Abstract
EDA is a facultative type III homology of human fibronectin encoded by an alternative spliced exon. The EDA+ and EDA- mRNA forms show a cell type specific distribution with their relative proportion varying during development, aging and oncogenic transformation. We have previously demonstrated that an 81 bp nucleotide sequence within the exon itself is essential for differential RNA processing. Fine mapping of cis acting elements within this region has been carried out to identify possible target sites for the modulation of alternative splicing. There are at least two short nucleotide sequences involved. Element A (GAAGAAGA) is a positive modulator for the recognition of the exon, its deletion results in constitutive exclusion of the EDA exon. Element B (CAAGG) is a negative modulator for exon recognition, its deletion results in constitutive inclusion of the EDA exon. This bipartite structure of the splicing enhancer is a novel feature of the mammalian exons.
Full text
PDF
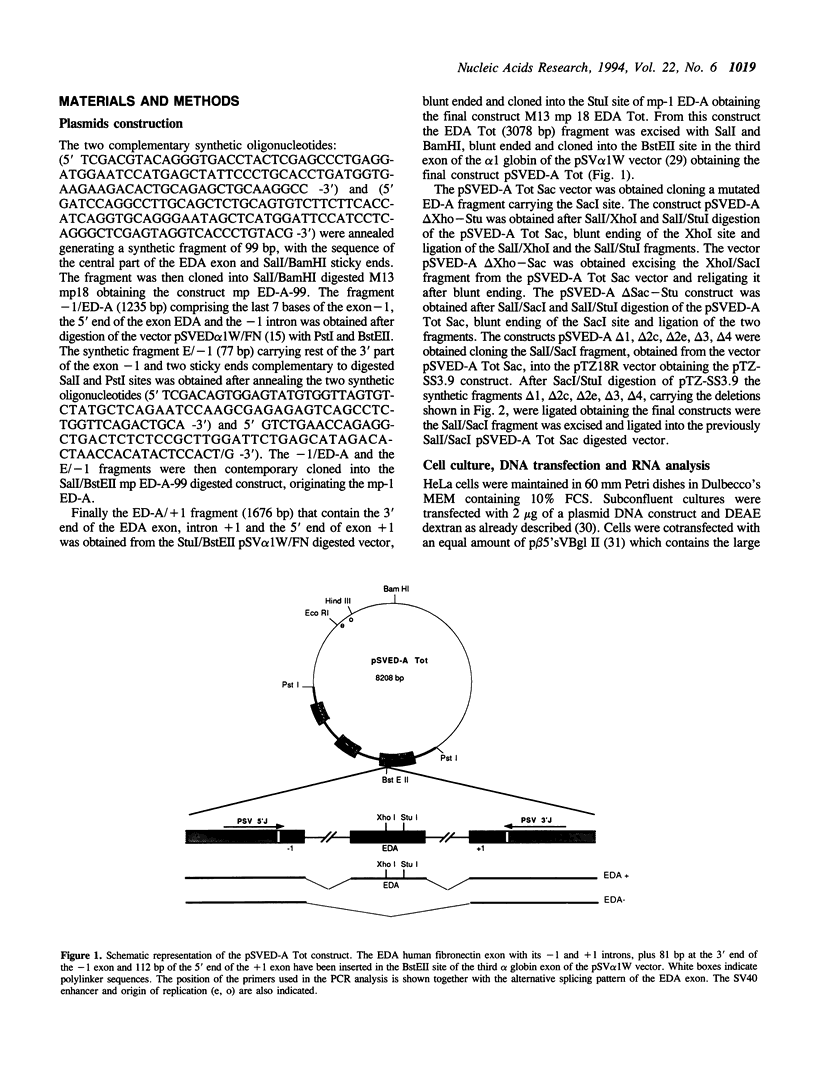

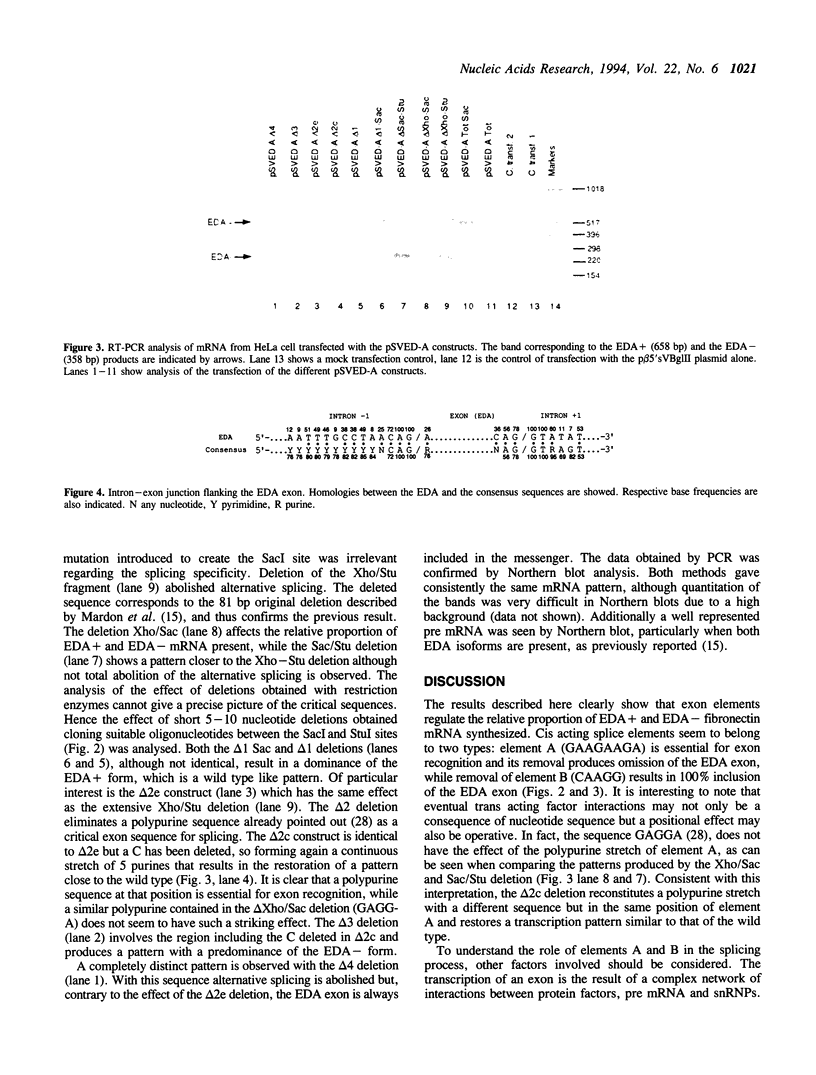

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barone M. V., Henchcliffe C., Baralle F. E., Paolella G. Cell type specific trans-acting factors are involved in alternative splicing of human fibronectin pre-mRNA. EMBO J. 1989 Apr;8(4):1079–1085. doi: 10.1002/j.1460-2075.1989.tb03476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993 Sep 2;365(6441):82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Gibbs R. A., Nguyen P. N., Edwards A., Civitello A. B., Caskey C. T. Multiplex DNA deletion detection and exon sequencing of the hypoxanthine phosphoribosyltransferase gene in Lesch-Nyhan families. Genomics. 1990 Jun;7(2):235–244. doi: 10.1016/0888-7543(90)90545-6. [DOI] [PubMed] [Google Scholar]
- Goux-Pelletan M., Libri D., d'Aubenton-Carafa Y., Fiszman M., Brody E., Marie J. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 1990 Jan;9(1):241–249. doi: 10.1002/j.1460-2075.1990.tb08101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Grosveld G. C., de Boer E., Shewmaker C. K., Flavell R. A. DNA sequences necessary for transcription of the rabbit beta-globin gene in vivo. Nature. 1982 Jan 14;295(5845):120–126. doi: 10.1038/295120a0. [DOI] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Roscigno R. F., Mulligan G. J., Finn L. A., Weber K. S. Identification of two distinct intron elements involved in alternative splicing of beta-tropomyosin pre-mRNA. Genes Dev. 1990 Jan;4(1):98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hoffman B. E., Grabowski P. J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3' splice site by a network of interactions spanning the exon. Genes Dev. 1992 Dec;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Umezawa K., Vibe-Pedersen K., Baralle F. E. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985 Jul;4(7):1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A. R., Vibe-Pedersen K., Baralle F. E. Human fibronectin: cell specific alternative mRNA splicing generates polypeptide chains differing in the number of internal repeats. Nucleic Acids Res. 1984 Jul 25;12(14):5853–5868. doi: 10.1093/nar/12.14.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S., Reed R. A functional association between the 5' and 3' splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993 Jun;7(6):1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolella G., Henchcliffe C., Sebastio G., Baralle F. E. Sequence analysis and in vivo expression show that alternative splicing of ED-B and ED-A regions of the human fibronectin gene are independent events. Nucleic Acids Res. 1988 Apr 25;16(8):3545–3557. doi: 10.1093/nar/16.8.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Patel R. S., Fonda D., Hynes R. O. Multiple sites of alternative splicing of the rat fibronectin gene transcript. EMBO J. 1987 Sep;6(9):2573–2580. doi: 10.1002/j.1460-2075.1987.tb02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993 Jul 16;74(1):105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Valcárcel J., Singh R., Zamore P. D., Green M. R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993 Mar 11;362(6416):171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Vibe-Pedersen K., Kornblihtt A. R., Baralle F. E. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J. 1984 Nov;3(11):2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Tanaka K., Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993 Mar;7(3):407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Neugebauer K. M., Lane W. S., Roth M. B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993 Apr 9;260(5105):219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zardi L., Carnemolla B., Siri A., Petersen T. E., Paolella G., Sebastio G., Baralle F. E. Transformed human cells produce a new fibronectin isoform by preferential alternative splicing of a previously unobserved exon. EMBO J. 1987 Aug;6(8):2337–2342. doi: 10.1002/j.1460-2075.1987.tb02509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



