Abstract
Almost all studies of the immune system of animals with metamorphosis have focused on either larval or on adult immunity, implicitly assuming that these traits are either perfectly correlated or evolutionarily independent. In this study, we use 80 crosses among 21 Drosophila melanogaster lines to investigate the degree and constancy of genetic correlation in immune system activity between larvae and adults. The constitutive transcription of Diptericin, a gene encoding a defensive anti-microbial peptide, was controlled by the same genetic factors in larvae and adults, with variation in expression determined exclusively by non-additive genetic effects. This contrasted with another peptide-encoding gene, Drosomycin, in which larval transcription was highly variable and determined by additive effects but adult transcription genetically invariant. We found no evidence for a fitness cost to the transcription of these genes in our study. The shared genetic control of larval and adult Diptericin transcription stands in contrast to predictions of the adaptive decoupling hypothesis, which states that distinct life-stages should permit the independent evolution of larval and adult phenotypes. Importantly, genetic correlations between larval and adult immunities imply that parasite pressure on one life-stage can drive the evolution of immunity (and resistance) in the other life-stage.
Keywords: Immunity, Life-stage, genetic correlation
Introduction
The insect immune system is currently the subject of intense study, attracting the attention of researchers from fields as diverse as evolutionary ecology, control of insect-vectored human disease, management of agricultural insect pests and fundamental immunology (e.g. Rolff & Siva-Jothy 2003; Bulmer et al. 2009; Vilmos & Kurucz 1998; Welchman et al. 2009). Much of this work has been focused on holometabolous insects (i.e. insects with morphologically distinct larval and adult stages). However, almost all studies only focus on the immune function of one of the life-stages, either the adult or the larva, often because the insect is being used as a model (e.g. for vertebrate immunity) in a context where the totality of the insect life is not relevant or because only one of the two life-stages is “important” in interactions with humans (e.g. only adult mosquitoes transmit malaria, and only larval gypsy moths defoliate forests). Considering each life-stage at the exclusion of the other is reasonable if larval and adult immune phenotypes are genetically independent. However, if immune capacity in larvae and adults were genetically correlated, then selective pressures that are exerted on one life-stage could drive the evolution of immunity in the other life-stage. It would thus not be fully informative to consider each life-stage in isolation.
The adaptive decoupling hypothesis predicts that larval and adult traits should be independent of each other, specifically positing that metamorphosis has evolved to unlink distinct functional and morphologic stages and allow their independent adaptation to distinct environments or niches (Moran 1994). Evidence for, and against, this theory is scarce and inconsistent. On the one hand, one study on tree-frogs has shown that larval and adult size can be genetically correlated (Watkins 2001). On the other hand, theory is supported by the absence of correlation between larval and adult phenotypes for clam shell growth (Hilbish et al. 1993) and heat tolerance in Drosophila buzatii (Loeschcke & Krebs 1996). In this latter desert-inhabiting species, the authors showed that artificial selection for increased resistance to heat in one life-stage had no influence on the evolution of the same trait in the other life-stage. To our knowledge, there has to date been no study investigating the evolutionary genetic relationship between immunity in larvae and adults. Collecting such data is however critical: genetic links, by coupling the evolution of larval and adult resistance, could have broad consequences for epidemiology and the evolution of the interaction between the host and its numerous parasites.
It is unclear whether we should expect larval and adult immunity to be controlled by the same genetic factors. One the one hand, the traits of Dipteran larvae and adults -including immunity – can be genetically correlated. Indeed, selecting lines of Aedes aegypti mosquitoes for faster larval development also leads to the evolution of lower adult immune capacity, as revealed by the decreased ability of adult females to melanize sephadex beads (Koella & Boete 2002). On the other hand, we know in Drosophila melanogaster at least one gene involved in larval immune signaling is dispensible in adults (Petersen et al. 1999). Bacterial challenge does not elicit the production of antimicrobial peptides (AMPs, effector molecules that defend insects against bacterial and fungal pathogens (Lemaitre & Hoffmann 2007)) in D. melanogaster larvae mutant for the transcription factor Serpent (Petersen et al. 1999). However, the same mutation does not prevent adults from mounting an immune response, illustrating a genetic difference in the regulation of immune systems of adults and larvae. Here, we investigate potential phenotypic differences in the genetic architecture of larval and adult immunity in Drosophila melanogaster using controlled crosses and a quantitative genetics approach. This methodology is not only powerful at identifying the constraints that determine evolutionary trajectories, but also quantifies the non-additive genetic components of trait control (i.e. the phenotypic effects attributable to specific combinations of alleles), and therefore informs on the potential for response to selection (Lynch & Walsh 1998).
In this work, we followed a half-diallel design (Lynch & Walsh 1998) crossing males from 5 lines to females from 16 other lines. Larval and adult immune gene expression was tested in the progeny (F1) of each of these 80 crosses. We assayed constitutive immune system activity (i.e. in the absence of experimental immune challenge) in flies by measuring transcription levels of genes encoding two AMPs, Diptericin A and Drosomycin (as in Fellous & Lazzaro 2010). The expression of these two peptide genes during infection is regulated by the two major humoral immune signalling pathways in Drosophila, the Toll and Imd pathways (Lemaitre & Hoffmann 2007). Although we are measuring constitutive expression of the two genes, their expression levels should give a general inference of antimicrobial immune levels. Transcription levels of defense genes are thought to reflect immune system activity and, as such, are a frequently used measure of invertebrate immune quality (e.g. Peng et al. 2005; Freitak et al. 2007; Wigby et al. 2008). Upon bacterial infection, D. melanogaster genotypes that express constitutively express antimicrobial peptide genes at the highest level are the best at controlling bacterial proliferation (Sackton et al. 2009) and, in Aedes aegypti, the constitutive up-regulation of several immunity genes associates with better resistance to bacterial and filarial pathogens (Kambris et al. 2009). By comparison with other immunological techniques, assaying constitutive immunity by measuring AMP gene expression in our study has the benefit that it can be performed in an identical fashion in adults and larvae. To test the hypothesis that investment into the immune system may come at a cost expressed in other traits (Zuk & Stoehr 2002), we also measured two life-history traits. We recorded the speed of larval development - using the age at adult emergence - and the size of the adult by individual dry weight.
Materials and Methods
The 21 isofemales lines of Drosophila melanogaster that we used for the crosses were founded from 21 single females caught in the Ithaca, NY, USA, region 4 years before the beginning of the experiment (c. 100 generations of inbreeding). Following a half-diallel design, we crossed males from 5 lines with females from 16 lines (i.e. 80 distinct crosses) and recorded the phenotypes of their F1 offspring. All F1 offspring in this crossing structure are completely outbred. We took great care to keep the rearing conditions of these 21 parental lines identical and un-crowded during the 2 generations prior to the experiment. For each cross, we put together 7 or 8 virgin males and 13 to 15 virgin females that were 3 days old (±1 day). These adults were kept together for 24h on standard medium (defined below) before they were allowed to lay eggs on grape-juice medium for 24h. This medium stimulates oviposition and, because of its dark color, allowed us to easily spot and capture single first-instar F1 larvae. After 24h on the grape-juice medium, parents were put back on standard yeast medium where they remained for 2 days until they were offered the opportunity to lay eggs on grape-juice medium for another 24h. We collected the F1 larvae one day after the end of oviposition period and transferred groups of 20 larvae into vials containing standard medium. In total, we collected 8 groups of 20 larvae per cross, which were placed on 8 trays that each contained 1 vial of larvae from every cross. These trays will be subsequently referred to as “replicate” trays. Three of these replicate trays were used to assay larval immune gene expression, three were used to assay adult immune gene expression and 2 were used to assay the life-history traits.
From the vials that were devoted to the study of larval immunity, we collected 6 foraging, late third-instar larvae. In the vials that were devoted to the study of adult immunity, 6 females were sampled 5 days after the emergence of the last adult in the vial. Immune phenotypes were measured in unsexed larvae and exclusively female adults. This design is conservative with respect to discovering phenotypic correlations across life-stages if there are sex differences in immune gene expression and has minimal impact on our results and conclusions. Larvae and adults were frozen at −80°C less than 30 minutes post-sampling. The vials devoted to life-history trait measurement were checked up to three times every day for the presence of newly emerged adults. All adult males and females from each vial were transferred to a new vial containing fresh media upon emergence, such that all adults were housed with their vial-mate siblings for the duration of the study. We froze (−20°C) 5 males and 5 females from each of these vials 5 days after the emergence of the last adult. These adults were then dried overnight in a oven at 60°C. Groups of males and females from the same vial were weighed separately to the nearest 0.0001 mg so as to estimate the average individual dry weight per sex. Throughout the experiment, all flies were kept in an incubator at 21°C, with 12h of light per 24h, on standard food medium containing 0.059 g of glucose, 0.0785g of dead yeast, 0.0094 g of agar and 0.94 mL of water per mL of medium, as well as 0.3% propionic acid and 0.03% phosphoric acid to inhibit microbial growth (Fellous & Lazzaro 2010).
Quantification of gene transcription
We used quantitative PCR (qPCR) to quantify the constitutive expression of Diptericin and Drosomycin, standardized to the expression of a housekeeping gene, rp49. We used standard protocols for RNA extraction, reverse transcription, and qPCR amplification (Fiumera et al. 2005; McGraw et al. 2007). Briefly, total RNA of each group of larvae or adults collected from a given vial was extracted in 96-well plates using RNeasy 96 kits (Qiagen), and oligo-dT primed cDNA was synthesized by reverse transcription (reagents from Promega). This cDNA served as the template for the qPCR reaction. AMP transcript abundance was measured on an ABI Prism 7000 (Applied Biosystems) according to the manufacturer’s protocols, with amplification products visualized using fluorescent probes (Diptericin A) or SYBRgreen (Drosomycin and rp49). Gene expression was quantified as a function of the number of PCR cycles necessary to reach a fluorescence threshold (CT value; see Statistical Methods below). The amplification plot of every reaction was carefully examined; we excluded reactions whose amplification slopes indicated inefficient amplification (Lutfalla & Uze 2006). The final data set consisted of 348 reactions for Diptericin, 453 for Drosomycin and 474 for rp49. The primer sequences used in qPCR amplification are as follows. Drosomycin: Forward 5′ CTGCCTGTCCGGAAGATACAA 3′, Reverse 5′ TCCCTCCTCCTTGCACACA 3′; these primers are specific to the Drosomycin gene and would not amplify any Drosomycin paralogues (numbered from 1 to 6) (Tian et al., 2008). Diptericin: Forward 5′ GCGGCGATGGTTTTGG 3′, Reverse 5′ CGCTGGTCCACACCTTCTG 3′ and Probe 5′ TTTGCAGTCCAGGGTC 3′; these primers are specific to the Diptericin A gene and would not amplify its Diptericin B paralogue. Rp49: Forward 5′ AGGC CCAAGATCGTGAAGAA 3′, Reverse 5′ GACGCACTC TGTTGTCGATACC 3′. Dissociation curves were recorded for qPCRs of all three genes to ensure that nonspecific priming and amplification was not occurring.
Statistical methods
In our first approach to the data, we analyzed each trait separately. We then focused on the links between traits. For immunity traits, our response variables were the CT values of Diptericin and Drosomycin after removing the effects of experimental variables that were not of biological interest (e.g. RNA extraction efficiency). To achieve this, we first modeled immune gene expressions using as explanatory factors the CT of the housekeeping gene rp49, the identity of the replicate tray, the identity of the RNA extraction plate, the life-stage and the interaction between the life-stage (larvae or adults) and the CT of the housekeeping gene. Replicate tray and plate identities were treated as random factors. This first model accounts for stochasticity and non-biological variance in the data collection by allowing us to scale nucleic acid extraction yield using the expression of a housekeeping gene (similar to the ΔΔCT method (Lutfalla & Uze 2006)) and allowing the simultaneous control of experimental covariates that were not of biological interest. We used the residuals from these initial models in all further analyses of genetic variation in antimicrobial gene expression. To test for genetic contributions to immune gene expression phenotypes, these residuals were then analyzed as the response variable in a series of mixed-effect models (REML method) that contained the identity of the maternal and paternal lines, the life-stage at which the trait was measured, and all interactions among these factors. More complex models evaluating genetic effects and experimental covariates on raw expression data yielded results similar to those obtained with the approach described here (data not shown). The life-history traits of individual age at emergence and adult dry weight were analyzed with similar models using raw data (i.e. not residuals). These models also included the identity of the vial and the replicate tray in which the flies had developed, as well as the sex of the individual and the interactions of sex with parental genotype. Maternal and paternal factors and all interactions that contained them were treated as random factors, their significance was estimated by comparing the difference of the −2 log-likelihood of the models with and without the factor to a chi-square distribution with one degree of freedom (Pinheiro & Bates 2000; Crawley 2002). The significances of fixed factors were tested with F tests. Homoscedasticity and normality of the residuals were confirmed to comply with the assumptions of analysis of variance.
We were specifically interested in testing the genetic correlation in immune traits across life-stages. To determine the genetic correlation between larval and adult expression of Diptericin A we followed Astles et al. (2006) method #5 (Astles et al. 2006) (Eq. 1). We calculated the ratio of the genetic variance components explaining gene expression across life-stages (i.e. the effects of the maternal and paternal lines and their interaction) divided by the variance components of the interactions between life-stage and the genetic factors.
| (1) |
We calculated separately the inter-life-stage correlation based on pure additive genetic effects (i.e. effects of the maternal and paternal lines only) and the correlation including non-additive genetic effects (i.e. including the interactions between maternal and paternal lines). We did not calculate similar inter-life-stage correlation for Drosomycin expression because we found no detectable genetic variation for this trait at the adult stage.
In order to test for correlations among all traits, including between life history and immune traits, and between larval and adult expression of Diptericin, we additionally used family means and the framework of the linear model. We first calculated the full-family means for larval expression of Diptericin and Drosomycin, adult expression of Diptericin, male and female age at emergence, and individual dry-weight. For each relationship tested, we first regressed full-family means of trait A to trait B (model: Trait B = Trait A). This analysis had the advantage of being statistically powerful and takes into account non-additive components of phenotypic variance. However, it may introduce of form of pseudo-replication when additive effects are involved because distinct crosses involve the same maternal or paternal lines. When the first model was significant we therefore carried out a second, more conservative analysis: we included in the model the identities of the maternal and paternal lines, but not their interaction, as random factors (model: Trait B = Maternal line + Paternal line + Trait A). This model reveals the additional variance in Trait B explained by Trait A after the additive effects of the parental lines on Trait B are taken into account.
All analyses were carried out with JMP 6.0.3 (SAS Institute 2006).
Results
a. Analysis of single traits
Immune gene activity
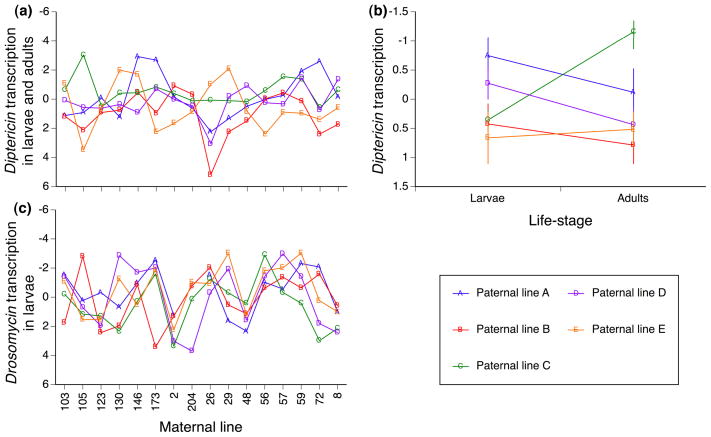
Among the 80 crosses, mean Diptericin transcription varied by 8 Ct units in larvae and 7.5 Ct units in adults indicating an approximate 250 fold variation in gene activity. Gene expression was most strongly predicted by the interaction between the maternal and the paternal lines (Table 1, Fig. 1a), which explained 10.5% of the total variance and more than 50% of the genetic variance. The influence of the parental genotype combination was similar on larvae and adults, as indicated by the non-significant three-ways interaction with life-stage (p> 0.1). There were only mild main effects of the maternal and paternal lines that were consistent over life-stages: these two terms respectively explained 3.7% and 1.4% of the genetic variance (proportion of the phenotypic variance that is explained by genetic factors). However, the influence of paternal genotype on larvae and adults was not always the same, as shown by the significant interaction between paternal line and life-stage (Table 1, Fig 1b). This interaction is driven by a single parental line “C”, whose larval transcription was approximately twice that of adults (CT value lower by 1.1 units).
Table 1.
Final statistical models for the constitutive transcription intensity of immunity genes, the age at emergence and dry-weight. Strongly non-significant terms (P> 0.1) are not shown unless they are involved in higher-order interactions or control for the structure of the dataset (i.e. vial and replicate tray in the analyses of age at emergence and dry-weight). For Drosomycin expression, the main effect of the maternal line could not be calculated because of the great difference in genetic variances between larvae and adults.
| Trait | % Variance explained | D.F. (Num, Denom) | Test-statistic | P value | |
|---|---|---|---|---|---|
| Diptericin A transcription | Random factors | χ2 | |||
| Maternal line | 0.72 | 1 | 1.85 | 0.17 | |
| Paternal line | 0.26 | 1 | 7.20 | 0.007 | |
| Maternal * Paternal lines | 10.5 | 1 | 4.90 | 0.027 | |
| Paternal line * Life-stage | 7.2 | 1 | 7.19 | 0.003 | |
| Fixed factors | F | ||||
| Life-stage | 1, 4.4 | 0.05 | 0.84 | ||
| Drosomycin transcription | Random factors | χ2 | |||
| Maternal line | 0 | n.a. | |||
| Maternal line * Life-stage | 14.5 | 1 | 16 | <0.0001 | |
| Fixed factors | F | ||||
| Life-stage | 1, 15 | 0.0019 | 0.97 | ||
| Age at emergence | Random factors | χ2 | |||
| Vial | 23 | ||||
| Replicate tray | 9.81 | ||||
| Maternal line | 0.002 | 1 | 0.12 | 0.73 | |
| Paternal line | 3.91 | 1 | 10.8 | 0.001 | |
| Maternal line * Sex | 1.54 | 1 | 16 | <0.0001 | |
| Fixed factors | F | ||||
| Sex | 1, 15 | 146.2 | <0.0001 | ||
| Dry-weight | Random factors | χ2 | |||
| Vial | 6.72 | ||||
| Replicate tray | 0.63 | ||||
| Maternal line | 12.4 | 1 | 20 | <0.0001 | |
| Paternal line | 15.4 | 1 | 53 | <0.0001 | |
| Maternal line * Sex | 11.6 | 1 | 21 | <0.0001 | |
| Paternal line *Sex | 19.4 | 1 | 46 | <0.0001 | |
| Fixed factors | |||||
| Sex | 1, 5.5 | 436 | <0.0001 | ||
Fig 1.
effects of the genetic factors (maternal and paternal lines) on the offspring phenotypes for (a) Diptericin transcription averaged over larvae and adults, (b) Diptericin transcription averaged per paternal line and life-stage, (c) Drosomycin transcription in larvae. Diptericin transcription is significantly predicted by the interaction between maternal and paternal lines and the interaction between the paternal line and the life-stage. Drosomycin transcription in larvae is significantly predicted by the maternal line; Drosomycin transcription in adults is not shown as it was not genetically variable. Symbols indicate mean CT values corrected for the effect of a house-keeping gene and other experimental covariates (see methods); vertical bars in panel b indicate standard errors.
Drosomycin transcription patterns in larvae and adults were very different. The sole significant explanatory factor for variation in this trait was the interaction between the maternal line and the life-stage in which transcription was measured (Fig 1c, Table 1). Mean CT values for the progeny of each maternal line ranged over 7 CT units in the larvae indicating an approximate 125 fold variation in gene activity. But, variation in the adult did not exceed 1.5 CT units, 2.8 fold variation in transcript level. Indeed, there was no significant genetic variation among families in adult expression of Drosomycin (P> 0.1). Larval expression was significantly predicted by maternal line (χ2= 13.8, df= 1, P= 0.0002), but not paternal line (P> 0.1). Such difference between the effects of parental lines could in principle be due to “maternal effects” (phenotypic differences due to environmentally mediated variation in maternal provisioning) or to maternal genetic influence. Since we carefully controlled the maternal and grand-maternal environments the contribution is more likely to be genetic. The general conclusions of our study are unchanged regardless of the mechanism for the maternal contribution.
Life-history traits
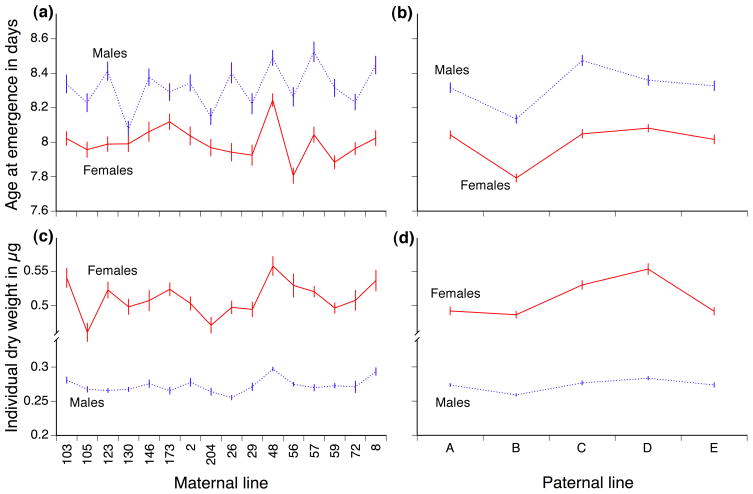
Of the 3200 first instar larvae that were transferred to the vials dedicated to life-history measurements, 3005 emerged as adults. Among the different crosses, the mean age at emergence varied from 7.8 to 8.9 days for male offspring and from 7.4 to 8.4 days for female offspring (Fig 2a). The paternal and the maternal line both influenced the speed of larval development, but there was no interaction between these two genetic factors, indicating that the primary genetic effects are additive. Maternal genotype had different effects on the development speed of male and female offspring, as shown by the significant interaction between maternal line and sex (Table 1, Figure 2a). This interaction was mostly due to one maternal line (line 130) whose male progeny developed particularly rapidly, although when this line is excluded the interaction is still significant (χ2= 4.05, df= 1, P= 0.044). Paternal genotype affected both male and female offspring equivalently. Together, vial identity and replicate tray explained 33% of the total phenotypic variance, but genetic factors explained only 5.5%, showing the importance of the microenvironment for age at emergence.
Figure 2.
effect of the paternal and maternal lines on males (dashed lines) and female (continuous lines) offspring for (a and b) age at emergence (c and d) adult dry weight. The interaction between the maternal and the paternal lines was never significant, but the interactions between parental lines and offspring sex were most often significant (see Table 1). Plotted values are family means; vertical bars indicate standard errors.
The mean dry weight of the flies from the different crosses varied from 0.24 to 0.32 μg for the males and from 0.42 to 0.62 μg for the females. The trait was significantly influenced by both maternal and paternal genotypes additively (Table 1), although the interaction between the maternal and paternal line was not significant (P> 0.1). Both maternal and paternal genotype significantly interacted with the sex of the offspring in predicting mass, showing that these additive genetic components had different effects on male and female offspring. Overall, genetic components explained about 60% of the total variance in offspring mass while vial and replicate trays only explained 7%, indicating that our experimental structure was well able to detect genetic effects over minor microenvironmental variations.
b. Correlations in immune gene activity across life-stages
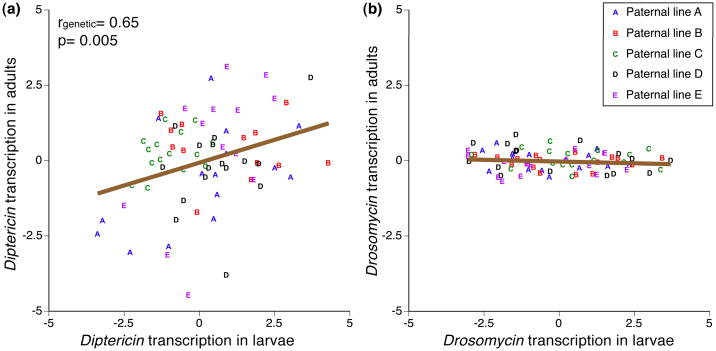
To determine whether adult and larval transcription of Diptericin were significantly correlated, we followed the methodology described by Astles et al. (2006). We quantified the genetic correlation between larval and adult constitutive expression of this gene in two fashions: we first included all genetic components (additive, dominance and epistasis) then restricted the analysis to additive effects only. We found a strong inter life-stage correlation r= 0.65 when all genetic components were included in the analysis (Fig 3a). However, the correlation was only r= 0.091 when we only included additive genetic factors, as expected from the highly significant interaction between maternal and paternal genotype in predicting Diptericin expression described above. We continued this analysis by using full-family means and linear regressions. The regression of full-family means of adult expression against larval values indicated a significant relationship (F1,68 = 8.61, P= 0.0047), confirming the genetic correlation observed above. A second, more conservative model that included maternal and the paternal genotypes as random effects (see the Statistical Methods section of Materials and Methods) gave a similar result (F1,68= 8.62, P= 0.0045), and confirmed the importance of non-additive factors in the correlation (P> 0.1 for maternal and paternal lines). Because of the lack of genetic variation for adult expression of Drosomycin (see above), it was not possible to perform a similar analysis of this trait.
Fig 3.
relationship between larval and adult transcription of (a) Diptericin and (b) Drosomycin. Symbols indicate the mean CT value of the offspring from the crosses corrected for experimental covariates. Larval and adult Diptericin transcription levels were significantly correlated, the correlation coefficient (rgenetic) is obtained from Eq. 1, p-value is from the linear regression between larval and adult values. Because adult Drosmycin transcription was not genetically variable its correlation with larval transcription was not calculated.
c. Correlations among distinct traits
Constitutive expression of both Diptericin and Drosomycin were significantly variable in larvae. We tested whether larval expression of these genes was genetically correlated, as might be expected if the genes were coordinately regulated or expressed in the same tissues. We used full-family means for this analysis, but found no relationship between these two traits (P> 0.1).
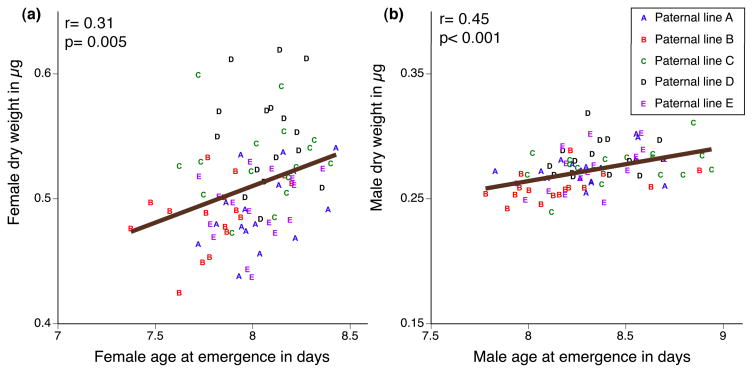
In insects, slower developing larvae generally produce larger, heavier adults. This expected relationship holds true in our experiment, as age at emergence and adult dry weight were positively correlated for both males and females (F1,78= 19.2, P< 0.001; F1,78= 8.57, P= 0.0045) (Fig 4a and 4b). For males, the regression remained significant when the parental genotypes were included in the analysis (F1,70= 10.8, P= 0.002), although this was not true for females (F1,66= 0.76, P= 0.38). Maternal and paternal lines were significant predictors of size and development time in offspring of both sexes (χ2> 3.9, df= 1, P< 0.05).
Fig 4.
relationship between age at emergence and adult dry weight for (a) females and (b) males. Symbols indicate mean offspring phenotype for each of the crosses. The coefficients of correlations (r) reported are obtained from the linear regression of dry weight to age at emergence.
It has been frequently hypothesized that constitutive expression of immune genes comes at a cost to other life history traits (Zuk & Stoehr 2002). To test this, we investigated the links between transcript numbers and our two life-history traits. The only significant relationship we discovered was a weak correlation between the dry weight of male offspring against the larval expression of Drosomycin (F1,77= 4.32; P= 0.041). Given the marginal significance of this result and the fact that Drosomycin expression was not related to the dry weight of female flies, we suspect this observation is the fruit of chance and multiple testing.
Discussion
We measured the larval and adult constitutive activity of 2 genes encoding antimicrobial peptides (AMPs) in offspring from 80 crosses of D. melanogaster lines. Larval and adult transcription levels of Diptericin were positively genetically correlated and controlled by non-additive genetic factors. We found additive genetic variation controlling the expression of Drosomycin in larvae, but there was no genetic variation for this trait in adults, indicating that genetic control of constitutive Drosomycin expression differs between life-stages and making a formal test for any genetic correlation impossible. The duration of larval development correlated positively with adult mass, but these traits did not correlate with the immune phenotypes we measured.
The existence of a genetic correlation between larval and adult transcription of Diptericin indicates that these two traits are controlled by the same genetic factors (i.e. they have a common genetic architecture). This observation contradicts the main assumption of the adaptive decoupling hypothesis (Moran 1994). This theory states that organisms with distinct life-stages benefit from having a complex life-cycle because it allows them the independent evolution of larval and adult phenotypes. Indeed, this hypothesis relies on the idea that, unlike Diptericin transcription in our study, larval and adult traits are genetically and ontologically independent. Our Diptericin result contrasts with the ones of a series of experiments on Drosophila buzzatii that showed the independent evolution of larval and adult tolerance to heat (Loeschcke & Krebs 1996) and other studies that did not find genetic correlation between larval and adult size in frogs and crustaceans (Hilbish et al. 1993; Johansson et al. 2010; but see Watkins 2001). In contrast, our data indicate that genetic control of Drosomycin expression differs across life-stages. Overall, the main prediction of the adaptive decoupling hypothesis, that traits in different life-stages can evolve independently, seems to be true for some traits but false for others. We had no a priori reason to expect different genetic correlations for Drosomycin and Diptericin expression (and in fact might have naively expected them to show the same pattern of correlation). It is unclear to us how to predict in general whether a given trait will be consistent with adaptive decoupling.
The constitutive expression of AMP genes may be partially determined by the size of the tissue in which it is synthesized. The positive relationship between larval and adult transcription for Diptericin could thus indicate that such synthesizing organs are of similar proportional size in larvae and adults. Our observation that Drosomycin expression was genetically variable in larvae but not in adults may indicate a tight regulation in adults (e.g. by stabilizing selection) or primary expression in tissues present in one of the life-stages only (e.g. reproductive tissue; (Ferrandon et al. 1988)). We previously reported different phenotypic plasticity in larval and adult AMP expression in response to the same larval dietary stimulus (Fellous & Lazzaro 2010). Adult constitutive transcription of AMPs is thus probably independent of the size of organs already formed in pre-metamorphosis larvae. Variation in AMP expression at the whole-fly level may also reflect up-stream variation in the genes’ regulatory cascades. Variation in such trans-regulation can result in a higher-order control of investment into immune function.
Resources allocation theory predicts that investment into immune function must trade-off with other life-history traits (Stearns 1992; Zuk & Stoehr 2002). Indeed, in several cases immune capacity has been shown to correlate negatively with other fitness traits such as competitive ability and fertility (e.g. Kraaijeveld & Godfray 1997; McKean et al. 2008). Such costs of immunity can be expressed in life-stages other than the one in which immune function is measured (Koella & Boete 2002). In our experiment, we measured the speed of larval development and mass of the adults as estimates of fitness. Neither trait correlated genetically with adult or larval expression of Diptericin and Drosomycin. At the same time, we did not find any positive genetic correlation between constitutive AMP expression and fitness measures, as might be spuriously expected if the genotypes differed in general vigor (Fry 1993). This general lack of correlations may have originated in at least three reasons. It is possible that constitutive expression of AMPs may be of very little cost and there may truly be no fitness tradeoff. Alternatively, it is possible that our life history traits provided poor estimates of fitness, and that the measurement of other traits would have revealed a trade-off. Finally, the appearance of trade-offs may depend on the experimental environment (Stearns 1992; Kraaijeveld & Godfray 1997; Zuk & Stoehr 2002), and the generally un-stressful conditions in our experiment may have prevented us from detecting a trade-off. We also note that the strong sensitivity of age at emergence on micro-environmental variation (see Results) may have concealed a true relationship between AMP expression and fitness. While we cannot formally distinguish among these potential explanations with the present data, we can say conclusively that our data do not support any hypothesized fitness tradeoff between constitutive immunity and other life history metrics in our experimental structure.
We find strong evidence that variation in constitutive expression of Diptericin is genetically correlated across life-stages. Shared genetic controls of larval and adult immunity could broadly impact the epidemiology and evolution of infectious diseases of both larvae and adults, even when the diseases themselves are specific to one of the two life-stages. The frequency and intensity of parasitic attacks on either life-stage could shape the selective pressures exerted on the immune system as a whole, and consequently impact the evolution of defense in both life-stages, as well as the epidemiology of infection in both life-stages. These effects could be quite complicated, particularly if there are different adaptive premiums on host investment into immune function across the two life-stages or if evolved resistance to one pathogen results in susceptibility to another (Cotter et al. 2004; Wilfert et al. 2007).
In order for phenotypic traits to respond to selection, however, phenotypic variation must stem from additive genetic variation (Lynch & Walsh 1998). Even though larval and adult transcription of Diptericin were genetically correlated, the relationship was not based on additive genetic variation. Instead the phenotypes depended on the specific combination of the maternal and paternal genotypes, indicating the importance of epistasis or dominance for the expression of this gene in this population. By contrast, larval expression of Drosomycin was exclusively determined by additive genetic variation. Previous studies on insect immunity revealed substantial additive genetic variation for immune function and exemplified evolutionary response to selection on host resistance to pathogens (Rolff et al. 2005; Wilfert & Schmid-Hempel 2008; Wegner et al. 2009). The sole control of immune function by epistasis or dominance may thus be the exception rather than the rule, and would certainly depend on the specific population and traits under study. In summary, we believe that the scenario of parasite pressure on one life-stage driving the evolution of immunity (and resistance) in the other is realistic but probably not universal. Whether coupled evolution of resistance to larval and adult pathogens happens would be determined by the nature of the immune components involved in the insect-pathogen interactions and on the genetic composition of the focal population.
Conclusions
Our experiment showed that variation in larval and adult immune traits can be controlled by the same genetic factors across life-stages. This observation is at odds with the main prediction of the adaptive decoupling hypothesis, which states that distinct life-stages permit the independent evolution of larval and adult phenotypes. Shared genetic control of larval and adult immunity has potentially broad consequences for insect-pathogen dynamics: under this scenario, the evolution of pathogen-resistance in one life-stage could be influenced by the epidemiological dynamics of parasites specific to the other life-stage. The transcriptional activities of the two immune genes that we studied relied on different genetic architectures, with only one exhibiting shared genetic control in larvae and adults. This illustrates the need to understand in some detail the immune components involved in a given host-pathogen system before predicting whether coupled evolution of larval and adult immune defenses may occur.
Acknowledgments
We thank Richard Preziosi, Patrice David, Celine Deveaux and Georges Lutfalla for help with the analyses, Aurelié Coulon and Miguel Rosado for help in the lab and Louis Lambrechts and two anonymous reviewers for comments on the manuscript. The work was funded by grants R01 AI062995 from the National Institutes of Health and DEB-0415851 from the National Science Foundation.
References
- Astles PA, Moore AJ, Preziosi RF. A comparison of methods to estimate cross-environment genetic correlations. J Evol Biol. 2006;19:114–122. doi: 10.1111/j.1420-9101.2005.00997.x. [DOI] [PubMed] [Google Scholar]
- Bulmer MS, Bachelet I, Raman R, Rosengaus RB, Sasisekharan R. Targeting an antimicrobial effector function in insect immunity as a pest control strategy. PNAS. 2009;106:12652–12657. doi: 10.1073/pnas.0904063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter SC, Kruuk LEB, Wilson KO. Costs of resistance: genetic correlations and potential trade-offs in an insect immune System. J Evol Biol. 2004;17:421–429. doi: 10.1046/j.1420-9101.2003.00655.x. [DOI] [PubMed] [Google Scholar]
- Crawley MJ. Statistical computing: an introduction to data analysis using S-plus. Wiley; Chichester, UK: 2002. [Google Scholar]
- Fellous S, Lazzaro BP. Larval food quality affects adult (but not larval) immune gene expression independent of effects on general condition. Mol Ecol. 2010;19:1462–1468. doi: 10.1111/j.1365-294X.2010.04567.x. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Jung AC, Criqui MC, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart JM, Hoffmann JA. A drosomycin–GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. The EMBO Journal. 1988;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 2005;169:243–257. doi: 10.1534/genetics.104.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitak D, Wheat CW, Heckel DG, Vogel H. Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusia ni. BMC Biol. 2007:5. doi: 10.1186/1741-7007-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry JD. The General Vigor Problem - Can Antagonistic Pleiotropy Be Detected When Genetic Covariances Are Positive. Evolution. 1993;47:327–333. doi: 10.1111/j.1558-5646.1993.tb01224.x. [DOI] [PubMed] [Google Scholar]
- Hilbish TJ, Winn EP, Rawson PD. Genetic variation and covariation during larval and juvenile growth in Mercenaria mercenaria. Mar Biol. 1993;115:97–104. [Google Scholar]
- Johansson F, Lederer B, Lind MI. Trait Performance Correlations across Life Stages under Environmental Stress Conditions in the Common Frog, Rana temporaria. PLoS ONE. 2010;5:e11680. doi: 10.1371/journal.pone.0011680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune Activation by Life-Shortening Wolbachia and Reduced Filarial Competence in Mosquitoes. Science. 2009;326:134–136. doi: 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koella JC, Boete C. A genetic correlation between age at pupation and melanization immune response of the yellow fever mosquito Aedes aegypti. Evolution. 2002;56:1074–1079. doi: 10.1111/j.0014-3820.2002.tb01419.x. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld AR, Godfray HC. Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature. 1997;389:278–80. doi: 10.1038/38483. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The Host Defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Loeschcke V, Krebs RA. Selection for Heat-Shock Resistance in Larval and in Adult Drosophila buzzatii: Comparing Direct and Indirect Responses. Evolution. 1996;50:2354–2359. doi: 10.1111/j.1558-5646.1996.tb03623.x. [DOI] [PubMed] [Google Scholar]
- Lutfalla G, Uze G. Performing Quantitative Reverse-Transcribed Polymerase Chain Reaction Experiments. Methods Enzymol. 2006;410:386–400. doi: 10.1016/S0076-6879(06)10019-1. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates; Massachusetts: 1998. [Google Scholar]
- McGraw LA, Fiumera AC, Ramakrishnan M, Madhavarapu S, Clark AG, Wolfner MF. Larval rearing environment affects several post-copulatory traits in Drosophila melanogaster. Biol Lett. 2007;3:607–610. doi: 10.1098/rsbl.2007.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean K, Yourth C, Lazzaro B, Clark A. The evolutionary costs of immunological maintenance and deployment. BMC Evol Biol. 2008;8:76. doi: 10.1186/1471-2148-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Adaptation and Constraint in the Complex Life Cycles of Animals. Ann Rev Ecol Syst. 1994;25:573–600. [Google Scholar]
- Peng J, Zipperlen P, Kubli E. Drosophila Sex-Peptide Stimulates Female Innate Immune System after Mating via the Toll and Imd Pathways. Curr Biol. 2005;15:1690–1694. doi: 10.1016/j.cub.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Petersen UM, Kadalayil L, Rehorn KP, Hoshizaki DK, Reuter R, Engstrom Y. Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. Embo J. 1999;18:4013–22. doi: 10.1093/emboj/18.14.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed-effects Models in S and S-PLUS. Springer; 2000. [Google Scholar]
- Rolff J, Armitage SAO, Coltman DW. Genetic constraints and sexual dimorphism in immune defense. Evolution. 2005;59:1844–1850. [PubMed] [Google Scholar]
- Rolff J, Siva-Jothy MT. Invertebrate ecological immunology. Science. 2003;301:472–5. doi: 10.1126/science.1080623. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Clark AG. Genotype and Gene Expression Associations with Immune Function in Drosophila. PLoS Genet. 2009;6:e1000797. doi: 10.1371/journal.pgen.1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC. The evolution of life histories. Oxford University Press; Oxford; New York: 1992. [Google Scholar]
- Tian C, Gao B, Rodriguez MdC, Lanz-Mendoza H, Ma B, Zhu S. Gene expression, antiparasitic activity, and functional evolution of the drosomycin family. Molecular Immunology. 2008;45:3909–3916. doi: 10.1016/j.molimm.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Vilmos P, Kurucz É. Insect immunity: evolutionary roots of the mammalian innate immune system. Immunology Letters. 1998;62:59–66. doi: 10.1016/s0165-2478(98)00023-6. [DOI] [PubMed] [Google Scholar]
- Watkins TB. A quantitative genetic test of adaptive decoupling across metamorphosis for locomotion and life-history in the pacific tree frog, Hyla regilla. Evolution. 2001;55:1668–1677. doi: 10.1111/j.0014-3820.2001.tb00686.x. [DOI] [PubMed] [Google Scholar]
- Wegner KM, Berenos C, Schmid-Hempel P. Host genetic architecture in single and multiple infections. J Evol Biol. 2009;22:396–404. doi: 10.1111/j.1420-9101.2008.01657.x. [DOI] [PubMed] [Google Scholar]
- Welchman DP, Aksoy S, Jiggins F, Lemaitre B. Insect Immunity: From Pattern Recognition to Symbiont-Mediated Host Defense. Cell Host & Microbe. 2009;6:107–114. doi: 10.1016/j.chom.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Wigby S, Domanitskaya EV, Choffat Y, Kubli E, Chapman T. The effect of mating on immunity can be masked by experimental piercing in female Drosophila melanogaster. J Ins Physiol. 2008;54:414–420. doi: 10.1016/j.jinsphys.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P. The genetic architecture of immune defense and reproduction in male Bombus terrestris bumblebees. Evolution. 2007;61:804–815. doi: 10.1111/j.1558-5646.2007.00079.x. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Schmid-Hempel P. The genetic architecture of susceptibility to parasites. BMC Evol Biol. 2008:8. doi: 10.1186/1471-2148-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk M, Stoehr AM. Immune defense and host life history. Am Nat. 2002;160:S9–S22. doi: 10.1086/342131. [DOI] [PubMed] [Google Scholar]