Abstract
Proteins of the small ubiquitin-related modifier (SUMO) family are conjugated to proteins to regulate such cellular processes as nuclear transport, transcription, chromosome segregation and DNA repair. Recently, numerous insights into regulatory mechanisms of the SUMO modification pathway have emerged. Although SUMO-conjugating enzymes can discriminate between SUMO targets, many substrates possess characteristics that facilitate their modification. Other post-translational modifications also regulate SUMO conjugation, suggesting that SUMO signaling is integrated with other signal transduction pathways. A better understanding of SUMO regulatory mechanisms will lead to improved approaches for analyzing the function of SUMO and substrate conjugation in distinct cellular pathways.
Introduction
Post-translational modification of proteins by members of the small ubiquitin-related modifier (SUMO) protein family regulates diverse cellular processes, including transcription, replication, chromosome segregation and DNA repair1, 2. The SUMO proteins share structural similarities with ubiquitin, and their conjugation to substrates occurs through a related enzymatic cascade involving the sequential action of an E1 activating enzyme, an E2 conjugating enzyme and an E3 protein ligase3, 4. Substrate modification by sumoylation can alter protein-protein interactions, change protein intracellular localization or direct changes in the activities of the protein to which SUMO is attached.
Here, we discuss recently discovered mechanisms that control and regulate the process of SUMO conjugation in the cell. Whereas global levels of SUMO modification can be altered by affecting the activities of the single E1 and E2 enzymes of the SUMO pathway (Box 1), we focus on mechanisms that contribute to the regulation of SUMO conjugation with respect to extrinsic factors or substrate modifications that alter or influence specificity. This includes a discussion of the roles of SUMO consensus motifs and SUMO-interacting motifs (SIMs), as well as the roles of additional post-translational modifications, such as phosphorylation, ubiquitylation and acetylation, that alter or regulate the process of SUMO modification.
Box 1. Control of global SUMO conjugation.
The small ubiquitin-related modifier (SUMO) conjugation system can be regulated by altering the expression levels or activities of enzymes in the pathway. At the level of transcription, calcium-induced keratinocyte differentiation is associated with upregulated gene expression of SUMO genes and the SUMO pathway machinery104. An overall increase in SUMO conjugation activity has also been observed in response to various stimuli, including heat shock and high levels of both oxidative and ethanol stresses105, 106. Conversely, at lower concentrations of reactive oxygen species, the SUMO pathway is inhibited by formation of a disulfide bridge between the catalytic Cys residues of the E1 activating enzyme and E2 conjugating enzyme107. Stability of the E1 and E2 enzymes can also tune the activities associated with the SUMO pathway. Chicken adenovirus GAM1 is reported to control E1 turnover by binding to the E1 enzyme and recruiting Cullin-RING ubiquitin ligases to target its subunits for degradation108, 109. GAM1 also decreases E2 enzyme levels, although the mechanism of this is not known108. Similarly, infection with Listeria monocytogenes is reported to cause proteasome-independent degradation of ubiquitin-like conjugating enzyme (UBC9) and proteasome-dependent degradation of SUMO conjugates110.
Other binding partners and small molecules can exert effects on the SUMO conjugation machinery. Ginkgolic acid is reported to block E1~SUMO thioester formation111, whereas the protein RSUME was reported to enhance E2~SUMO thioester formation112. E2-binding proteins, including the RING finger protein MEL18 and two SUMO-like domain-containing proteins – the yeast DNA repair protein Rad60 and human nuclear factor-interacting protein NIP45 – may regulate SUMO modification by sequestering the conjugation machinery57, 58, 113. The E2 enzyme can also be modified by SUMO within its amino-terminal helix, which has been shown to alter E2 specificity to particular SUMO substrates29.
The SUMO pathway
The process of activating and attaching a SUMO to substrates results in the formation of an isopeptide bond between the carboxy-terminal carboxyl group of the SUMO and an ε-amino group of a substrate acceptor Lys residue1–4. As with many other ubiquitin and ubiquitin-like proteins (Ubl proteins), all eukaryotic SUMO proteins are translated as immature precursors that must first be processed by a protease to generate the mature form. The mature form has a C-terminal diglycine motif that is required for efficient adenylation by a heterodimeric SUMO E1 enzyme (Fig. 1). Once formed, the SUMO adenylate is attacked by a conserved Cys on the E1 enzyme to form an E1~SUMO thioester and then transferred to a conserved Cys on a SUMO E2 enzyme, thereby generating an E2~SUMO thioester. Although the SUMO E2 can directly interact with some SUMO substrates to transfer the SUMO to substrate acceptor Lys residues, E3 protein ligases often facilitate this process through two mechanisms. They can recruit the E2~SUMO thioester and substrate into a complex to promote specificity or, in the case of substrates that interact directly with the E2 enzyme, they can enhance conjugation by stimulating the ability of the E2 enzyme to discharge SUMO to substrates. Akin to dephosphorylation, which serves to restore the function of a protein before phosphorylation, SUMO conjugation is reversible through the activities of proteases, including Ubl-specific proteases and sentrin-specific proteases (Ulps and SENPs) (Box 2) that deconjugate SUMO from substrates.
Figure 1. The SUMO conjugation cycle.
Small ubiquitin-related modifier (SUMO) undergoes processing by ubiquitin-like protein-specific proteases (Ulps) and sentrin-specific proteases (SENPs) to its mature form (step 1), revealing a carboxy-terminal Gly-Gly motif. SUMO is then adenylated by the SUMO-activating enzyme subunit 1 (SAE1)-ubiquitin-like activating enzyme subunit 2 (UBA2) E1 complex in an ATP·Mg2+-dependent reaction and transferred to the catalytic Cys of the UBA2 subunit (step 2). Following activation, SUMO is transferred to the catalytic Cys of the E2 conjugating enzyme, ubiquitin-like conjugating enzyme 9 (UBC9) (step 3). It can then catalyze conjugation to a substrate in an E3 ligase-independent manner through recognition of SUMO consensus motifs (ΨKXE) that contain a Lys acceptor residue (step 4). In addition, SUMO ligases can facilitate SUMO transfer through distinct mechanisms. The E3 ligase may coordinate the E2~SUMO thioester in an optimal conformation for catalysis without directly contacting the substrate, as is the case for Ran-binding protein 2 (RanBP2)11 (step 5). The Siz and PIAS (protein inhibitor of activated STAT) E3 ligases contact the E2 and SUMO through their SP-RING and Siz/PIAS carboxy-terminal domain (SP-CTD) domains, respectively (step 6). In both steps 5 and 6, substrate specificity is derived from the E2 enzyme. The Siz/PIAS family proteins also contain a PINIT domain that can contact the substrate, as is the case for the substrate proliferating cell nuclear antigen (PCNA)65, 68(step 7). Substrate specificity imparted by E3-substrate interactions are thought to be particularly important for directing conjugation to non-consensus Lys residues. Substrates modified by SUMO can contact SUMO-binding proteins through their SUMO-interacting motifs (SIMs) (step 8). Deconjugation is performed by Ulp and SENP proteases and free SUMO may be recycled for another round of conjugation (step 9).
Box 2. Proteases.
The family of proteases that catalyze small ubiquitin-related modifier (SUMO) processing and deconjugation includes two ubiquitin-like protein-specific proteases in yeast (Ulp1 and Ulp2) and six sentrin-specific proteases (SENPs) in humans (SENP1-3 and SENP5-7). Family members differ in their subcellular localization and can undergo changes in localization in a cell cycle dependent manner. For example, Ulp1 is localized to the nuclear periphery throughout interphase through tethering to nuclear pore complexes by karyopherins, but it enters the nucleoplasm during mitosis114, 115. By contrast, Ulp2 is localized to the nucleus by multiple nuclear localization signals and remains there throughout the cell cycle116. As with Ulp1, SENP2 is localized to the nuclear pore complex117, whereas SENP1, SENP6 and SENP7 are present in the nucleoplasm118–120 and SENP3 and SENP5 are observed in the nucleolus121, 122.
SUMO proteases also differ with respect to SUMO isoform or SUMO chain specificity. In yeast, Ulp1 is the major SUMO protease and is required for cell cycle progression123, whereas Ulp2 is responsible for deconjugation of SUMO chains124. Both SENP1 and SENP2 have been reported to deconjugate all SUMO isoforms equally well, but SENP1 displays a slight preference for processing SUMO1125, whereas SENP2 displays a slight preference for SUMO2126, 127. In addition, SENP3 and SENP5-7 have all been shown to preferentially deconjugate SUMO2- and SUMO3-modified proteins and SUMO chains119, 122, 128.
The levels of SUMO proteases change in response to a number of stimuli. SENP1 undergoes transcriptional upregulation in prostate cancer129 and forms reversible crosslinks that inhibit its activity under conditions of oxidative stress130. Conversely, increased levels of reactive oxygen species stabilize SENP3, whereas phosphorylation mediated by the ADP-ribosylation factor GTP-binding protein (ARF) tumor suppressor can decrease SENP3 levels through the ubiquitin-proteasome pathway131, 132. SENP2 has also been reported to undergo proteasome-mediated degradation133. In addition to regulating SUMO levels by protease turnover, phosphorylation can impair protease function, as Ulp2 is inhibited during mitosis by Cdc5-dependent phosphorylation134.
Yeast and invertebrates have a single SUMO-encoding gene, whereas vertebrate genomes have at least four SUMO isoforms, SUMO1-4, each encoded by distinct genes. However, it remains unclear whether SUMO4 is processed or conjugated to cellular proteins5, 6. SUMO2 and SUMO3 are often referred to as SUMO2/3 because they share 97% identity and because antibodies cannot distinguish the two isoforms. By contrast, SUMO2/3 shares only ~50% sequence identity with SUMO1, a larger difference in sequence similarity than that between human ubiquitin and the Ubl protein NEDD8 (sequence similarity ~55%), which require distinct enzymes for activation and conjugation3. Consistent with differences observed at the primary sequence level, SUMO1 and SUMO2/3 are conjugated to distinct substrates in vivo and also differ in their ability to form SUMO chains. Chain formation is attributed to the observation that SUMO2/3, as well as SUMO proteins from Saccharomyces cerevisiae and Schizosaccharomyces pombe, possess Lys residues near their amino termini that serve as SUMO acceptor sites, a feature that is not readily apparent in SUMO1. The biological relevance of SUMO chains remains an active area of research, although SUMO chains have been implicated in several processes, which are discussed below.
Substrate specificity in the SUMO pathway has remained somewhat of an enigma because the pathway relies on a single E2 enzyme, ubiquitin-like conjugating enzyme 9 (UBC9), and only a few E3 ligases have been identified3. The use of a single E2 and a few E3 enzymes in the SUMO conjugation pathway is in stark contrast to the ubiquitin pathway, which uses tens of E2 enzymes in unique combinations with hundreds of E3 enzymes to regulate substrate selection. As discussed below, the SUMO E2 is unique among E2 enzymes in its ability to specifically recognize and conjugate SUMO to some substrates in the absence of an E3 ligase, whereas ubiquitin-conjugating enzymes generally require an E3 ligase to impart substrate specificity7. However, SUMO conjugation is almost always enhanced in the presence of SUMO E3 ligases.
Known SUMO E3 enzymes include proteins that contain SIMs, as exemplified by the E3 ligase domain of Ran-binding protein 2 (RanBP2)8, and a family of SP-RING-containing proteins, including the Siz and protein inhibitor of STAT (PIAS) proteins9. Members of the Siz/PIAS family of E3 ligases include Siz1, Siz2, methyl methanesulfonate-sensitivity protein 21 (Mms21) and molecular zipper protein 3 (Zip3) in yeast and PIAS1, PIAS3, PIASxα, PIASxβ and PIASy in humans. These ligases include an SP-RING domain and are thought to function in an analogous manner to ubiquitin RING E3 enzymes through the binding and co-localization of substrates and the E2~SUMO thioester, which brings them into close proximity to promote SUMO transfer (Fig. 1). As noted above, the SUMO E3 ligase domain of RanBP2 is distinct from canonical RING-type or HECT-type E3 ligases. In this case, the E3 domain includes tandem elements that each possess E3 ligase activity10, and these domains include within them motifs that bind SUMO and the E2, as an E2~SUMO thioester, to stimulate the E2 enzyme to discharge SUMO11. As such, the E3 functions to coordinate the E2~SUMO thioester in an orientation that is optimal for catalysis and for productive E2-mediated substrate interactions (Fig. 1). Interestingly, this E3 does not require direct interaction with the substrate, although the location of RanBP2 at the nuclear pore complex (NPC) could facilitate interactions with substrates that pass through the NPC. Several other proteins have been reported as E3 enzymes and include human polycomb 2 homolog (PC2), histone deacetylase 4 (HDAC4), topoisomerase I-binding RING finger protein (TOPORS) and Ras homolog enriched in striatum (RHES)12–15, although the molecular details of how they promote SUMO modification remain less clear. Of the E3 ligases that have been characterized, some display substrate preferences whereas others display SUMO isoform-specific conjugation of particular targets.
The number of SUMO proteases rivals the number of E3 enzymes in any particular organism and they have a pivotal role in regulating the SUMO pathway. Although less clear, it is thought that substrate specificity of SUMO proteases is achieved because these enzymes exhibit distinct subcellular localizations and display specificities for some substrates as well as SUMO isoforms16 (Box 2).
Consensus motifs for SUMO conjugation
Many SUMO-modified proteins identified contain an acceptor Lys within a ψKX(D/E) consensus motif, where ψ is a large hydrophobic residue17. These residues directly interact with the SUMO E2 UBC9 (Fig. 2) and thus have a critical role in regulating the stability of interactions between the E2 enzyme and the substrate18. Structures of complexes between UBC9 and substrates containing SUMO consensus motifs demonstrate that the consensus motif adopts an extended conformation, wherein the acceptor Lys fits into a hydrophobic groove on UBC9, and electrostatic and hydrogen bonding interactions between UBC9 and substrate residues that flank the Lys residue facilitate recognition of the consensus motif19, 20. It is also worth noting that residues on the E2 surface not only position the Lys within the E2 active site but suppress the Lys pKa to increase the rate of catalysis21. Because the SUMO consensus motif adopts an extended conformation for productive interactions with the E2 enzyme, the context of this motif within the substrate is a crucial determinant that regulates its ability to be modified by SUMO. Consistent with this, most validated SUMO consensus sites occur in extended loops or intrinsically disordered regions of the substrate outside of its globular fold.
Figure 2. SUMO consensus motifs and SUMO-interacting motifs.
a | Amino acid sequence alignment of the canonical small ubiquitin-related modifier (SUMO) consensus motif, inverted consensus motif, hydrophobic cluster motif, phosphorylation-dependent SUMO motif (PDSM) and negatively charged amino acid-dependent SUMO motif (NDSM). Ψ represents a hydrophobic amino acid and K is the Lys modified by SUMO. b | Electrostatic potential surface representation of ubiquitin-like conjugating enzyme 9 (UBC9) adjacent to the Ran GTPase-activating protein 1 (RanGAP1) consensus motif (Protein Databank (PDB) code 2GRN). A model for the PDSM motif shown in stick representation was constructed by combining the RanGAP1 consensus motif with a phosphorylated peptide obtained from a complex between PIN1 and a phosphorylated RNA polymerase II carboxy-terminal domain peptide (PDB code 1F8A). The phosphorylated serine of the PDSM interacts with a basic patch on the surface of UBC9. Lys residues on UBC9 that are important for phosphate recognition are labeled. c | Sequence alignment of selected SUMO-interacting motifs (SIMs). Residues of the hydrophobic core sequence are highlighted in blue. Acidic residues and phosphorylated Ser residues are marked in pink and green, respectively. d | Electrostatic surface representation of human SUMO1 bound to a stick representation of the SIM of Ran-binding protein 2 internal repeat 1 (RanBP2 IR1; PDB code 1Z5S). SUMO1 and the Lys and Arg residues thought to be important for contacting phospho-SIMs and acidic residue flanking SIMs are labeled. Positive, apolar and acidic electrostatic potential on UBC9 and SUMO1 surfaces is indicated as blue, white and red, respectively. Molecular graphic representations of the structure were generated using PyMOL135. PIAS, protein inhibitor of activated STAT; PML, promyelocytic leukemia protein; TDG, thymine DNA glycosylase; USP25, ubiquitin-specific protease 25.
In addition to canonical four amino acid SUMO consensus motifs, longer sequences that include both SUMO consensus motifs and additional elements have been identified in some SUMO substrates. These include phosphorylation-dependent SUMO motifs (PDSMs) and negatively charged amino acid-dependent SUMO motifs (NDSMs) (Fig. 2a). PDSMs occur in proteins that are substrates for modification by Pro-directed kinases22 and comprise a SUMO consensus motif located adjacent to a phosphorylation site, ψKX(D/E)XXSP. In all known cases, phosphorylation increases levels of SUMO conjugation both in vitro and in vivo. This effect was first noted in studies of heat shock factors, in which stress induces phosphorylation and results in increased levels of SUMO modification on the PDSM Lys residue23. This mechanism is also used by transcription factors, such as peroxisome proliferator-activated receptor γ (PPARγ) and myocyte-specific enhancer factor 2A (MEF2A)24, 25. In these instances, phosphorylation-dependent SUMO conjugation converts these transcription factors from activators to repressors, which highlights a crucial role for PDSMs and SUMO in signal transduction. SUMO conjugation is enhanced by phosphorylated PDSMs because the phosphorylated Ser side chain interacts with a basic patch on the E2 surface, extending interactions with the E2 enzyme beyond recognition of the SUMO consensus motif26 (Fig. 2b). This mechanism is probably shared with proteins that contain NDSMs, which comprise negatively charged residues that are C-terminal to the SUMO consensus site in the place of the phosphorylation site of PDSMs, although NDSMs may interact with a different subset of Lys residues on the UBC9 surface26, 27.
As noted above, consensus motifs come in many flavors and do not necessarily adhere to strict sequence or geometric requirements. For instance, a Lys residue that is readily modified by SUMO exists in an α-helix in both mammalian E2-25K (a ubiquitin E2) and UBC9 enzymes28, 29. Although it remains unclear how the SUMO E2 binds to E2-25K before conjugation, one possibility is that the substrate presents the Lys in the context of a three-dimensional epitope that mimics a canonical SUMO motif. In addition to interactions with the substrate, it is known that the SUMO E2 and SUMO can form a complex through non-covalent interactions that are distant from the E2 active site30–32. This observation is perhaps analogous to non-covalent interactions between ubiquitin and E2 enzymes that are observed in the ubiquitin pathway33, suggesting that the presence of SUMO on a substrate could influence subsequent interactions with E2~SUMO thioester complexes, for instance during SUMO chain formation.
In most studies, SUMO consensus motifs adopt similar polarity in their interactions with the E2 enzyme. However, recent studies have uncovered new motifs for SUMO conjugation, including inverted consensus motifs and motifs with an N-terminal hydrophobic cluster34 (Fig. 2a), highlighting the need for further investigation into the full diversity of SUMO modification sites. These alternative motifs, which include the acceptor Lys, probably impart further specificity to E2-substrate interactions through direct interaction with the E2.
Regulation by SIMs
Whereas SUMO consensus motifs mediate interactions between the E2 and substrate, SIMs mediate non-covalent interactions between SUMO and SIM-containing proteins35. Initially identified through two-hybrid screening and biophysical studies36–38, SIMs are generally characterized by a short stretch of hydrophobic amino acids, (V/I)X(V/I)(V/I), which are flanked by acidic residues (Fig. 2c). When in complex with SUMO, the SIM adopts a parallel or anti-parallel β-strand conformation that extends the SUMO β-sheet to allow the hydrophobic side chains of the SIM to interact with a hydrophobic pocket on the SUMO surface11, 39 (Fig. 2d). Although poorly characterized, the position of acidic residues that border the SIM probably determine the polarity of SIM-SUMO interactions through salt bridging or hydrogen bonding interactions with conserved basic residues on the SUMO surface. SIMs have been uncovered in a wide range of proteins, including SUMO enzymes, SUMO substrates, SUMO-binding proteins and SUMO-targeted ubiquitin ligases35, 40. This section discusses how SIMs contribute to function in each of these protein classes.
SIMs in SUMO substrates and SUMO-binding proteins
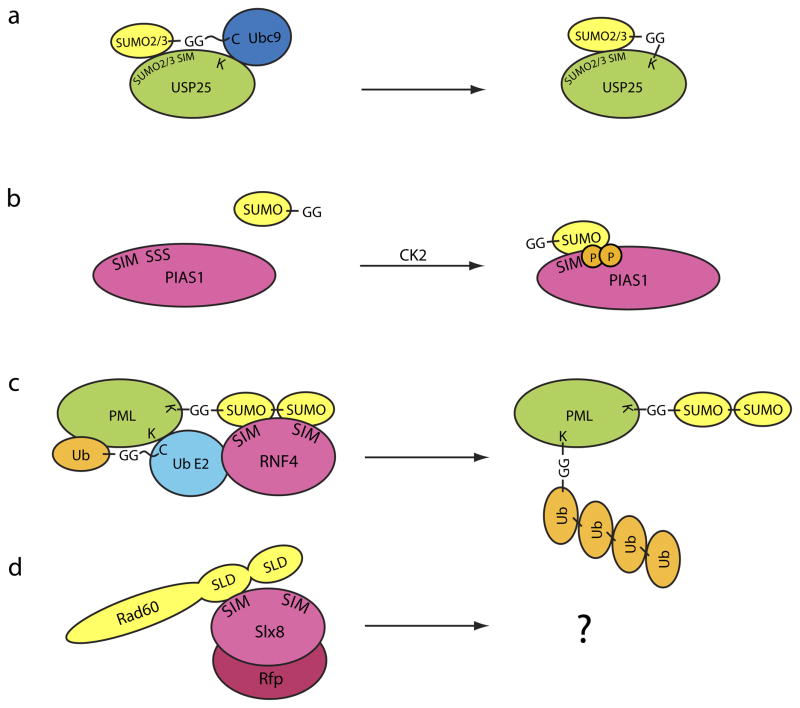
SIMs can mediate SUMO modification of a number of substrates, resulting in changes in their activity and/or localization. This process is exemplified by the base excision repair enzyme thymine DNA glycosylase (TDG), promyelocytic leukemia protein (PML) and the transcription factor DAXX, and has been reviewed previously2, 35, 41. Although the SIMs in these proteins do not display specificity for a particular SUMO isoform, this is not always the case42, 43. Ubiquitin-specific protease 25 (USP25) contains a SIM that contributes to preferential modification of this protein by SUMO2/344 (Fig. 3a). Although this SIM is required for efficient modification by SUMO2/3, its location within USP25 does not seem to be important for this function. SUMO2/3 specificity has also been reported for extended SIMs that are present in the transcriptional regulators MCAF1 and CoREST1, although these proteins do not seem to be substrates for SUMO modification45, 46. The basic architecture of the SIM motif has been characterized in complexes with SUMO, but the molecular determinants that underlie the ability of the SIM to recognize a particular SUMO isoform await further characterization.
Figure 3. Roles of SUMO-interacting motifs.
a | A specific small ubiquitin-related modifier (SUMO)-interacting motif (SIM) on ubiquitin-specific protease 25 (USP25) results in its modification by SUMO2 and/or SUMO3. b | Phosphorylation of SIMs that contain adjacent Ser residues (phospho-SIMs) by casein kinase 2 (CK2) activates non-covalent interactions between SUMO and protein inhibitor of activated STAT 1 (PIAS1). c | SIMs present on SUMO-targeted ubiquitin ligases recognize SUMO chains on promyelocytic leukemia protein (PML). This results in ubiquitylation of Lys residues on PML by a ubiquitin E2 conjugation enzyme (Ub E2) and the ligase, RING finger protein 4 (RNF4), followed by proteasome-mediated degradation. d | A SUMO-like domain (SLD) present in the yeast DNA repair protein Rad60 is recognized by SIMs on the SUMO-targeted Slx8-RING finger protein (Rfp) ubiquitin E3 ligase complex, although the consequences of these interactions are unclear.
In addition to canonical SIMs, a subclass of SIMs contain Ser residues that are proximal to the SIM and that are also sites of phosphorylation (Fig. 2c, 3b). Phospho-SIMs have been characterized in PML, EXO9 and in PIAS proteins47. In these cases, Ser residues C-terminal to the SIM are phosphorylated by casein kinase 2 (CK2), a ubiquitously expressed, constitutively active kinase that functions in multiple cellular pathways, including that of the transcription factor nuclear factor-κB (NF-κB) and Wnt signaling48. Phosphorylation by CK2 enhances non-covalent interactions with SUMO through contacts with multiple Lys residues on the SUMO surface47. The presence of Ser residues adjacent to SIMs has also been observed for homeodomain-interacting protein kinases and the ubiquitin-like activating enzyme subunit 2 (UBA2) subunit of the SUMO E136. However, it remains unclear whether phosphorylation of these or other substrates is required to enhance interactions with a SUMO protein.
SUMO-targeted ubiquitin ligases
SUMO-targeted ubiquitin ligases use SIMs to interact with SUMO-modified proteins and proteins that contain SUMO-like domains (SLDs), which are structurally related to SUMO despite low sequence similarity. First characterized in yeast, the heterodimeric SUMO-targeted ubiquitin ligases Slx8–RING finger protein (Rfp) in S. pombe and Slx5–Slx8 in S. cerevisiae are important for the maintenance of global levels of SUMO conjugates in the cell49–52. In fission yeast, Slx8 is a RING finger ubiquitin ligase, whereas Rfp1 and Rfp2 contain tandem SIMs that mediate SUMO binding and a RING-like domain that is important for interaction with Slx8. The budding yeast homolog Slx5–Slx8 maintains this heterodimeric architecture51, although higher eukaryotes encode a single protein (RING finger protein 4 (RNF4) in humans) that contains both SIMs and a functional RING domain49, 50. It seems likely that RNF4 functions as a dimer, based on what is known about Slx8–Rfp, Slx5–Slx8 and other heterodimeric RING E3 ligases.
The presence of multiple SIMs in these ubiquitin ligases is noteworthy because tandem SIMs could recognize two or more SUMO molecules in a SUMO chain53–55 (Fig. 3c). Consistent with this hypothesis, the first SUMO-targeted ubiquitin ligase-interacting proteins to be identified included the yeast DNA repair protein Rad60 and the human nuclear factor-interacting protein NIP45, proteins that contain tandem SLDs56 (Fig. 3d). SLDs have been reported to interact directly with components of the SUMO conjugation machinery, including mouse UBC9 and Uba2, Ubc9 and the E3 SUMO ligase Pli1 in S. pombe57, 58. It remains unclear whether interactions between SLDs and SUMO-targeted ubiquitin ligases depend on SIMs49, 57, 58 and SLD-containing proteins have not been verified as in vivo targets of these ubiquitin ligases. However, Rad60 is an in vitro substrate for Slx8–Rfp149. In vivo targets of SUMO-targeted ubiquitin ligases include SUMO-conjugated forms of human PML54, 55, hypoxia-inducible factor 2-α (HIF2α)59 and budding yeast modifier of transcription 1 (Mot1)60, although other recent data suggest that these ligases can recognize and ubiquitylate target proteins in a manner that is dependent on their SIMs but independent of substrate modification by SUMO61.
SIMs in the SUMO conjugation apparatus
SIMs have been identified in SUMO conjugation enzymes, including the SUMO E1 and several SUMO E3 ligases. Two distinct SIMs have been characterized within the C-terminal extension of the SUMO E1 UBA2 subunit, one at the C-terminus that was recently characterized by crystallography62 and one within the body of the C-terminal extension that was characterized by NMR37. Although E1 SIM-SUMO interactions share similarities with respect to other SUMO-SIM complexes62, the functional significance of the E1 SIMs remain unclear because the entire C-terminal extension is dispensable for human E1 activity in vitro and for function of the S. cerevisiae E1 in vivo63.
SIMs are commonly observed in SUMO E3 enzymes. Two SIMs exist in the E3 ligase domain of RanBP2, commonly termed IR1 and IR2. Although different functions have been attributed to these SIMs64, it remains unclear whether both are required for E3 activity. In the one case in which an atomic structure is known, the IR1 SIM mediates non-covalent interactions with SUMO1 (Fig. 2c,d) in a complex that contains SUMO1-modified Ran GTPase-activating protein 1 (RanGAP1) and the SUMO E2 UBC911. Importantly, deletion of the SIM resulted in decreased E3 ligase activity on other SUMO substrates, suggesting that non-covalent interactions are important not only for interactions with SUMO-conjugated RanGAP1 but also for interactions with a charged E2~SUMO thioester complex during the conjugation of other substrates11.
Members of the Siz/PIAS E3 family also contain SIMs in their C-terminal domains. In Siz1 this motif is not required for E3 activity, although its presence elicits a slight enhancement of SUMO modification of both consensus and non-consensus Lys residues65. For PIAS1, non-covalent interactions with SUMO occur through a C-terminal phospho-SIM that is phosphorylated by CK2 (Fig. 3b)47. Phosphorylation of Ser residues in the phospho-SIM presumably functions as a switch that allows the E3 enzyme to interact with SUMO, although it remains unclear whether this interaction is important for E3 ligase activity or for localization of the E3 to discrete foci through interactions with other SUMO-conjugated proteins.
Recent work has also uncovered two SIMs in PC2 that are thought to contribute to SUMO binding and E3 activity in vivo66, 67. As discussed above for other SIMs, Pc2 and Siz/PIAS E3 SIMs are flanked by acidic residues that probably assist in stabilizing electrostatic interactions with basic residues on the SUMO surface.
Regulation of E3 ligase activity
Some SUMO substrates interact specifically with the E2 enzyme in reactions that can be enhanced by the activity of E3 ligases. However, other substrates require specific E3 enzymes for SUMO modification that can also contribute to SUMO isoform selection in higher eukaryotes and to the spatial and temporal regulation of SUMO modification. This section addresses the general activities of ligases that contribute to SUMO modification, as well as those that impart substrate or SUMO isoform specificity.
Activities of SUMO ligases
In the absence of an E3 ligase, substrate specificity can be derived through E2-substrate interactions and, in some cases, interactions between the E2~SUMO thioester and SIMs. This is conceptually analogous to ligase-independent ubiquitin conjugation, which can occur on substrates that contain ubiquitin-binding domains7. SUMO E3 ligases are thought to catalyze SUMO transfer through two mechanisms. First, E3 enzymes can bind to the E2~SUMO thioester to coordinate it into a productive orientation for catalysis, thereby enhancing SUMO conjugation by promoting E2-dependent substrate interactions in reactions that do not require the E3 enzyme to directly interact with the substrate (Fig. 1). Second, the E3 enzyme may directly interact with the substrate, thereby recruiting the E2~SUMO thioester and substrate into a complex to facilitate conjugation. For members of the Siz/PIAS ligase family, the PINIT domain has been shown in some cases to be important for substrate binding65, 68, indicating that specificity is derived through a combination of substrate interactions with the E2~SUMO thioester and the E3 enzyme (Fig. 1).
Control of ligase activity and localization
Spatial and temporal regulation of the E3 enzyme controls the SUMO modification of some substrates. In yeast, cell cycle dependent localization of Siz1 controls the extent of SUMO modification of septins69. In this case, the nuclear transport receptor karyopherins mediate nuclear localization of Siz1 during interphase and transport it to the cytoplasm at the onset of mitosis, where it accumulates at septin rings formed during cell division at the bud neck of budding yeast69. Along with cell cycle dependent regulation of E3 enzymes, other post-translational modifications can alter E3 activity. For example, the homeodomain-interacting protein kinase 2 (HIPK2), a Pro-directed kinase and SUMO substrate, phosphorylates PC2, thereby enhancing the E3 activity of PC2 toward HIPK270. Notably, this phosphorylation occurs within a PDSM-like site, VKPETP, in which the Lys residue is modified by SUMO and Thr residue is phosphorylated, although this region of the protein does not appear to be required for E2 binding71. Conversely, phosphorylation of TOPORS, which is reported to be both a SUMO and ubiquitin ligase, elicits an inhibitory effect on SUMO modification. Whereas phosphorylation of TOPORS by PLK1 is required for its ubiquitin E3 activity on the tumor suppressor p53, phosphorylation decreases its SUMO E3 activity toward the same substrate72. It is unclear whether these examples illustrate a general mechanism of phosphorylation-mediated tuning of SUMO E3 ligase activity.
Automodification by SUMO has also been reported to alter SUMO E3 ligase activity. In the case of PIASy, SUMO modification of this E3 enhances its activity toward the transcription factor TCF4 in vivo, although the increase in activity may be indirect because automodification also causes changes in PIASy localization73. Although it is unclear whether automodification represents a general mechanism to regulate SUMO E3 ligases, it is attractive to consider because E3 automodification is a common theme for the regulation of E3 enzymes in the ubiquitin pathway74. In another example, ubiquitin E3 enzymes, such as human SIAH2 and TRIM32, have been shown to mediate the proteasomal degradation of PIAS proteins. Whereas SIAH2-mediated degradation seems to occur under normal cellular conditions75, TRIM32 facilitates the degradation of PIAS proteins in response to nitrosative stress. For example, S-nitrosylation of PIAS3 increases its affinity for TRIM32 and thereby promotes PIAS3 ubiquitination76. Just as ubiquitin ligases have been shown to regulate SUMO E3 enzymes, the reverse is also true. In response to genotoxic stress, PIAS1 and PIAS4 localize to sites of DNA damage along with other components of the DNA damage response, such as the breast cancer type 1 susceptibility protein (BRCA1)–BRCA-associated RING domain protein 1 (BARD1) ubiquitin ligase complex77, 78. In this case, PIAS proteins are predicted to facilitate SUMO modification of BRCA1, increasing its ubiquitin E3 ligase activity77, 78.
Substrate and SUMO isoform specificity of E3 ligases
The presence of SUMO E3 ligases contributes to the global levels of SUMO conjugates in the cell, but it is also clear that some E3s exhibit substrate specificity. In budding yeast, the two main E3 enzymes, Siz1 and Siz2, contribute to global levels of SUMO conjugates and in many instances share overlapping substrate specificity68. This overlap is probably due, in part, to the local concentrations of the E3 enzymes in the cell and their ability to stimulate SUMO transfer in reactions in which specificity is primarily dictated by substrate interactions with the E2 rather than the E3. However, some E3 ligases do exhibit specificity, and they are sometimes required for SUMO modification of substrates, including cases in which modification occurs on non-consensus site Lys acceptors.
An example of both types of E3 activity is illustrated by SUMO modification of proliferating cell nuclear antigen (PCNA), the processivity factor for eukaryotic DNA polymerases, in budding yeast. PCNA is modified in vitro and in vivo on two Lys residues, Lys127 and Lys16479, 80. Lys127 occurs within a canonical SUMO consensus motif and, although its modification is stimulated by Siz1, it can also be modified by Siz2 in the absence of Siz1. Siz2 is the primary E3 ligase for Lys127 modification in vivo81. Conversely, Lys164 is only modified in the presence of Siz1. In this case, specificity is dependent on the Siz1 PINIT domain, which is presumed to interact directly with a loop on PCNA to position Lys164 near the E2-SUMO thioester65, 68. Although the mechanisms underlying substrate specificity for specific E3-substrate interactions are less clear, other examples do exist. MEF2A is SUMO-modified by PIASx, but not in the presence of other PIAS proteins82, and DNA topoisomerase IIα is SUMO-modified in the presence of RanBP2 but not PIASy83.
These examples illustrate some specific E3-substrate pairings. However, the choice of which SUMO isoform to use during modification adds another layer of complexity to E3-imparted specificity. For instance, some E3 enzymes have been shown to modify particular substrates with a certain SUMO isoform – for example, PIASy conjugates SUMO1, not SUMO2/3, to NF-κB essential modulator (NEMO)84 and RanBP2 stabilizes SUMO modified RanGAP1 at the NPC, but only when it is modified with SUMO185, 86. Despite the fact that RanGAP1 was the first SUMO substrate to be identified, the mechanism underlying its SUMO1-specific modification remained unclear until recently. RanGAP1 can be modified equally well by both SUMO1 and SUMO2 in vitro87. However, in vivo, SUMO2/3-modified RanGAP1 is readily deconjugated by SUMO proteases, leading to low steady state levels of RanGAP1-SUMO2/3. Protection of RanGAP1-SUMO1 is mediated by formation of a stable complex between RanGAP1-SUMO1, UBC9 and RanBP2 and it is thought that this complex is less stable with RanGAP1-SUMO2 owing to the lower affinity of RanBP2 for the SUMO2 isoform.
This principle, namely that substrate-SUMO conjugates are protected from proteases when modified with a particular SUMO isoform, may underlie SUMO isoform selection under steady state conditions as exhibited by other substrates.
Regulation by substrate phosphorylation
Phosphorylation contributes to diverse outcomes in the SUMO pathway. The consequences of phosphorylating SUMO enzymes, phospho-SIMs and extended PDSMs have been discussed above. This section focuses on substrate phosphorylation and its role in mediating SUMO conjugation.
Positive regulation of substrate modification
Substrate phosphorylation has been shown to enhance SUMO modification for several SUMO substrates and phosphorylation-dependent SUMO conjugation motifs are a key example of this (see above). Phosphorylation of substrates adjacent to canonical SUMO consensus motifs can also have a positive effect on SUMO conjugation. In response to genotoxic stress, phosphorylation of PML by an extracellular signal-regulated kinase (ERK1 or ERK2) or HIPK2 enhances its SUMO modification88–90. Similarly, in response to retinoic acid, phosphorylation of the nuclear receptor TR2 by ERK2 leads to its SUMO-modification and association with PML bodies91. In both cases, phosphorylation does not occur within canonical PDSMs or phospho-SIMs. For human PML, phosphorylation by HIPK2 occurs on residues 8, 36 and 3889, but it is SUMO-modified on a non-consensus Lys at position 65 in addition to two SUMO consensus motifs at Lys160 and Lys49092. For mouse TR2, phosphorylation occurs on residues 203, 208 and 210, but only Thr210 phosphorylation enhances SUMO conjugation. In this case, the site of modification by SUMO in TR2 is not known but it is clear that phosphorylation sites are not immediately adjacent to any Lys residues. Based on these observations, it would appear that phosphorylation might lead to differential localization or conformational changes that expose the SUMO modification site to the SUMO conjugation apparatus.
Negative regulation of substrate modification
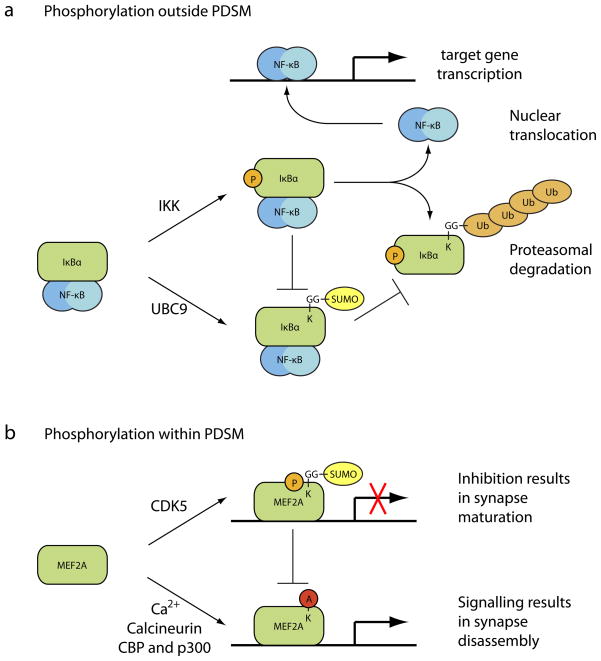
Phosphorylation of the NF-κB inhibitor α (IκBα) and the transcriptional regulators JUN, FOS and p53 has been reported to inhibit their modification by SUMO, thereby stimulating transcriptional activity93–96 (Fig. 4a). In addition, phosphorylation of the human cytomegalovirus kinase IE72 inhibits its ability to be SUMO conjugated97. Phosphorylation also results in decreased sumoylation of SATB1, a DNA-binding protein that is involved in chromatin remodeling98. In this case, phosphorylation was shown to inhibit interactions between SATB1 and PIAS proteins, thereby preventing PIAS-mediated SUMO modification. SUMO-conjugated SATB1 is shuttled to PML bodies, where it undergoes caspase-mediated cleavage, so phosphorylation functions as a switch to control these downstream events98.
Figure 4. Phosphorylation and SUMO modification regulate transcription factors.
a | Transcription factors, including nuclear factor-κB (NF-κB) inhibitor α (IκBα), are regulated through post-translation modifications. Phosphorylation of IκBα by the IκB kinase (IKK) outside a phosphorylation-dependent small ubiquitin-related modifier (SUMO) motif (PDSM) inhibits its SUMO modification by ubiquitin-like conjugating enzyme 9 (UBC9) and promotes modification by ubiquitin, targeting IκBα for degradation and releasing the transcription factor NF-κB to turn on target gene transcription. SUMO modification of IκBα in the absence of phosphorylation inhibits transcription by blocking IκBα turnover and release of NF-κB. b | Phosphorylation of the transcription factor myocyte-specific enhancer factor 2A (MEF2A) by cyclin-dependent kinase 5 (CDK5) within a PDSM enhances its modification by SUMO, inhibiting transcription and resulting in synapse maturation. Calcium signaling results in dephosphorylation by the calcium-dependent phosphatase calcineurin and acetylation of MEF2A by the histone acetyl transferases CREB-binding protein (CBP) and p300, converting it from a transcriptional repressor to a transcriptional activator and causing synapse disassembly.
Alternative Lys modifications
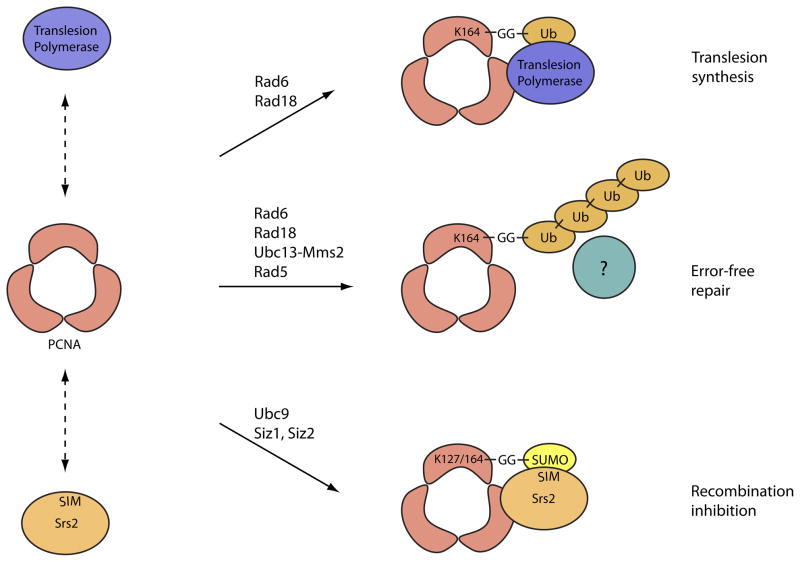
Although SUMO conjugation, ubiquitylation and acetylation are distinct Lys modifications that can occur within any single substrate, these modifications have been observed on the same Lys residue in some substrates. For example, IκBα can be modified by ubiquitin and SUMO on the same Lys residue93. In this case, SUMO modification was shown to inhibit phosphorylation-induced ubiquitylation and subsequent proteasomal degradation of IκBα (Fig. 4a). PCNA provides another example of ubiquitin and SUMO modifications occurring on the same Lys residue79 (Fig. 5). In yeast, SUMO modification of PCNA at Lys164 and Lys127 during S phase recruits the anti-recombinogenic ATP-dependent DNA helicase Srs2 in a manner that is dependent on a SIM located at its C-terminus99. The additional interaction with SUMO presumably increases the abundance of Srs2 at the replication fork to prevent unwanted recombination through disruption of Rad51 filaments. For PCNA, ubiquitylation on Lys164 does not result in proteasomal degradation, but rather initiates post-replication repair100. While monoubiquitylated PCNA participates in the recruitment of translesion DNA polymerases to promote error-prone repair, Lys63-linked polyubiquitylation promotes error-free repair, which may occur through the formation of sister chromatid junctions that are resolved by complexes of the ATP-dependent helicase Sgs1, DNA topoisomerase 3 (Top3) and RecQ-mediated genome instability protein 1 (Rmi1)101, 102. Unlike IκBα, SUMO and ubiquitin could coexist on yeast PCNA because it is a trimer of three identical subunits.
Figure 5. Recruitment to PCNA by ubiquitin-like proteins.
Post-translational modification of the processivity factor for eukaryotic DNA polymerases, proliferating cell nuclear antigen (PCNA), by ubiquitin-like modifiers results in the recruitment of specific proteins that otherwise display weak interactions with PCNA. In response to DNA damage, monoubiquitylation of PCNA by the yeast E2 conjugating enzyme Rad6 and the E3 ligase Rad18 recruits translesion polymerases and results in error-prone damage bypass. After monoubiquitylation, polyubiquitylation of PCNA by an E2 complex – comprising ubiquitin-conjugating enzyme 13 (Ubc13) and the ubiquitin-conjugating enzyme variant Mms2 – and the ubiquitin E3 ligase Rad5 results in error-free repair, although the specific factors recruited are not clear. PCNA is also modified by small ubiquitin-related modifier (SUMO) on Lys164 during S phase by the E2 enzyme ubiquitin-like conjugating enzyme 9 (Ubc9) and the SUMO E3 ligase Siz1, and on Lys127 by Ubc9, Siz1 and Siz2. This results in recruitment of the ATP-dependent DNA helicase Srs2 to inhibit recombination during replication.
Switches between SUMO-modification and acetylation have been reported. These competing modifications control signaling pathways in the cell. PDSM-mediated SUMO modification of MEF2A inhibits the transcriptional activity of MEF2A (see above, Fig. 4b) and, as a result, promotes synapse maturation in neurons25, 82. MEF2A can also be acetylated on the same Lys that is targeted for SUMO modification and acetylation of MEF2A is associated with an increase in MEF2A transcriptional activity25 (Fig. 4b). In this pathway, calcium signaling results in dephosphorylation of MEF2A by calcineurin, thereby inhibiting its SUMO modification through the PDSM-dependent mechanism. Likewise, calcium/calmodulin-dependent kinases phosphorylate histone deacetylases, resulting in their nuclear export to promote MEF2A acetylation. Hence, calcium signaling orchestrates a SUMO to acetyl switch that regulates transcription of MEF2 target genes. A similar SUMO to acetyl switch has been described for the protein HIC1103.
Conclusions
The SUMO conjugation pathway contributes to the regulation of a wide variety of cellular processes and further roles for SUMO are likely to be revealed. Although the basic mechanisms underlying E2-mediated SUMO conjugation of SUMO consensus sites are well defined, details regarding substrate selection in most other cases remains less clear, especially when it involves SUMO modification of Lys residues that differ from canonical SUMO consensus sites. It seems that Siz/PIAS E3 ligases participate in this process65, but questions remain with respect to how they bind and target specific substrates. Furthermore, although RanBP2 does not require direct interactions with the substrate to promote its E3 ligase activity11, this E3 ligase activity is localized to the NPC, suggesting that specificity could be achieved by other components of the NPC. Also, some SIMs display specificity toward a particular SUMO isoform44, but the preferences are often slight and the molecular basis for SUMO isoform specificity has not been determined. As such, it remains key for the community to continue work to identify and clarify mechanisms that underlie substrate specificity and SUMO isoform selection.
Proteins with SLDs have now been shown to bind SIMs and components of the SUMO conjugation machinery57. Although some inhibit SUMO chain formation through E2 binding58, it seems likely that other mechanisms exist to control the levels of SUMO conjugates in cells. As such, it will be important to determine whether the levels or activities of these proteins are regulated in response to stimuli, thereby altering cellular processes through global or local changes in SUMO modification.
Phosphorylation is a well established post-translational modification that is used to regulate signal transduction and it is now emerging as a regulatory mechanism to control SUMO modification of transcription factors. However, it remains unclear whether phosphorylation serves a broader purpose in the regulation of SUMO modification of other protein classes with differing cellular functions. Given the increasing recognition of SUMO as an important player in eukaryotic biology, illuminating the mechanisms that underlie the regulation of the SUMO conjugation cascade should have profound consequences on our understanding of the SUMO pathway and its effects on a host of cellular processes.
Acknowledgments
J.R.G. and C.D.L. were supported in part by NIH R01 GM065872.
Glossary Terms
- Ubiquitin
A protein with a β-grasp fold that can be conjugated post-translationally to proteins through an E1-E2-E3 enzyme cascade, most often to Lys residues within the substrate.
- E1
An enzyme that activates ubiquitin and ubiquitin-like proteins (including SUMO) through sequential adenylation and thioester bond formation with their carboxy-termini.
- E2
An enzyme that forms a thioester bond with ubiquitin (Ub) and ubiquitin-like (Ubl) proteins (including SUMO) following transfer from an E1 enzyme. E2 enzymes are referred to as Ub- or Ubl-conjugating enzymes because they can conjugate the Ub or Ubl protein directly to a substrate. However, E2 enzymes often require E3 ligases for proper function.
- E3
Often referred to as a protein ligase, an E3 enzyme either facilitates transfer of ubiquitin or ubiquitin-like proteins (including SUMO) from the E2 conjugating enzyme to a substrate or catalyzes thioester bond transfer between the E2 and E3 Cys before substrate conjugation, as is the case for HECT E3s.
- Ulps and SENPs
(Ubiquitin-like protein-specific proteases and sentrin-specific proteases). A family of Cys proteases that process SUMO to its mature form and deconjugate SUMO from modified substrates.
- RanBP2
(Ran-binding protein 2). A component of the nuclear pore complex that possesses SUMO E3 protein ligase activity. A domain within RanBP2 interacts directly with SUMO-modified Ran GTPase-activating protein 1 (RanGAP1) in a stable complex with ubiquitin-like conjugating enzyme 9.
- SP-RING
A domain present in the Siz and PIAS family of SUMO E3s that is structurally similar to the RING domains of ubiquitin E3 ligases, which coordinate two zinc ions and interact with E2~ubiquitin thioesters to facilitate ubiquitin transfer to a substrate. By contrast, SP-RING domains coordinate a single zinc ion and work in conjunction with the Siz/PIAS carboxy-terminal domain to activate the E2~SUMO thioester during conjugation.
- Siz/PIAS family
A family of SUMO E3 ligases that contain an SP-RING and Siz/PIAS carboxy-terminal domain that coordinate the E2~SUMO thioester. These proteins also contain conserved SAP and PINIT domains that have been implicated in substrate specificity by directing protein-DNA or protein-protein interactions.
- PDSM
A phosphorylation-dependent SUMO motif that is composed of an extended consensus motif, ψKX(D/E)XXSP, where S is the Ser that is phosphorylated. Phosphorylation of the Ser enhances E2 enzyme binding and modification of the Lys in the motif.
- RanGAP1
(Ran GTPase-activating protein 1). A Ran GTPase-activating protein and the first SUMO substrate identified. The carboxy-terminal domain of RanGAP1 is both necessary and sufficient for modification by SUMO. This domain includes a SUMO consensus motif and additional elements that interact directly with the E2 enzyme.
- PINIT domain
A domain in the Siz/PIAS E3 ligase family, named after a conserved five amino acid PINIT motif. The PINIT domain is located amino-terminal to the SP-RING and Siz/PIAS carboxy-terminal domain and is thought to be a substrate-binding domain.
References
- 1.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nature Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo D, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 6.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 7.Hoeller D, et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 9.Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nature Struct Mol Biol. 2004;11:984–991. doi: 10.1038/nsmb834. References 8 and 10 identify an E3 ligase domain within RanBP2 that is unrelated to RING- or HECT-type E3 ligases. [DOI] [PubMed] [Google Scholar]
- 11.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. This structure demonstrates how RanBP2 may function by orienting the E2~SUMO thioester for catalysis without contacting the substrate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weger S, Hammer E, Heilbronn R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005;579:5007–5012. doi: 10.1016/j.febslet.2005.07.088. [DOI] [PubMed] [Google Scholar]
- 15.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 18.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 19.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. References 17–19 define the sequence and mode of interaction between SUMO consensus motifs and the SUMO E2 conjugating enzyme UBC9. [DOI] [PubMed] [Google Scholar]
- 20.Lin D, et al. Identification of a substrate recognition site on Ubc9. J Biol Chem. 2002;277:21740–21748. doi: 10.1074/jbc.M108418200. [DOI] [PubMed] [Google Scholar]
- 21.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nature Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 22.Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Hietakangas V, et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashita D, et al. The transactivating function of peroxisome proliferator-activated receptor gamma is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells. 2004;9:1017–1029. doi: 10.1111/j.1365-2443.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- 25.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. Defines a SUMO to acetyl switch on the MEF2A transcription factor that regulates postsynaptic differentiation. [DOI] [PubMed] [Google Scholar]
- 26.Mohideen F, et al. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nature Struct Mol Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. Characterization of a basic patch on UBC9 that is required for PDSM recognition and its SUMO modification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichler A, et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nature Struct Mol Biol. 2005;12:264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 29.Knipscheer P, et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duda DM, et al. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J Mol Biol. 2007;369:619–630. doi: 10.1016/j.jmb.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Matic I, et al. Site-Specific Identification of SUMO-2 Targets in Cells Reveals an Inverted SUMOylation Motif and a Hydrophobic Cluster SUMOylation Motif. Mol Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. Together with references 37 and 38, this work defines a SUMO-interacting motif that mediates non-covalent binding to SUMO. [DOI] [PubMed] [Google Scholar]
- 37.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 39.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 40.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nature Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 42.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, et al. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Sekiyama N, et al. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem. 2008;283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 46.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Duncan JS, Litchfield DW. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta. 2008;1784:33–47. doi: 10.1016/j.bbapap.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 49.Prudden J, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Y, et al. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 52.Uzunova K, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. References 49–52 identify a conserved family of ubiquitin ligases that regulate cellular levels of SUMO-conjugated proteins. [DOI] [PubMed] [Google Scholar]
- 53.Mullen JR, Brill SJ. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem. 2008;283:19912–19921. doi: 10.1074/jbc.M802690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 55.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nature Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 56.Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics. 2005;6:22. doi: 10.1186/1471-2105-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. Molecular mimicry of SUMO promotes DNA repair. Nature Struct Mol Biol. 2009;16:509–516. doi: 10.1038/nsmb.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekiyama N, et al. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins. 2010;78:1491–1502. doi: 10.1002/prot.22667. [DOI] [PubMed] [Google Scholar]
- 59.van Hagen M, Overmeer RM, Abolvardi SS, Vertegaal AC. RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2alpha. Nucleic Acids Res. 2010;38:1922–1931. doi: 10.1093/nar/gkp1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol. 2009;29:1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie Y, Rubenstein EM, Matt T, Hochstrasser M. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 2010;24:893–903. doi: 10.1101/gad.1906510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tatham MH, et al. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nature Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 65.Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell. 2009;35:669–682. doi: 10.1016/j.molcel.2009.07.013. This study shows that the PINIT domain of Siz1 is essential for redirecting SUMO modification from a consensus Lys to a non-consensus Lys on PCNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merrill JC, et al. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS ONE. 2010;5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang SH, Sharrocks AD. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol Cell Biol. 2010;30:2193–2205. doi: 10.1128/MCB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reindle A, et al. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci. 2006;119:4749–4757. doi: 10.1242/jcs.03243. [DOI] [PubMed] [Google Scholar]
- 69.Makhnevych T, Ptak C, Lusk CP, Aitchison JD, Wozniak RW. The role of karyopherins in the regulated sumoylation of septins. J Cell Biol. 2007;177:39–49. doi: 10.1083/jcb.200608066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roscic A, et al. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol Cell. 2006;24:77–89. doi: 10.1016/j.molcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Kagey MH, Melhuish TA, Powers SE, Wotton D. Multiple activities contribute to Pc2 E3 function. EMBO J. 2005;24:108–119. doi: 10.1038/sj.emboj.7600506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang X, et al. Plk1-mediated phosphorylation of Topors regulates p53 stability. J Biol Chem. 2009;284:18588–18592. doi: 10.1074/jbc.C109.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ihara M, Yamamoto H, Kikuchi A. SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol Cell Biol. 2005;25:3506–3518. doi: 10.1128/MCB.25.9.3506-3518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 75.Depaux A, Regnier-Ricard F, Germani A, Varin-Blank N. A crosstalk between hSiah2 and Pias E3-ligases modulates Pias-dependent activation. Oncogene. 2007;26:6665–6676. doi: 10.1038/sj.onc.1210486. [DOI] [PubMed] [Google Scholar]
- 76.Qu J, et al. Nitric oxide destabilizes Pias3 and regulates sumoylation. PLoS ONE. 2007;2:e1085. doi: 10.1371/journal.pone.0001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 78.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. This study was the first to show that PCNA is modified by SUMO and that ubiquitin and SUMO modifications elicit different responses to DNA damage. [DOI] [PubMed] [Google Scholar]
- 80.Windecker H, Ulrich HD. Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J Mol Biol. 2008;376:221–231. doi: 10.1016/j.jmb.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Parker JL, et al. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008;27:2422–2431. doi: 10.1038/emboj.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shalizi A, et al. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J Neurosci. 2007;27:10037–10046. doi: 10.1523/JNEUROSCI.0361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dawlaty MM, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nature Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 85.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. References 85 and 86 show that SUMO is a post-translational modifier that can alter protein localization and interactions. [DOI] [PubMed] [Google Scholar]
- 87.Zhu S, et al. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol Cell. 2009;33:570–580. doi: 10.1016/j.molcel.2009.02.008. This study demonstrates that RanGAP1-SUMO conjugates are protected from proteases when modified by SUMO1 but not SUMO2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gresko E, et al. PML tumor suppressor is regulated by HIPK2-mediated phosphorylation in response to DNA damage. Oncogene. 2009;28:698–708. doi: 10.1038/onc.2008.420. [DOI] [PubMed] [Google Scholar]
- 90.Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389–401. doi: 10.1016/s1535-6108(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 91.Gupta P, et al. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc Natl Acad Sci U S A. 2008;105:11424–11429. doi: 10.1073/pnas.0710561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kamitani T, et al. Identification of three major sentrinization sites in PML. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 93.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 94.Muller S, et al. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 95.Lin JY, Ohshima T, Shimotohno K. Association of Ubc9, an E2 ligase for SUMO conjugation, with p53 is regulated by phosphorylation of p53. FEBS Lett. 2004;573:15–18. doi: 10.1016/j.febslet.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 96.Bossis G, et al. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. SUMO-1 modification of human cytomegalovirus IE1/IE72. J Virol. 2002;76:2990–2996. doi: 10.1128/JVI.76.6.2990-2996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan JA, Song J, Chen Y, Durrin LK. Phosphorylation-dependent interaction of SATB1 and PIAS1 directs SUMO-regulated caspase cleavage of SATB1. Mol Cell Biol. 2010;30:2823–2836. doi: 10.1128/MCB.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. This study showed that Srs2 is recruited to PCNA in a SUMO-dependent manner (see also reference 79) [DOI] [PubMed] [Google Scholar]
- 100.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 101.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 102.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 103.Stankovic-Valentin N, et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deyrieux AF, Rosas-Acosta G, Ozbun MA, Wilson VG. Sumoylation dynamics during keratinocyte differentiation. J Cell Sci. 2007;120:125–136. doi: 10.1242/jcs.03317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 106.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 108.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 109.Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 110.Ribet D, et al. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 2010;464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fukuda I, et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 112.Carbia-Nagashima A, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 113.Zhang J, Goodson ML, Hong Y, Sarge KD. MEL-18 interacts with HSF2 and the SUMO E2 UBC9 to inhibit HSF2 sumoylation. J Biol Chem. 2008;283:7464–7469. doi: 10.1074/jbc.M707122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor DL, Ho JC, Oliver A, Watts FZ. Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J Cell Sci. 2002;115:1113–1122. doi: 10.1242/jcs.115.6.1113. [DOI] [PubMed] [Google Scholar]
- 115.Panse VG, Kuster B, Gerstberger T, Hurt E. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nature Cell Biol. 2003;5:21–27. doi: 10.1038/ncb893. [DOI] [PubMed] [Google Scholar]
- 116.Kroetz MB, Su D, Hochstrasser M. Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Mol Biol Cell. 2009;20:2196–2206. doi: 10.1091/mbc.E08-10-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 118.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 119.Mukhopadhyay D, et al. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J. 2009;421:223–230. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- 121.Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem. 2000;267:6423–6427. doi: 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 122.Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 123.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. This study indentified the first SUMO protease, Ulp1. [DOI] [PubMed] [Google Scholar]
- 124.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 125.Shen LN, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J. 2006;397:279–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 127.Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nature Struct Mol Biol. 2006;13:1060–1068. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- 128.Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem. 2008;283:32045–32055. doi: 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bawa-Khalfe T, Cheng J, Wang Z, Yeh ET. Induction of the SUMO-specific protease 1 transcription by the androgen receptor in prostate cancer cells. J Biol Chem. 2007;282:37341–37349. doi: 10.1074/jbc.M706978200. [DOI] [PubMed] [Google Scholar]
- 130.Xu Z, et al. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 2008;22:127–137. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- 131.Huang C, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–2762. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kuo ML, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378–3387. doi: 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- 133.Itahana Y, Yeh ET, Zhang Y. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol Cell Biol. 2006;26:4675–4689. doi: 10.1128/MCB.01830-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- 135.DeLano W. The PyMOL molecular graphics system. 2002. [Google Scholar]