not been well studied in rodents, transcriptional regulatory cis-elements in the Crp promoter, including HNF-1α binding sites, are highly conserved between rodents and humans. Our findings demonstrate that Crp production in mice is also suppressed by nutrient-restricting conditions, such as low glucose/low serum in culture, fasting and DR, through the formation of the Sirt1-HNF-1α complex. By regulating Crp and other HNF-1α target genes, Sirt1 may contribute to the anti-aging effects of DR.
Sirt1 might not be the only sirtuin that could mediate the nutrient-sensitive transcriptional regulation because nutrient-restricting conditions still decrease Crp expression in Sirt1-deficient liver and hepatocytes (data not shown). Although the reason for this observation is currently unclear, we suspect that Sirt6 might be the most likely candidate that could compensate for the lack of Sirt1 in mediating this nutrient-sensitive transcriptional regulation (Kawahara et al. 2009; Zhong et al. 2010). Nonetheless, because Sirt1 and nutrient restriction together function to regulate HNF-1α-mediated transcriptional regulation in primary hepatocytes, Sirt1 must have a role in the regulation of physiological responses to nutrient-restricting conditions in mammals.
In conclusion, the present study provides critical insight into how Sirt1 controls a tissue-specific transcriptional network in response to energy-limiting conditions. Characterization of the signals by which nutrient restriction activates Sirt1 may yield new targets for pharmacologic intervention to mimic the anti-aging effects of DR.
Experimental procedures
Promoter dataset assembly
Orthologous genes were identified by HomoloGene (Wheeler et al. 2006) or by reciprocal best BLAST hit in each species. Mouse, human, and rat promoters were collected from the University of California Santa Cruz (UCSC) genome browser and Ensembl using the RefSeq genes when available or the longest mRNA species to identify the transcriptional start site. 10,000 bases of promoter sequences were collected unless it was interrupted by the coding region of the adjacent upstream gene. Up to 5000 bp of 5’ UTR sequences were included. Repetitive sequences were masked with RepeatMasker (Chen 2004). Conserved and putative regulatory promoter regions were identified by wconsensus (Hertz & Stormo 1999).
Statistical analysis of binding matrices
The TRANSFAC database of binding matrices (release 6.0) was consolidated to include only the highest quality matrices for each mammalian transcription factor. Statistical methods employed to measure enrichment of binding sites using PBAL, PBAY and CHN were previously described (Wang & Stormo 2003; Hu et al. 2004). These statistics were summed to create a score for ranking as shown in Table 1.
Production of recombinant GST-HNF-1α and His-Sirt1 proteins
BL21(DE3)pLysS cells were transformed with His-tagged Sirt1, pGEX-2T, and pGEX-2T-HNF-1α plasmids. Transformed BL21(DE3)pLysS cells were grown overnight at 37 °C in 5 ml of Terrific broth containing 75 µg/ml kanamycin (and 37 µg/ml chloramphenicol for His-Sirt1). Cells were spun down, resuspended in 500 ml of the same media, and grown at 37 °C to an OD600 of 0.7. Recombinant proteins were induced by 1.5 mM isopropyl-D-thiogalactopyranoside (Sigma, MO). After inducing for 1 h at 37 °C, cells were spun down and resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5% glycerol, 1 mM DTT, 1 mM EDTA, 0.1% Triton X-100, complete EDTA-free protease inhibitor tablet [Roche]) with 150 µg/ml lysozyme. Cells were lysed in a French press at 10,000 psi for 2 min, and the lysate was cleared at 10,000g for 15 min. The His-tagged Sirt1 was purified with 2 ml Ni-NTA resin (Qiagen, CA) by binding for 1 h at 4 °C, washing with lysis buffer, wash buffer 1 (lysis buffer with 40 mM imidazole), wash buffer 2 (lysis buffer with 36 mM imidazole) and eluting with 150 mM imidazole-containing buffer. GST and GST-HNF-1α were purified with 500 µl of a 1:1 slurry of glutathione (GSH)-sepharose 4B resin (Amersham) in lysis buffer by binding for 1 h at 4 °C, washing with GST wash buffer (lysis buffer with 10% glycerol), and eluting with GST wash buffer plus 10 mM reduced glutathione. Protein purity and yield was assessed by SDS-PAGE and staining with Coomassie. GST, GST-HNF-1α, and His-Sirt1 were desalted into binding buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% NP-40) using a HiPrep 26/10 desalting column (Amersham).
In vitro interaction
Immediately after purification, 1.4 µg each of GST, GST-HNF-1α, and His-Sirt1 were mixed in various combinations in 2.8 ml of binding buffer for 1.5 h at 4 °C. 100 µl of a 50% slurry of GSH-Sepharose 4B resin in binding buffer was added, and the mixture was rocked overnight at 4 °C. Beads were washed three times with 1 ml binding buffer, once in 1 ml binding buffer containing 2.5 µg/ml BSA, and once more in 1 ml binding buffer, and eluted with 100 µl 2× Laemmli buffer by boiling for 5 min. Eluates were analyzed by western blot.
Co-immunoprecipitation
HepG2 cells were transfected with pSG5, pTS158 (pSG5-HNF-1α) (Divine et al. 2003), pCS2-FLAG-HNF-1α, pBluescript-Sirt1-minigene (Revollo et al. 2004), or pBluescript using Superfect (Qiagen) according to the manufacturer’s protocol. 24 hr later, cells were treated with normal media (DMEM containing 25mM glucose, 10% FBS, penicillin/streptomycin) or low glucose (0.5 mM) and/or low serum (0.5%) media for 24 hr. Cells were harvested in cold PBS, pelleted at 1000 g, and lysed in IP buffer (PBS, 0.5% NP-50, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, 10 µM trichostatin A [Upstate Biotechnology], 10 mM Nicotinamide, 1 mM DTT, complete EDTA-free protease inhibitor tablets [Amersham]) by shearing 5 times with a 27 G needle and 5 times with a 30 G needle. Livers were homogenized in IP buffer. Lysates were cleared by centrifugation at 20,000 g for 5 min at 4°C. Protein concentration was measured by Bradford assay. For Flag immunoprecipitation, extracts were combined with 40 µl of a 1:1 slurry of washed anti-Flag M2-conjugated agarose beads (Sigma) in IP buffer, and rocked overnight at 4°C. For Sirt1 immunoprecipitation, protein A beads were washed 3 times in PBS, blocked 1 hr in 0.5 mg/ml BSA in PBS at 4°C, washed with IP buffer plus 0.1 mg/ml BSA, and resuspended 1:1 in IP buffer plus 0.1 mg/ml BSA. Extracts were pre-cleared with 100 µl of blocked protein A beads for 1 hr at 4°C. BSA was added to the extracts to a final concentration of 0.1 mg/ml. 4 µg anti-Sirt1 (AS-16, Sigma) or non-specific rabbit IgG was added, and the extracts were rocked 1 hr at 4°C. 50 µl of a 1:1 slurry of blocked protein A beads was added to the extract, and the mixture was rocked overnight at 4°C. Beads were washed 5 times in IP buffer (FLAG IP) or IP buffer plus 0.1 mg/ml BSA (Sirt1 IP), and bound proteins were eluted with 3X Laemmli buffer and boiling for 4 min.
Time course and addition of inhibitors
HepG2 cells were transfected with pCS2-FLAG-HNF-1α and pBluescript-Sirt1-minigene as described. 24 hr later, cells were rinsed with PBS and cultured in media containing low glucose/low serum (0.5mM glucose, 0.5% FBS) for 24 hr. EX527 (10 µM final conc, Tocris Bioscience) and CHX (10 µg/ml, Sigma) were added at the start of low glucose/low serum as indicated. For the time course, cells were harvested at 0, 1, 3, 6, 12, and 24 hours after the start of the low glucose/low serum condition. Immunoprecipitation was performed as described.
Western blotting
Western blotting was performed as previously described. Primary antibodies included anti-Sirt1 (1:5000, Upstate Biotechnology), anti-Flag M2 (1:1000, Sigma), and anti-HNF-1α (1:250, BD Biosciences). Secondary antibodies included the horseradish peroxidase-conjugated anti-rabbit IgG (1:10,000, Amersham), and anti-mouse IgM (1:10,000 Amersham).
Hepatocyte isolation and Sirt1 manipulation
Primary hepatocytes were isolated from male mice by hepatic portal collagenase perfusion as previously described (Klauning et al. 1981). Cells were maintained in DMEM (25mM glucose), 10% FBS, and penicillin-streptomycin unless otherwise noted. To overexpress Sirt1, primary hepatocytes were cultured overnight and then infected with Sirt1- or GFP-expressing adenoviruses (Lehman et al. 2000; Alcendor et al. 2004) for one hour. For qRT-PCR, 4–6 hours after infection, cells were rinsed with PBS and cultured in normal (25 mM glucose, 10% FBS) or low glucose/low serum (0.5 mM glucose, 0.5% FBS) media for 48 hr. For EX527 treatment, the inhibitor or its solvent (DMSO) was added to primary hepatocyte cultures at the start of the low glucose/low serum condition.
Luciferase assays
For Sirt1 overexpression, primary hepatocytes were infected with the Sirt1-expressing adenovirus as described above. Cells were subsequently transfected with 0.8 µg of the luciferase reporter pGL3-mCrp-1777 and an HNF-1α expression construct (pTS148) or empty vector control (pSG5) (Divine et al. 2003). 24 hr later, cells were washed with PBS and cultured in normal (25 mM glucose, 10% FBS) or low glucose/low serum (0.5 mM glucose, 0.5% FBS) media for 48 hours as indicated. Luciferase activity was measured with the Luciferase Assay System (Promega) according to the manufacturer’s protocol. Luciferase activity levels were normalized to protein amounts.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as adapted from the Misonix ChIP protocol, 2005. Briefly, protein-DNA complexes were cross-linked by incubating the cells with 5 mM DTBP in PBS on ice for 30 min, quenching with a solution containing 100 mM Tris, pH 8 and 150 mM NaCl, then incubating with 1% paraformaldehyde for 5 min and quenching in 125 mM glycine. Cells were harvested and lysed in sonication buffer for 20 min on ice. Sonication with a Branson 250/450 Sonifier (Branson Ultrasonics Corp) at output 3, 50% duty for a total of 10 min alternating rest sheared the DNA to an average size of ~400 base pairs. After centrifugation, the supernatant was pre-cleared by incubating with salmon sperm DNA/Protein A Agarose beads (Upstate). Immunoprecipitation was performed with anti-Sirt1 antibody (AS-16, Sigma), anti-histone H4 antibody (Upstate), anti-acetyl-Histone H3 (Lys9) antibody (Upstate), anti-acetyl-Histone H4 (Lys16) antibody (Upstate), or rabbit IgG control followed by incubation with salmon sperm DNA/Protein A Agarose beads. Crosslinks were reversed, and immunoprecipitated DNA was precipitated and purified. Standard PCR amplification was performed to amplify Crp promoter regions.
Quantitative real-time PCR
Total RNA was extracted from cultured cells or liver tissue using the RNeasy Kit (Qiagen). cDNA was synthesized using the Omniscript RT Kit (Qiagen). Quantitative real-time PCR was performed and analyzed using an Applied Biosystems 7900HT Real-Time PCR System (Applied Biosystems). PCR conditions were: 10 min at 95°C, then 40 cycles of 15 sec at 95°C, 1 min at 60°C, 30 sec at 72°C. Relative expression levels (normalized to Gapdh) were determined using the comparative CT method.
Animals
Fasting and DR experiments were performed as described previously (Satoh et al. 2010). Animal care and use procedures were approved by the Washington University Animal Studies Committee and consistent with the Washington University Policy on Animal Care.
Statistical analysis
Statistical analyses were performed using unpaired Student’s t test for two groups and one-way ANOVA with the Tukey-Kramer post hoc test for more than two groups. In all analyses, the α level was set at 0.05.
Supplementary Material
Complex formation of Sirt1 and HNF-1α in cells cultured in low serum and low glucose media. HepG2 cells were transfected with the indicated expression vectors and cultured for 24 hrs in media with low serum (0.5%) and/or low glucose (0.5 mM). Extracts were immunoprecipitated with anti-Myc-conjugated agarose beads, and the eluted immune complexes were immunoblotted with the indicated antibodies.
Acknowledgements
We thank Laura Staloch and Ted Simon for pSG5, pTS158 (pSG5-HNF-1α), pGEX-2T and pTS358 (pGEX-2T-GST-HNF-1α), Kristen Kroll for pCS2-myc and pCS2-Flag, Alok Agrawal for pCrp-300, Seung-Soon Im and Yong-ho Ahn for pGT2-1112 and pGT2-166, and Daniel Kelly and Jun-ichi Sadoshima for GFP and Sirt1 adenoviruses. We also thank Zhouji Chen for primary hepatocyte isolation, Ashley Nenninger and the laboratory of David Cistola for their help, and members of the Imai laboratory for their advice and discussions. This work was supported by grants from the National Institutes of Health (HG00249) to G.D.S. and T.W., and the National Institute on Aging (AG024150), the American Diabetes Association, and the Ellison Medical Foundation to S.I. S.I. serves as a scientific advisory board member for Sirtris, a GSK company.
Footnotes
Author contributions
S.I. designed, directed, and coordinated the project; A.A.G and T.W. conducted bioinformatic analyses and assessed the results with G.D.S. and S.I.; A.A.G. and C.S.B designed and performed experiments and analyzed the results with S.I.; The manuscript was written by A.A.G. and S.I. and commented by all other authors.
References
- Al-Regaiey KA, Masternak MM, Bonkowski M, Sun L, Bartke A. Long-lived Growth Hormone Receptor Knockout Mice: Interaction of Reduced IGF-1/insulin Signaling and Caloric Restriction. Endocrinology. 2004;146:851–860. doi: 10.1210/en.2004-1120. [DOI] [PubMed] [Google Scholar]
- Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ. Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Chapter 4. Curr. Protoc. Bioinformatics. 2004;(Unit 4):10. doi: 10.1002/0471250953.bi0410s05. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, De Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divine JK, McCaul SP, Simon TC. HNF-1α and endodermal transcription factors cooperatively activate Fabpl: MODY3 mutations abrogate cooperativity. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G62–G72. doi: 10.1152/ajpgi.00074.2003. [DOI] [PubMed] [Google Scholar]
- Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, van Deursen J, Gores GJ, Chen J, Lou Z, Chini EN. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Invest. 2010;120:545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc. Natl. Acad. Sci. USA. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- Garten A, Petzold S, Korner A, Imai S, Kiess W. Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol. Metab. 2009;20:130–138. doi: 10.1016/j.tem.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang T, Stormo GD, Gordon JI. RNA interference of achaete-scute homolog 1 in mouse prostate neuroendocrine cells reveals its gene targets and DNA binding sites. Proc. Natl. Acad. Sci. USA. 2004;101:5559–5564. doi: 10.1073/pnas.0306988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurme M, Kivimaki M, Pertovaara M, Lehtimaki T, Karhunen PJ, Jylha M, Hervonen A, Eklund C. CRP gene is involved in the regulation of human longevity: a follow-up study in Finnish nonagenarians. Mech. Ageing Dev. 2007;128:574–576. doi: 10.1016/j.mad.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Imai S. Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr. Pharm. Des. 2009a;15:20–28. doi: 10.2174/138161209787185814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S. SIRT1 and caloric restriction: an insight into possible trade-offs between robustness and frailty. Curr. Opin. Clin. Nutr. Metab. Care. 2009b;12:350–356. doi: 10.1097/MCO.0b013e32832c932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Imai S, Guarente L. Sirtuins: A universal link between NAD, metabolism, and aging. In: Guarente L, Partridge L, Wallace D, editors. The Molecular Biology of Aging. New York: Cold Spring Habor Laboratory Press; 2007. pp. 39–72. [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Klauning J, Goldblatt P, Hinton D, Lipsky M, Chacko J, Trump B. Mouse liver cell culture. 1. Hepatocyte isolation. In Vitro. 1981;17:913–925. doi: 10.1007/BF02618288. [DOI] [PubMed] [Google Scholar]
- Lee YH, Sauer B, Gonzalez FJ. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1α knockout mouse. Mol. Cell. Biol. 1998;18:3059–3068. doi: 10.1128/mcb.18.5.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. j. Am. Soc. Nephrol. 2000;11 Suppl 16:S140–S143. [PubMed] [Google Scholar]
- Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/EBPalpha transcriptional complex. J. Biol. Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, Imai S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 2001a;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Screenan S, Munoz KN, Philipson L, Pontoglio M, Yaniv M, Polonsky KS, Stoffel M. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001b;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Stoffel M. Molecular etiologies of MODY and other early-onset forms of diabetes. Curr. Diab. Rep. 2002;2:125–134. doi: 10.1007/s11892-002-0071-9. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J. Biol. Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Wang T, Stormo GD. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Helmberg W, Kapustin Y, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD(+) levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lee SM, Shannon S, Gao B, Chen W, Chen A, Divekar R, McBurney MW, Braley-Mullen H, Zaghouani H, Fang D. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 2009;119:3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen HZ, Liu JJ, Jia YY, Zhang ZQ, Yang RF, Zhang Y, Xu J, Wei YS, Liu DP, Liang CC. SIRT1 suppresses activator protein-1 transcriptional activity and cyclooxygenase-2 expression in macrophages. J. Biol. Chem. 2010;285:7097–7110. doi: 10.1074/jbc.M109.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complex formation of Sirt1 and HNF-1α in cells cultured in low serum and low glucose media. HepG2 cells were transfected with the indicated expression vectors and cultured for 24 hrs in media with low serum (0.5%) and/or low glucose (0.5 mM). Extracts were immunoprecipitated with anti-Myc-conjugated agarose beads, and the eluted immune complexes were immunoblotted with the indicated antibodies.