Abstract
During protein synthesis, mRNA and tRNA undergo coupled translocation through the ribosome in a process that is catalyzed by elongation factor EF-G. Based on cryo-EM reconstructions, counterclockwise and clockwise rotational movements between the large and small ribosomal subunits have been implicated in a proposed ratcheting mechanism to drive the unidirectional movement of translocation. We have used a combination of two fluorescence-based approaches to study the timing of these events: Intersubunit FRET measurements to observe relative rotational movement of the subunits and a fluorescence quenching assay to monitor translocation of mRNA. Binding of EF-G·GTP first induces rapid counterclockwise intersubunit rotation, followed by a slower, clockwise reversal of the rotational movement. Comparison of the rates of these movements reveals that mRNA translocation occurs during the second, clockwise rotation event, corresponding to the transition from the hybrid state to the classical state.
Introduction
During protein synthesis, mRNA and tRNAs are moved through the ribosome by the dynamic process of translocation. Sequential movement of tRNAs from the A (aminoacyl) site to the P (peptidyl) site to the E (exit) site is coupled with movement of their associated codons in the mRNA. Each of the three tRNA binding sites is shared between the small and large ribosomal subunits; the mRNA and anticodon stem-loop (ASL) domains of the tRNAs bind to the small subunit, and the acceptor ends of the tRNA bind to the large subunit1–5. This arrangement reflects the way in which the tRNAs move. Following peptide bond formation, the acceptor ends of the tRNAs first move on the large subunit, resulting in the A/P and P/E hybrid binding states, in which the anticodon ends of the peptidyl-tRNA and deacylated tRNA remain bound to the A and P sites, respectively, of the small subunit, while their acceptor ends move into the P and E sites of the large subunit6. In the second step of translocation, the ASLs of the tRNAs, along with their associated mRNA codons, are translocated from the small subunit A and P sites to the P and E sites, respectively, resulting in movement of the tRNAs from the A/P and P/E hybrid states to the classical P/P and E/E states (Fig. 1). The second step of translocation has a strong dependence on elongation factor G (or EF-2, in eukaryotes). Although formation of the hybrid state in the first step of translocation can occur spontaneously6, it is also favored by binding of EF-G7–8.
Figure 1.
Hybrid-state model of translocation and experimental design. (A) Schematic depiction of intersubunit rotation and tRNA movement in pre-translocation ribosomes. Movement of mRNA is not depicted here. Spontaneous, non-productive forward and reverse intersubunit rotation, coupled to fluctuations between the classical and hybrid states of tRNA binding can occur in the absence of EF-G. (B) In the presence of EF-G, intersubunit rotation is coupled to translocation of tRNA on the 30S subunit. In the first step of translocation, EF-G can either induce hybrid state formation in non-rotated, classical-state ribosomes or bind directly to rotated, hybrid-state ribosomes. In the second step, tRNAs are translocated on the 30S subunit. (C) Experimental design. Intersubunit rotation was measured by changes in the FRET signal between a donor dye (blue) attached to protein S6 on the 30S subunit and an acceptor dye (orange) attached to protein L9 on the 50S subunit. Counterclockwise rotation, accompanying formation of the hybrid-state intermediate, results in a decrease in FRET; reverse, clockwise movement increases the efficiency of energy transfer. mRNA translocation was measured by quenching of the fluorescence of a pyrene dye (yellow) attached to position +9 of the mRNA as it enters the ribosome.
Division of translocation between two steps on separate subunits suggested the possibility that intersubunit movement plays a role. In cryo-EM reconstructions of ribosome complexes containing bound EF-G, it was observed that the small subunit was rotated in a counterclockwise* direction compared with its usual orientation relative to the large subunit9–11. It was proposed that this intersubunit movement is part of a two-step ratchet mechanism responsible for translocation of mRNA and tRNA9,12. According to this model, in the first step, counterclockwise rotation is favored by EF-G·GTP binding. In the second step, GTP hydrolysis results in translocation of mRNA and tRNA followed by EF-G release and reverse, clockwise rotation of the 30S into the non-rotated conformation9. A strong test of the ratchet model was carried out in experiments in which rotational movement between the subunits was restricted by formation of an intersubunit disulfide bridge between ribosomal proteins S6 and L2; formation of this cross-link resulted in a complete and specific block in translocation that could be reversed by disruption of the cross-link13.
The possibility that the ratchet mechanism involves hybrid-state tRNA intermediates was suggested by the observation that, in the rotated conformation of the ribosome, the deacylated tRNA had moved from its classical P/P state into an orientation predicted to be a P/E hybrid state11. Indeed, based on a combination of intersubunit FRET measurements and chemical probing experiments, the rotated state was found to be indistinguishable from the hybrid state14. Further support for unification of the hybrid-state and ratchet mechanisms has come from more recent cryo-EM studies, which have identified ribosomes in the rotated conformation containing tRNAs bound in both the A/P and P/E hybrid states15–16.
Single-molecule FRET experiments have revealed that pre-translocation ribosome complexes containing deacylated tRNA in the P site fluctuate spontaneously back and forth between the rotated and non-rotated conformations in the absence of EF-G or GTP17. This spontaneous movement (which does not result in translocation) appears to be coupled to fluctuations of tRNAs between the hybrid and classical states17–22 (Fig. 1A). In the presence of EF-G and GTP, the second step of translocation moves the peptidyl-tRNA into the P/P state; this fixes the ribosome in the classical, non-rotated conformation17,19 (Fig. 1B), because the strong specificity of the large subunit E site for a deacylated tRNA acceptor end23 prevents peptidyl-tRNA from entering the P/E state.
Despite considerable progress in recent years, we are far from a complete understanding of the molecular mechanism of translocation. For example, it remains to be shown whether intersubunit rotation follows, precedes or is coupled to tRNA and mRNA translocation. And, since EF-G has been shown to shift the equilibrium toward the rotated, hybrid-state conformation of the ribosome 7–9,11,17,19,21, the question arises as to whether release of EF-G from the ribosome is required for clockwise reversal of the movement into the non-rotated conformation. Here, we address these questions with pre-steady-state kinetic experiments, using FRET to follow intersubunit movement of the ribosome and a fluorescence quenching assay to monitor translocation of mRNA in E.coli ribosomes (Fig. 1C). We find that mRNA translocation occurs during the second step of intersubunit movement, in which clockwise rotation restores the hybrid-state ribosome to the classical state. Moreover, in contrast to the original proposal of the ratchet model9, this productive, reverse movement of the 30S subunit requires neither GTP hydrolysis nor release of EF-G from the ribosome.
Results
Intersubunit rotation is accompanied by mRNA translocation
To follow intersubunit movement during translocation, we used a previously developed FRET assay19. Fluorophores were attached to specific cysteine residues introduced by directed mutagenesis into ribosomal proteins S6 (D41C), S11 (E75C), and L9 (N11C) (Figure 1C)14,19. Proteins S6 or S11 labeled with donor (Alexa-488) dye and protein L9 labeled with acceptor (Alexa-568) dye were incorporated into 30S and 50S subunits, respectively, by in vitro reconstitution as described previously19. We have previously demonstrated that conditions that favor formation of the hybrid state result in an increase in S11-L9 FRET and an anti-correlated decrease in S6-L9 FRET, both of which are indicative of counterclockwise rotation of the 30S subunit19. Several lines of evidence have shown that the observed FRET changes are caused by intersubunit rotation, rather than any local structural dynamics of ribosomal proteins or changes in the environment and/or orientation of the fluorophores14,17,19.
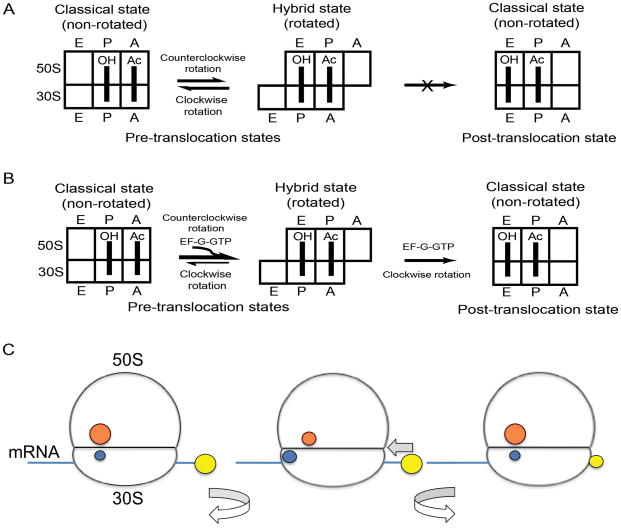
Pre-translocation complexes were formed by binding deacylated elongator tRNAMet to the P site and the peptidyl-tRNA analogue N-Ac-Phe-tRNAPhe to the A site of fluorescently labeled ribosomes (either S6-D(donor)/L9-A(acceptor) or S11-D/L9-A) in the presence of a short, unlabeled mRNA. Our previous single-molecule FRET measurements showed that, under similar experimental conditions, the hybrid-state and classical conformations of the ribosome are in equilibrium with each other and the majority of ribosomes are in the rotated, hybrid-state conformation17. When pre-translocation ribosome complexes containing fluorophores attached to proteins S6 and L9 (S6-D/L9-A) were mixed with EF-G and GTP using a stopped-flow fluorimeter, we observed a rapid increase in acceptor fluorescence (Fig. 2A). This increase in FRET indicates movement of S6 toward L9 and, as was established previously19, reverse, clockwise rotation of the ribosome into the non-rotated conformation. Conversely, mixing of S11D/L9A ribosomes with EF-G·GTP resulted in a corresponding decrease in FRET (Fig. 2A), also indicative of clockwise rotation of the 30S19. Kinetic traces for acceptor fluorescence for both S6-D/L9-A and S11-D/L9-A ribosomes are fit well by a single-exponential function giving apparent rate constants of 4.5 ± 1.2 s−1 and 4.1 ± 1 s−1, respectively (Table 1).
Figure 2.
Kinetics of intersubunit rotation and mRNA translocation. (A,C) Kinetics of intersubunit rotation followed by FRET (acceptor fluorescence) (B,D) Kinetics of mRNA translocation measured by quenching of fluorescence of pyrene attached to the mRNA24. (A,B) Pre-translocation ribosomes containing deacylated elongator tRNAMet in the P site and N-Ac-Phe-tRNAPhe in the A site were rapidly mixed with EF-G·GTP. (A) Intersubunit FRET signal (acceptor fluorescence) for S6-D/L9-A (red) and S11-D/L9-A (green) ribosomes. (B) Quenching of fluorescence of pyrene-labeled mRNA in wild-type (blue) and S6-D/L9-A (red) ribosomes. (C,D) Pre-translocation ribosomes containing initiator tRNAfMet in the P site and N-Ac-Phe-tRNAPhe in the A site were rapidly mixed with EF-G·GTP. (C) Intersubunit FRET signal (acceptor fluorescence) for S6-D/L9-A ribosomes (red). (D) Quenching of fluorescence of pyrene-labeled mRNA in wild-type (blue) and S6-D/L9-A (red) ribosomes. Single-exponential fits for FRET traces and double-exponential fits for mRNA quenching are shown by black lines.
Table 1.
Rates of intersubunit movement and mRNA translocation
| P-site tRNA | Rate of clockwise rotation (s−1) | Rate of mRNA translocation in unlabeled wild-type ribosomes | Rate of mRNA translocation in S6D/L9A ribosomes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| k1 (s−1) | k2 (s−1) | A1/(A1+A2) | kav (s−1) | k1 (s−1) | k2 (s−1) | A1/(A1+A2) | kav (s−1) | ||
| tRNAMet | 4.5±1.2 (4.1±1.0 for S11D/L9A) | 6.9±1.4 | 0.7±0.1 | 0.57±0.06 | 4.2±0.8 | 8.0±1.3 | 0.5±0.2 | 0.67±0.02 | 5.5±1.0 |
| tRNAfMet | 1.1±0.2 | 1.1±0.4 | 0.20±0.09 | 0.47±0.09 | 0.6±0.2 | 1.5±0.5 | 0.20±0.05 | 0.45±0.03 | 0.8±0.2 |
| tRNAfMet 1 mM Spc | 0.04±0.01 | 0.10±0.02 | 0.012±0.002 | 0.30±0.04 | 0.04±0.01 | 0.12±0.02 | 0.012±0.002 | 0.39±0.03 | 0.05±0.01 |
| tRNAfMet 0.2 mM Hyg | 0.005±0.001 | 0.033±0.010 | 0.003±0.001 | 0.25 ±0.12 | 0.010±0.005 | 0.029±0.008 | 0.002±0.001 | 0.19±0.03 | 0.007±0.003 |
Pre-translocation complexes containing tRNAMet or tRNAfMet bound to the P site and N-Ac-Phe-tRNAPhe in the A site were mixed with EF-G in the presence of GTP as described in Methods. Approximately ten time traces were accumulated for each experiment. Rate constants averaged from two to five experiments are presented in the table. Rates of intersubunit rotation were determined by single-exponential fits of the kinetic traces. k1 and k2 are the rate constants of double-exponential fits of the mRNA translocation data; A1 and A2 are their respective amplitudes; A1/(A1+A2) is the relative contribution of the faster phase to the total amplitude of pyrene quenching. Weighted average values (kav) for mRNA translocation rates were calculated by combining the rate constants derived from the two-exponential fits: kav = (k1A1 + k2A2)/(A1 + A2). Spc, spectinomycin; Hyg, hygromycin B.
The kinetics of mRNA translocation were followed by quenching of the fluorescence of a pyrene dye attached to the 3′ end of the mRNA as it moves within the ribosome24 (Fig. 1C). Pre-translocation complexes were assembled with pyrene-labeled mRNA, deacylated tRNAMet, N-Ac-Phe-tRNAPhe and either fluorescently-labeled S6-D/L9-A ribosomes or unlabeled wild-type ribosomes. When these complexes were mixed with EF-G·GTP, rapid quenching of pyrene fluorescence was observed, indicative of mRNA translocation (Fig. 2B). Comparison of the kinetic traces for pyrene quenching and intersubunit FRET shows that the translocation of mRNA and clockwise rotation occur on the same time scale.
As has been reported previously25–27, the kinetics of mRNA translocation are clearly biphasic and are best fitted by the sum of two exponentials, corresponding to apparent rate constants k1 and k2 (Fig. 2B, Suppl. Fig. 1 and Table 1); the faster rate constant k1 accounts for 55–70% of the amplitude of the change in fluorescence (Table 1). Even though the biphasic character of fluorescence changes associated with mRNA translocation is well documented25–27, the physical basis of this phenomenon remains unclear. The rate of intersubunit rotation falls between k1 and k2, matching the value of kav, the weighted average rate constant calculated as the sum of k1 and k2 normalized to their respective contributions to the total amplitude of fluorescence change [kav = (k1*A1 + k2*A2)/(A1 + A2)]. The amplitude (but not the rate) of the fluorescence change was notably smaller in S6-D/L9-A ribosomes than in unlabeled ribosomes. This is not surprising, since only about 50–70 % of reconstituted, fluorescently-labeled ribosomes are able to bind tRNA and mRNA, which is also the fraction of labeled ribosomes that are active in translocation19. The rate of mRNA translocation for fluorescently-labeled ribosomes (5.5 ± 1.1 s−1) was similar to that measured for wild-type, unlabeled ribosomes (4.2 ± 1.1 s−1), indicating that the presence of the labels does not interfere with translocation.
To further test the relationship between intersubunit movement and mRNA translocation, we next asked whether conditions that slow the rate of mRNA translocation also decrease the rate of intersubunit rotation. It was previously reported that the rate of translocation is slowed by a factor of 2–4 when the P site of the pre-initiation complex is filled with initiator tRNAfMet rather than elongator tRNAMet24,28. Accordingly, we measured the rate of translocation of pre-translocation complexes containing deacylated initiator tRNAfMet bound in the P site and N-Ac-Phe-tRNAPhe in the A site of S6-D/L9-A ribosomes. The presence of initiator tRNAfMet slowed the rate of clockwise intersubunit rotation from 4.5 s−1 to 1.1 s−1 (Fig 2C and table 1) and mRNA translocation from 5.5 s−1 to 0.8 s−1 (Fig. 2D and table 1), providing further evidence for coupling of mRNA translocation to intersubunit rotation.
Direct observation of both steps of intersubunit rotation
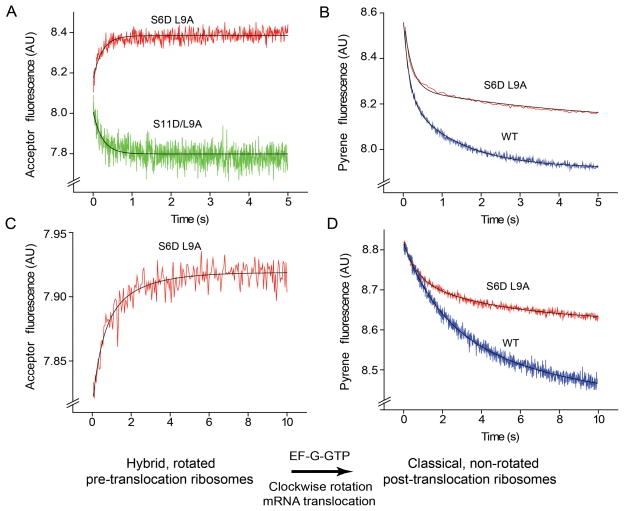
In the experiments described above, we were unable to resolve the first (counterclockwise) step from the second (clockwise) step of intersubunit rotation. This could be explained by insufficient differences between the rates of the two intersubunit rotational steps, complicated by the fact that the majority of pre-translocation ribosomes were already in the rotated, hybrid-state conformation before mixing with EF-G. To address this question, we asked whether slowing translocation with antibiotics would allow us to resolve the two intersubunit rotational steps, as suggested by the results of Pan et al8 and Peske et al 26. We slowed translocation using the inhibitors spectinomycin and hygromycin B, which inhibit rates of translocation by a factor of ~ 30 and 300, respectively, by distinctly different mechanisms26,29–30, without affecting EF-G binding, rates of GTP hydrolysis, phosphate release or hybrid-state formation8,26. Pre-translocation complexes containing deacylated initiator tRNAfMet in the P site and N-Ac-Phe-tRNAPhe in the A site of S6-D/L9-A or unlabeled ribosomes were pre-incubated with either 2 mM spectinomycin or 0.4 mM hygromycin B and mixed with EF-G·GTP (Fig. 3). Using the pyrene quenching method, mRNA translocation was slowed by a factor of ~10- and ~100 by spectinomycin and hygromycin B, respectively (Fig. 3B and D). In agreement with the findings of Peske et al.26, the amplitude of the slow phase of the biphasic mRNA translocation reaction increased to 60–70% in the presence of spectinomycin and 80–85% in the presence of hygromycin B at the expense of the rapid phase (Table 1). Intersubunit FRET measurements using the S6D/L9A construct showed an initial rapid decrease, indicating counterclockwise rotation, followed by a slow increase, corresponding to reverse, clockwise rotation (Fig. 3A and C). The slow phase of the FRET traces can be fit to a single exponential giving apparent rate constants for clockwise rotation of 0.04 s−1 and 0.005 s−1 for the spectinomycin and hygromycin B complexes, respectively. These rates are similar to the kav rates observed for pyrene quenching, and about two orders of magnitude slower than the rate of the first rotational step (estimated to be approximately 1.4 and 2.4 s−1 for the hygromycin B and spectinomycin complexes, respectively). These results clearly show that mRNA translocation takes place during the second, clockwise step of intersubunit rotation.
Figure 3.
Resolution of the first and second steps of intersubunit rotation in the presence of antibiotics. (A,C) Kinetics of intersubunit rotation and (B,D) mRNA translocation in the presence of (A,B) spectinomycin and (C,D) hygromycin B. Pre-translocation ribosomes containing deacylated initiator tRNAfMet in the P site and N-Ac-Phe-tRNAPhe in the A site were pre-incubated with either (A,B) spectinomycin or (C-D) hygromycin B and rapidly mixed with EF-G·GTP. The FRET signal (acceptor fluorescence) for S6-D/L9-A ribosomes is shown in red; quenching of pyrene-labeled mRNA in wild-type and S6-D/L9-A ribosomes is shown in blue and green, respectively. Single-exponential (intersubunit FRET) and double-exponential (mRNA translocation) fits are shown by black lines.
If mRNA translocation is indeed coupled to the second, clockwise rotational step of EF-G-catalyzed translocation, blocking the second step should allow counterclockwise rotation but without reversal of rotation or movement of mRNA. Accordingly, we separated the two steps by reacting a pre-translocation complex with EF-G and GDP in the presence of fusidic acid. In contrast to EF-G·GTP, EF-G·GDP does not induce translocation, but in the presence of fusidic acid favors the hybrid-state conformation of the ribosome 7. The result was a rapid decrease in FRET, indicating counterclockwise rotation into the hybrid-state conformation (Fig. 4). No subsequent increase in FRET or quenching of the pyrene-labeled mRNA were observed, consistent with previous reports that GDP does not support mRNA translocation7–8,31; instead, a small increase in pyrene fluorescence was detected (Fig. 4) that reflects binding of EF-G to the pre-translocation ribosome24. This result further supports our conclusion that counterclockwise intersubunit rotation into the hybrid state precedes clockwise rotation and mRNA translocation.
Figure 4.
Stabilization of the rotated, hybrid-state conformation by binding EF-G·GDP in the presence of fusidic acid. Pre-translocation ribosomes containing initiator tRNAfMet in the P site and N-Ac-Phe-tRNAPhe in the A site were rapidly mixed with EF-G·GDP and fusidic acid. The intersubunit FRET signal (acceptor fluorescence) for S6-D/L9-A ribosomes is shown in red. Quenching of pyrene-labeled mRNA fluorescence in wild-type and S6-D/L9-A ribosomes is shown in blue and green, respectively.
The second step of rotation does not require EF-G release
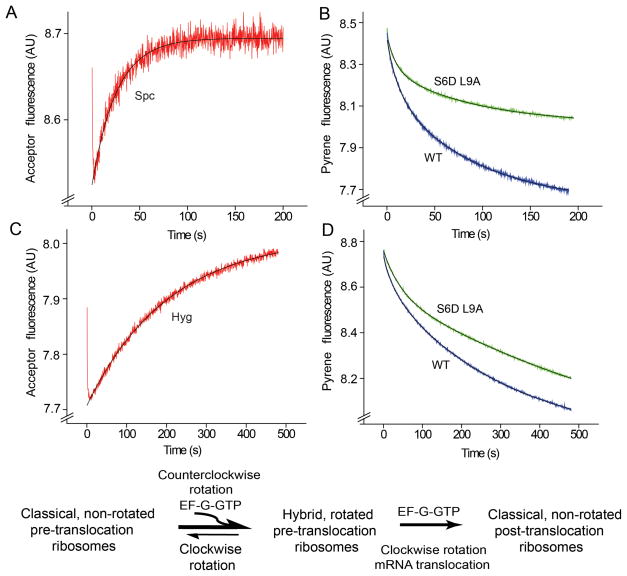
Since EF-G shifts the equilibrium toward the rotated, hybrid-state conformation of the ribosome 7–9,11,17,19,21, we asked whether release of EF-G is required for the reverse rotational movement into the classical state. Release of EF-G from the ribosome can be blocked either in the presence of the non-hydrolyzable GTP analogue GDPNP or by the antibiotic fusidic acid, both of which have been shown to inhibit EF-G release from the ribosome; fusidic acid blocks EF-G release following GTP hydrolysis without interfering with single-round translocation7,32–36. Pre-translocation S6-D/L9-A ribosomes, containing deacylated elongator tRNAMet in the P site and N-Ac-Phe-tRNAPhe in the A site, were rapidly mixed with EF-G·GDPNP or with EF-G·GTP and fusidic acid. In both cases, a rapid increase in intersubunit FRET was observed, indicating clockwise intersubunit rotation (Fig. 5A). This result unambiguously demonstrates that clockwise rotation does not require EF-G release or GTP hydrolysis. Measurements of mRNA translocation rates showed that, in agreement with previous reports35–36, fusidic acid does not inhibit single-round mRNA translocation (Fig. 5B, Table 2), while replacing GTP with GDPNP slowed down translocation only by a factor of ~ 2.5. The latter observation is consistent with the reduction by a factor of 3 in translocation rate observed in the presence of GDPNP in two previous reports8,35, but much less than the inhibition by a factor of 50 reported by Rodnina et al.37. Replacement of GTP by GDPNP slowed the rate of clockwise rotation to the same extent as mRNA translocation (by a factor of ~2.5), providing further evidence for coupling of mRNA translocation to the second rotational step.
Figure 5.
Effect of inhibition of EF-G release on the second (clockwise) step of intersubunit rotation and mRNA translocation. Pre-translocation ribosomes containing elongator tRNAMet in the P site and N-Ac-Phe-tRNAPhe in the A site were rapidly mixed with EF-G·GDPNP or EF-G·GTP and fusidic acid. (A) FRET signal (acceptor fluorescence) for S6-D/L9-A ribosomes. (B) Quenching of pyrene-labeled mRNA in wild-type ribosomes. EF-G·GDPNP traces are shown in blue and those for EF-G·GTP and fusidic acid are shown in red. Single-exponential (intersubunit FRET) and double-exponential (mRNA translocation) fits are shown by black lines.
Table 2.
Effect of inhibition of EF-G release on rates of intersubunit rotation and mRNA translocation
| Rate of clockwise rotation (s−1) | Rate of mRNA translocation in WT ribosomes | ||||
|---|---|---|---|---|---|
| k1 (s−1) | k2 (s−1) | A1/(A1+A2) | kav (s−1) | ||
| EF-G-GTP | 4.5±1.2 | 6.9±1.4 | 0.7±0.1 | 0.57±0.06 | 4.2±0.8 |
| EF-G-GTP + fusidic acid | 6.7±0.6 | 5.6±0.3 | 0.6±0.1 | 0.57±0.01 | 3.5±0.2 |
| EF-G-GDPNP | 1.7±0.2 | 2.7±0.4 | 0.4±0.1 | 0.55±0.04 | 1.6±0.3 |
Pre-translocation complexes containing tRNAMet in the P site and N-Ac-Phe-tRNAPhe in the A site were mixed with EF-G in the presence of GTP, GDPNP or GTP + Fusidic acid as described in Methods. Approximately ten time traces were accumulated for each experiment. Rate constants averaged from two to five experiments are presented in the table. k1, k2, A1, A2 and kav are as defined in Table 1.
Discussion
These studies show, for the first time, the occurrence of the two long-hypothesized separate and opposite rotational movements between the ribosomal subunits during EF-G-catalyzed translocation. Moreover, they show that the translocational movement of mRNA on the ribosome occurs during the second, clockwise rotational step of translocation. During the first, counterclockwise step, the tRNAs move from the classical A/A and P/P states to the hybrid A/P and P/E states6,8,17–18,21, without movement of mRNA. In the second step, intersubunit rotation is reversed, as the tRNAs move into the classical P/P and E/E states, accompanied by movement of the mRNA by one codon. Slowing the rate of mRNA translocation, using a variety of experimental strategies, also slows the rate of clockwise rotation to a similar degree, suggesting that these two processes are coupled (Figs. 2,3; Table 1).
Our experiments provide several critical tests of the ratchet model of translocation. Until now, stable complexes of EF-G bound to the ribosome containing a single deacylated tRNA (and, therefore, incapable of undergoing translocation) have been predominantly characterized by chemical probing, FRET, cryo-EM and crystallographic approaches. By slowing translocation, we were able to resolve the long-hypothesized transient stabilization of the pre-translocation ribosome in the rotated conformation by EF-G (Fig. 3). This result is consistent with the observation of transient stabilization of tRNAs in the hybrid state during translocation8, providing new evidence for coupling of hybrid state formation and intersubunit movement. More specifically, our experiments show that mRNA translocation occurs during the second (clockwise) step of intersubunit rotation. Furthermore, the kinetic traces for clockwise rotation can be fit to a single exponential (Figs. 2, 3); therefore, within the detection limits of our experiments, intersubunit rotation must occur without any stable rotational intermediates. This finding is consistent with our previous single-molecule FRET experiments, which demonstrated that spontaneous fluctuation of pre-translocation ribosomes between the classical and hybrid states involves only two rotational conformational states17.
Although the kinetics of intersubunit rotation can be fit to a single exponential, measurement of the rate of movement of mRNA is complicated by the biphasic character of the kinetics of quenching of the pyrene label. In spite of the widespread use of fluorescence changes in labeled mRNAs for measuring mRNA translocation24–25,28,35, no clear explanation of this biphasic behavior has so far emerged. Some papers attribute only the rate of the fast phase to translocation25–27, while others fit the data to a single exponential24,28,35,38. However, as we show here, slowing the rate of translocation by two different antibiotics or by replacement of P-site elongator tRNA with initiator tRNAfMet decreases the rates of both phases (Table 1), thus implicating both the fast and slow phases in translocation of mRNA, rather than in translocation-independent processes such as photobleaching. The existence of two populations of ribosomes that translocate at different rates26 seems an unlikely explanation, because of the monophasic behavior of the kinetics of clockwise rotation. Another possibility is that the biphasic kinetics reflect additional, kinetically distinct dynamic events, such as movement of the head of the 30S subunit10–11,39. Alternatively, quenching of the pyrene label may arise from two different molecular mechanisms, such as alternative stacking of the dye on different structures in the ribosome; however, the observation that a fluorescence enhancement assay, using a dye bound to a different position of the mRNA, also shows a similar biphasic behavior 26,38 would seem to argue against this possibility. Thus, we find no convincing explanation for the biphasic behavior of the currently used mRNA translocation assays. Nevertheless, it is clear that the observed fluorescence changes correlate with translocation of mRNA in the ribosome, and so are widely accepted as a standard method for measuring translocation rates 24–25,27–28,35,40–42. For example, another assay based on changes in the signal from fluorescently-labeled tRNAs (with either wybutine or proflavine labels in the anticodon or D-loop) reports very similar values for the kinetics of translocation 8,37,43–44. Conditions that are known to slow the rate of translocation, such as antibiotic inhibitors of translocation, increased Mg2+ concentration or binding deacylated tRNA or a tRNA anticodon stem-loop in the A site, decrease the rate of pyrene quenching consistently with other biochemical measurements of translocation 24,27–28,42. Mutations in rRNA and tRNAs that affect translocation according to the other biochemical assays also change the rate of pyrene quenching 25,27–28,41.
In any case, we observe close agreement between the monophasic kinetics of intersubunit rotation and the biphasic kinetics of mRNA translocation using a pseudo-first-order rate constant (kav) calculated as a weighted sum of the rates of the fast and slow rate phases. According to this empirical treatment, the rate of mRNA translocation(kav) was similar to the rate of clockwise rotation under all tested conditions, in which the respective rates of translocation varied by more than two orders of magnitude (Tables 1 and 2).
Inhibition of EF-G release by fusidic acid had no measurable effect on the rates of single-round mRNA translocation and clockwise rotation (Table 2; Fig. 5). Therefore, clockwise intersubunit rotation can occur unaffected by the presence of EF-G bound to the ribosome. In cryo-EM reconstructions of EF-G-ribosome complexes10–11, as well as in a recent crystal structure of EF-G bound to a non-rotated, classical-state ribosome45, domain IV of EF-G, which was previously shown to be essential for catalysis of translocation37,46, overlaps with the 30S subunit A site. This EF-G conformation is disallowed in pre-translocation ribosomes because of potential clash with the A-site tRNA. Therefore, EF-G must undergo a large-scale conformational change during mRNA translocation and clockwise movement of the small subunit; indeed, a disulfide cross-link restricting the conformational mobility of domain IV of EF-G abolished its translocation activity47.
Together with the results of previous findings, our studies show that translocation involves two steps of intersubunit movement: a counterclockwise rotation step in which the tRNAs move from classical A/A and P/P states to hybrid A/P and P/E states, followed by a reverse, clockwise rotation step in which the tRNAs move into the classical P/P and E/E states, accompanied by movement of the mRNA by one codon. Although our FRET measurements fail to provide evidence for more than two intersubunit rotational states, we cannot exclude the possibility that other intermediate conformational states of the ribosome play a role in the mechanism of this complex macromolecular process. Our ignorance of many crucial details, such as the molecular nature of the pawl of the ratchet, emphasizes how far we are from a true understanding of the mechanism of translocation.
Methods
Materials and sample preparation
tRNAfMet was purchased from MP Biomedicals; GTP, GDPNP, fusidic acid, spectinomycin, hygromycin B, tRNAPhe and tRNAMet were purchased from Sigma. GDP was further purified from contaminating GMP and GTP using ion-exchange chromatography as previously described19. Unlabeled mRNA (5′ GGC AAG GAG GUA AAA AUG UUU AAA CGU AAA UCU ACU 3′) and 3′ amino-modified mRNA (5′ GGC AAG GAG GUA AAA AUG UUU AAA 3′) were synthesized by Integrated DNA Technologies. 3′ amino-modified mRNA was conjugated with pyrene and subsequently purified according to published procedures24. fMet-tRNAfMet, N-Ac-Phe-tRNAPhe, and EF-G with a 6-His tag were prepared and purified as previously described6,28,48. Ribosomes from E. coli strains MRE600 (wild-type) and the fluorescently labeled 70S ribosome constructs S6-D/L9-A and S11-D/L9-A were prepared as described19,49–51.
Stopped-flow kinetic experiments
Ribosomal complexes were constructed and all stopped-flow experiments performed in buffer containing 20 mM Hepes·KOH (pH 7.5), 6 mM MgCl2, 150 mM NH4Cl, 6 mM β mercaptoethanol, 0.01% (w/v) Nikkol, 2 mM spermidine and 0.1 mM spermine. Pre-translocation complexes for FRET experiments were constructed by incubation of fluorescently-labeled 70S ribosomes (1 μM) with unlabeled mRNA (2 μM) and 2 μM deacylated tRNAMet (or deacylated tRNAfMet) for 20 minutes at 37°C followed by incubation with N-Ac-Phe-tRNAPhe (1.5 μM) for 30 minutes at 37°C.
For measurement of mRNA translocation kinetics, pre-translocation complexes were assembled similarly, except that unlabeled mRNA was replaced with pyrene-labeled mRNA (0.85 μM). In all experiments, pre-translocation ribosomes were mixed with EF-G and GTP (or GDPNP, GDP + fusidic acid, GTP + fusidic acid, depending on the experiment) using an Applied Photophysics stopped-flow fluorimeter. Final concentrations after mixing were: 35 nM ribosomes, 375 nM EF-G, 1 mM GTP, 0.2 mM GDP, 0.5 mM GDPNP, 0.1 mM fusidic acid. For the experiments in Fig. 4, pre-translocation complexes (70 nM) were pre-incubated with either spectinomycin (2 mM ) or hygromycin B (0.4 mM) for 5 minutes at 37°C before mixing with EF-G and GTP. For FRET experiments, the donor dye (Alexa-488) was excited at 490 nm and the acceptor (Alexa-568) emission was detected using a 590 nM long-pass filter (Applied Photophysics). For the mRNA translocation assay, the pyrene dye was excited at 343 nm and fluorescence emission detected using a 375 nm long-pass filter. Monochromator slits were adjusted to 9.3 nm. All stopped-flow experiments were done at 23°C. Time traces were analyzed using Pro-Data-Viewer software (Applied Photophysics).
Supplementary Material
Acknowledgments
These studies were supported by grant no. GM-17129 from the NIH and NSF grant no. MCB-0723300 (to H.F.N.), and a NATO-NSF postdoctoral fellowship to D.N.E. The authors thank Dan Herschlag, Marina Rodnina, Simpson Joseph, Andrei Korostelev and Laura Lancaster for critical discussions.
Footnotes
By “counterclockwise”, we mean to indicate the sense of movement of the 30S subunit (foreground), as viewed from the exterior of the 30S, holding the 50S subunit (background) fixed.
Authors’ contributions: D.N.E. and H.F.N. designed the research; D.N.E. performed the experiments; D.N.E. and H.F.N. analyzed the data and wrote the paper.
References
- 1.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 2.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 3.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 4.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 5.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–41. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 6.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–8. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 7.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–82. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–29. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–22. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, et al. Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 11.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–34. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 12.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci U S A. 2007;104:19671–8. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci U S A. 2007;104:4881–5. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–7. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 15.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–7. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924–7. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–8. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–40. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 20.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–7. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–17. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lill R, et al. Specific recognition of the 3′-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J Mol Biol. 1988;203:699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- 24.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–81. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 25.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2010;107:709–14. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–94. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 27.Shi X, Chiu K, Ghosh S, Joseph S. Bases in 16S rRNA important for subunit association, tRNA binding, and translocation. Biochemistry. 2009;48:6772–82. doi: 10.1021/bi900472a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13 :234–41. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borovinskaya MA, Shoji S, Fredrick K, Cate JH. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA. 2008;14:1590–9. doi: 10.1261/rna.1076908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol. 2007;2:545–52. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zavialov AV, Hauryliuk VV, Ehrenberg M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belitsina NV, Glukhova MA, Spirin AS. Elongation factor G-promoted translocation and polypeptide elongation in ribosomes without GTP cleavage: use of columns with matrix-bound polyuridylic acid. Methods Enzymol. 1979;60:761–79. doi: 10.1016/s0076-6879(79)60070-8. [DOI] [PubMed] [Google Scholar]
- 33.Bodley JW, Zieve FJ, Lin L, Zieve ST. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem Biophys Res Commun. 1969;37:437–43. doi: 10.1016/0006-291x(69)90934-6. [DOI] [PubMed] [Google Scholar]
- 34.Inoue-Yokosawa N, Ishikawa C, Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J Biol Chem. 1974;249:4321–3. [PubMed] [Google Scholar]
- 35.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28:2053–65. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–80. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 38.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–23. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 39.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 40.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006;23:865–74. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Feinberg JS, Joseph S. Ribose 2′-hydroxyl groups in the 5′ strand of the acceptor arm of P-site tRNA are not essential for EF-G catalyzed translocation. RNA. 2006;12:580–8. doi: 10.1261/rna.2290706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2010;6 :54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 44.Robertson JM, Paulsen H, Wintermeyer W. Pre-steady-state kinetics of ribosomal translocation. J Mol Biol. 1986;192:351–60. doi: 10.1016/0022-2836(86)90370-0. [DOI] [PubMed] [Google Scholar]
- 45.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martemyanov KA, Yarunin AS, Liljas A, Gudkov AT. An intact conformation at the tip of elongation factor G domain IV is functionally important. FEBS Lett. 1998;434:205–8. doi: 10.1016/s0014-5793(98)00982-x. [DOI] [PubMed] [Google Scholar]
- 47.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–5. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 48.Wilson KS, Noller HF. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–9. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 49.Culver GM, Noller HF. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–43. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. Measurement of internal movements within the 30 S ribosomal subunit using Forster resonance energy transfer. J Mol Biol. 2005;354:459–72. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Lieberman KR, et al. The 23 S rRNA environment of ribosomal protein L9 in the 50 S ribosomal subunit. J Mol Biol. 2000;297:1129–43. doi: 10.1006/jmbi.2000.3621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.