Abstract
The connection between intestinal microbiota and host physiology is increasingly becoming recognized. The details of this dynamic interaction, however, remain to be explored. Toll-like receptor 2 (Tlr2) is important for its role in bacterial recognition, intestinal inflammation, and obesity-related metabolic changes. Therefore, we sought to determine the epigenomic and metagenomic consequences of Tlr2 deficiency in the colonic mucosa of mice to gain insights into biological pathways that shape the interface between the gut microbiota and the mammalian host. Colonic mucosa from wild type (WT) and Tlr2−/− C57BL/6 mice was interrogated by microarrays specific for DNA methylation and gene expression. The mucosal microbiome was studied by next-generation pyrosequencing of bacterial 16S rRNA. The expression of genes involved in immune processes was significantly modified by the absence of Tlr2, a number of which correlated with DNA methylation changes. The epigenomic and transcriptomic modifications associated with alteration in mucosal microbial composition. Several bacterial species, including members of the Firmicutes were significantly different in abundance between WT and Tlr2−/− animals. This manuscript highlights the intimate interrelationships between expression of immune-related genes and immunity pathways in the host with compositional and functional differences of the mammalian microbiome.—Kellermayer, R., Dowd, S. E., Harris, R. A., Balasa, A., Schaible, T. D., Wolcott, R. D., Tatevian, N., Szigeti, R., Li, Z., Versalovic, J., Smith, C. W. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice.
Keywords: metagenomics, inflammatory bowel diseases, metabolic syndrome, epigenetics, Tlr2
The mutualistic relationship between the host and its intestinal microbiota (microbial community) plays a major role in mammalian immunomodulation and metabolism (1–4). Outstanding examples of this are Crohn's disease (CD) and ulcerative colitis (UC), chronic human disorders that are collectively designated as inflammatory bowel diseases (IBDs). Growing evidence supports the hypothesis that these entities develop secondary to a genetic predisposition for an exaggerated mucosal immune response against components of the intestinal microbiota, and that this process is modulated by environmental factors (5, 6). The microbiota and the host may simultaneously and/or interdependently respond to such factors. These responses are likely affected by the genetic composition of the host. With regard to this relationship, epigenetic mechanisms (molecular processes that can influence gene expression without a change in the genetic code) are environmentally responsive and have been implicated in the pathogenesis of IBDs (6, 7). An increasing number of observations reveal associations not only between genetic and epigenetic variation (8), but also between intestinal pathogens and mucosal epigenetic changes (9). However, these interactions have rarely been addressed in regard to IBD susceptibility genes, bacteria, and host-specific epigenomic modification, such as DNA methylation (methylation of cytosines in CpG dinucleotides; ref. 8). Meanwhile, environmentally sensitive nongenomic alterations are likely significant in the development of these diseases (6), and relevant intestinal epithelial epigenetic modification can occur in response to bacterial components (10).
Toll-like receptors are conserved transmembrane molecules expressed in a number of intestinal cells (epithelial cells, T and B lymphocytes, and macrophages) implicated in the complex network of microbial pattern recognition. They are acknowledged as central modulators of gut tissue integrity (11–13). Specifically, Toll-like receptor 2 (Tlr2) has been associated with IBDs, although its role as an intestinal inflammation regulator has been debated (14). In the meantime, acute and chronic models of murine colitis (15–17), in addition to canine (18) and human (19, 20) studies, underscore its significance in modulating mucosal immune responses relevant to IBDs. Notably, the R753Q (G/A) single-nucleotide polymorphism of TLR2 has been correlated with a severe phenotype of UC (21), and was shown to induce functional deficiency of TLR2 (22). Tlr2 has also been recognized to play a pivotal role in diet-induced murine metabolic syndrome. The potential role of the microbiota has been emphasized in this respect, but it has not been directly studied (23, 24).
Since alterations in mucosal pattern recognition might affect intestinal microbial composition, Tlr2-deficient mice have been tested for such changes. High-resolution density-gradient gel electrophoresis (DGGE) experiments assessing the variable regions of bacterial 16S ribosomal RNA gene (16S rRNA) in fecal samples either from Tlr2- or Tlr4-knockout mice failed to detect significant variation compared to controls (25, 26). Fecal microbiota, however, differs from mucosa-associated bacterial composition (27); the latter is likely more relevant for intestinal immunomodulation (28). In addition, recent observations with high-throughput pyrosequencing revealed altered gut microbiota in Tlr5−/− mice to drive systemic metabolic derangements (29). Therefore, technologies capable of providing increased resolution may be beneficial when addressing the microbiome in terms of Toll-like receptor deficiency.
Here, we interrogated the colonic mucosa of Tlr2−/− mice for epigenomic, transcriptomic, and metagenomic changes with high-throughput molecular methods to provide a proof of concept for significant microbiota shifts to associate with epithelial epigenetic changes influenced by the host genome.
MATERIALS AND METHODS
Animals and tissue collection
Tlr2 −/− male mice (generously provided by Dr. Jesus Vallejo, Baylor College of Medicine, Houston, TX, USA; originating from Dr. Shizuo Akira, ref. 30) and their wild-type (WT; C57BL/6) counterparts were housed and reared in our animal facility within the same room and fed standard irradiated, pelleted rodent diet (2920X; Harlan, Madison, WI, USA). For the purposes of the microbial studies, the animals were reared in the same room from birth to postnatal day 90 (P90). This age correlates with young adulthood in humans, when the onset of IBD peaks. The genotype of the animals was confirmed randomly. At P90, mice were sacrificed with CO2 asphyxiation between 11:00 AM and 3:00 PM without previous food restriction. Colonic mucosa was collected as described before (7). The protocol was approved by the Institutional Animal Care Committee for Baylor College of Medicine.
Histology
Mucosal scrapings were formaldehyde (10%) fixed, paraffin embedded, sectioned, and stained according to standard pathological procedures (31). Tlr2−/− and WT histology was confirmed by two pathologists (N.T. and R.S.).
Isolation and manipulation of nucleic acids
All procedures were performed on colonic mucosal scrapings. Genomic DNA was isolated with the QIAamp DNA Mini Kit (51304; Qiagen, Valencia, CA, USA) and bisulfite converted with the EZ DNA Methylation-Gold Kit (D5006; Zymo Research, Orange, CA, USA). Total RNA was isolated with the RNeasy Mini Kit (74106; Qiagen) and stored at −80°C. Reverse transcription was done with the Taqman Reverse Transcription Kit (N808-0234; Applied Biosystems, Branchburg, NJ, USA) according to the manufacturer's instructions.
Methylation-specific amplification microarray (MSAM)
MSAM was carried out as described previously (32); 2 independent Tlr2−/− vs. WT cohybridizations were performed (for raw and processed microarray data, see http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE21845). We designed a custom microarray (Agilent Technologies, Santa Clara, CA). SmaI/XmaI intervals between 100 and 2200 bp were identified in the mm9 assembly excluding chromosome Y. The resulting 41,237 intervals were uploaded to Agilent eArray (https://earray.chem.agilent.com/earray/) and HD-ChIP database probes overlapping the intervals were identified, allowing up to 4 probes/interval. In total, 95,386 probes covering 26,740 (65%) of the 100- to 2200-bp intervals, providing an average of 3.6 probes/interval, were included. Standard Agilent control probes (1565) along with 8121 probes overlapping 10–15 kb SmaI/XmaI intervals to act as further controls completed the 2 × 105,000 array design. The average signal intensity within each SmaI/XmaI interval was calculated. An interval was considered a hit if it showed a >1.6-fold change in both Tlr2−/− vs. WT comparisons. The same cutoff yielded outstanding validation in similar experiments (7). We used a list of all SmaI/XmaI intervals (1414 total) that did not change in methylation (<1.2 or >0.85 ratio in both arrays) for control purposes.
Pyrosequencing
DNA was amplified with traditional primer biotynilation following bisulfite conversion. A quantitative bisulfite pyrosequencing protocol was used for all methylation analyses with the utilization of the Pyro Q CpG program (Qiagen). Methylation measurements at both of the SmaI/XmaI sites of the gene-associated intervals were performed in the case of alanyl (membrane) aminopeptidase/CD13 [Anpep; +41 and+252 bp from transcription start site (TSS)]; and annexin A8 (Anxa8; −131 and +897 bp from TSS; primers in Table 1). The mean methylation of interferon-induced protein with tetratricopeptide repeats 2 (Ifit2) was measured and calculated from 2 CpG sites in the promoter region of the gene (−394, −384 bp from TSS), 65 and 75 bp downstream (the nearest) from the binding site of transcription factor IRF3, respectively (primers in Table 1). In case of SmaI/XmaI intervals where both informative CpGs changed in methylation, an algorithm was used to calculate and compare methylation ratios (32).
Table 1.
Pyrosequencing primers for Anpep- and Anxa8-associated SmaI/XmaI intervals, and Ifit2 promoter region
| Primer | Sequence |
|---|---|
| 5pAnpepF1 | GGGTTTTATTTTTTGTGAGGATAT |
| 5pAnpep R1-B | CCACCRACAAAACTATAATAATACACAC |
| 5pAnpep S1 | ATTTTTTGTGAGGATATAAGT |
| 3pAnpepF1 | GGGTGTGGTAGTTGTGTGTATTATTATAG |
| 3pAnpep R1-B | CTAATTCCAAAACTTACTTTCATCTACAA |
| 3pAnpep S1 | AGAATAGGAATGTAGAGAATTT |
| 5pAnxa8F1-B | GTTATAGTTTAAATATTTTTAGGGTAGG |
| 5pAnxa8R1 | CRCTTAACTCTAAACCCCTCCTAT |
| 5pAnxa8S1 | CCCACCCAAACACTA |
| 3pAnxa8F1 | TTAGTTTTAGGAAAATTTTTGTTGTTGTG |
| 3pAnxa8R1-B | TAATAACTCATCTCTTCCTACCTTCTAATC |
| 3pAnxa8S33 | GAGAAGTTGTAGGTGTAGATGTT |
| Ifit2F2 | GATTATTAGGAAATTGGAAATGAGTGA |
| Ifit2R2-B | AAACCAAACAAAATCCTTTTCTATCC |
| Ifit2S2 | ATTTTTGTTGTTTAGAATATTATAG |
B designates the biotinylated primer.
Expression microarray
Four Tlr2 −/− vs. WT whole genomic expression microarray (4×44k Whole Mouse Genome microarray, Quick-Amp 2-color labeling kit; Agilent Technologies) comparisons were performed (for raw and processed microarray data, see http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE21845). Probe intensities for each transcript were averaged, and Tlr2−/− vs. WT intensity ratios were calculated. Transcripts were considered a hit if those showed a >1.6-fold change in ≥2 of 4 microarray comparisons, with the remaining arrays not contradicting the increase or decrease of expression (i.e., ≥1 in case ≥2 other >1.6; ≤1 in case ≥2 other <0.625).
Gene expression
Quantitative real-time PCR was performed using the following TaqMan gene expression assays (Applied Biosystems): Anpep/CD13 (Mm 00476227_m1), β-actin (B-actin; Mm00607939_s1), tumor necrosis factor receptor superfamily (Fas; Mm00433237_m1), Ifit2 (Mm00492606_m1), galectin-2 (Lgals2; Mm00840285_m1), spondin-2 precursor (Spon2; Mm00513596_m1), and signal transducer and activator of transcription-1 (Stat1; Mm00439531_m1). Expression changes were determined as described before (7).
DNA extraction for microbial studies
After thawing, the colonic mucosal scrapings were centrifuged at 14,000 rpm for 30 s and resuspended in 500 μl RLT buffer (Qiagen) with β-mercaptoethanol. Sterile 5-mm steel beads (Qiagen) and 500 μl sterile 0.1-mm glass beads (Scientific Industries, Inc., Bohemia, NY, USA) were added for complete bacterial lyses in a TissueLyser (Qiagen), run at 30 Hz for 5 min. Samples were centrifuged briefly, and 100 μl of 100% ethanol was added to a 100-μl aliquot of the sample supernatant. This mixture was added to a DNA spin column, and DNA recovery protocols were followed as instructed in the QIAamp DNA Mini Kit (Qiagen), starting at step 5 of the tissue protocol. DNA was eluted from the column with 30 μl water, and samples were diluted accordingly to a final concentration of 20 ng/μl. DNA samples were quantified using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France) and confirmed with fluorometric analyses.
Massively parallel bacterial tag-encoded FLX-Titanium amplicon pyrosequencing (bTEFAP)
bTEFAP was performed as described previously (33). bTEFAP utilizes Titanium reagents and procedures (454 Life Sciences, Branford, CT, USA) and a 1-step PCR, mixture of Hot Start and HotStar high-fidelity taq polymerases (Qiagen), and amplicons originating from the 27F region, numbered in relation to Escherichia coli rRNA. The bTEFAP procedures were performed at the Research and Testing Laboratory (Lubbock, TX, USA; http://www.researchandtesting.com).
Bacterial diversity data analysis
All failed sequence reads, low-quality sequence ends, and tags were removed; sequences shorter than 300 bp were removed, and sequences were depleted of any nonbacterial ribosome sequences and chimeras using custom software described previously (33) and the Black Box Chimera Check B2C2 software (described and freely available at http://www.researchandtesting.com/B2C2.html). This process provided 4303 to 8871 filtered sequences in the individual mucosal DNA samples, which were queried using a distributed BLASTn .NET algorithm (34) against a database of high-quality 16S rRNA bacterial sequences derived from the National Center for Biotechnology Information. Database sequences were characterized as high quality based on similar criteria as described for RDP 9 (http://rdp.cme.msu.edu/; ref. 35). Using a .NET and C# analysis pipeline, the resulting BLASTn outputs were compiled, validated using taxonomic distance methods, and compiled as described previously (33). Mathematical modeling to estimate the total number of genera and species were conducted using Rarefaction, Shannon-index, and Chao1 within the Mothur analysis software (http://www.mothur.org; ref. 36), as has been described previously (37). Taxonomic trace-back analysis was also performed. Sequences with identity scores > 97% (<3% divergence) were resolved at the species level, between 95 and 97% at the genus level, between 90 and 95% at the family level, between 85 and 90% at the order level, between 80 and 85% at the class level, and below this to the phylum. Statistical tests, including principal components analysis (PCA), multiple analysis of variance (MANOVA), and hierarchal clustering to evaluate microbiome results, were performed using NCSS 2007 (NCSS, Kaysville, UT, USA). Student's t test comparisons between the WT and Tlr2−/− groups were adjusted for multiple testing by false discovery rate (FDR) determination (38). Taxonomic category differences with an FDR ≤ 5% were considered significant.
Statistical and bioinformatic analysis
Unpaired, 2-tailed t tests (except for correlation analyses, where 1-tailed t tests were employed), correlation, and odds ratio (OR) calculations were utilized in the group comparisons, where statistical significance was declared at P < 0.05. Error bars represent sem. Fatigo functional enrichment analysis (http://babelomics3.bioinfo.cipf.es/; ref. 39) was used to identify gene ontology classifications significantly over- or underrepresented among the gene lists (P<0.01 in this case); cited P values were adjusted for multiple testing. Data for enhancer and CTCF sites were downloaded from the Vista Enhancer Browser (http://enhancer.lbl.gov/frnt_page.shtml) and CTCFBSDB (http://insulatordb.uthsc.edu/help.php), respectively. Genomic coordinates were lifted over to mm9 using the University of California–Santa Cruz liftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). The search for overlaps with DNase hypersensitive sites utilized the University of Washington ENCODE DNaseI Hypersensitivity by Digital DnaseI data (John A. Stamatoyannopoulos, University of Washington, Seattle, WA, USA; http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg18&g=wgEncodeUwDnaseSeq). Data obtained from Caco2 cells with the HotSpot algorithm in two replicates (1 and 2) were used. The presence of enhancers, CTCF sites, and DNase-hypersensitive sites was assessed in genomic intervals encompassing the SmaI/XmaI intervals with altered methylation in the Tlr2−/− animals, and 2 kb upstream and downstream. CpG island association was declared if one or both of the SmaI/XmaI sites of a single interval were within a CpG island. The promoter region of a gene was arbitrarily declared as −5 kb upstream or +2 kb downstream from a TSS. All reported genomic coordinates are based on mm9 (http://genome.ucsc.edu).
RESULTS
Mucosal scrapings contain predominantly epithelial cells
To compare mucosal scrapings from Tlr2−/− and WT animals, the specimens were first histologically evaluated. In both study groups, >95% of the cells were epithelial (Supplemental Fig. S1). Therefore, the consecutively detected DNA methylation or gene expression changes must reflect Tlr2-deletion-dependent alterations within colonocytes rather than cellular composition modification of the mucosa secondary to the genetic defect.
Tlr2 deficiency induces colonic mucosal DNA methylation changes
To test the effects of Tlr2 deficiency on colonic mucosal epigenomics, a methylation-sensitive/insensitive restriction endonuclease isoschizomer (SmaI/XmaI)-based MSAM was utilized; 141 intervals had increased methylation, and 246 intervals had decreased methylation secondary to homozygous Tlr2 deletion (for processed array data, see http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE21845). Namely, ∼1.4% (387/26,740) of the interrogated genome was affected in Tlr2−/− animals. Since our earlier studies in murine colonic mucosa utilizing identical cutoffs for significance yielded an outstanding validation rate (7), we confirmed Tlr2-dependent epigenetic changes in 1 SmaI/XmaI interval with increased (Anpep associated; Fig. 1A), and 1 interval with decreased methylation (Anxa8 associated; Fig. 2) by bisulfite pyrosequencing. Gene ontology comparison between genes associated with differentially methylated intervals and genes linked to genomic intervals without a methylation change did not reveal significant (P<0.01) categories. This was likely due to the low number of affected intervals. In the meantime, searches for overlaps between DNase-hypersensitivity regions [OR=1.27, 95% confidence interval (CI): 1.004–1.605; and OR=1.86, 95% CI: 1.474–2.337] in two independent replicates, and CCCTC-binding factor (CTCF) sites (OR=1.88, 95% CI: 1.15–3.087) revealed enrichment for these in the Tlr2-dependent intervals with methylation change compared to controls. Similarly, CpG island (CpG-dense genomic regions, frequently found in gene promoters, the methylation of which usually inversely correlates with the expression of the associated gene; OR=3.96; 95% CI: 3.04 to 5.14), and gene promoter (OR=1.91; 95% CI: 1.52 to 2.41) association was increased in the Tlr2-dependent intervals highlighting structurally and functionally select loci. However, there was no difference in this regard in conjunction with enhancers.
Figure 1.
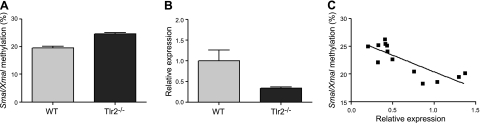
Tlr2-dependent DNA methylation changes at Anpep correlate with its expression. A) Methylation of the Anpep-associated SmaI/XmaI interval (calculated from both flanking sites; see Materials and Methods) increased in Tlr2−/− colonic mucosa (WT, n=8; Tlr2−/−, n=9; P<0.0001). B) Anpep expression decreased in Tlr2−/− colonic mucosa (WT, n=10; Tlr2−/−, n=11; P=0.017). C) Methylation at Anpep inversely correlated with expression (r=−0.8; n=13; P=0.008).
Figure 2.
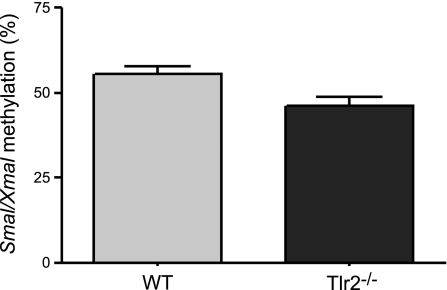
Methylation of the Anxa-associated 3′ SmaI/XmaI site is decreased in Tlr2−/− colonic mucosa (P=0.0154; n=10/group).
Colonic mucosal gene expression changes affect immune processes in Tlr2−/− mice
Following the realization of Tlr2-dependent epigenetic changes, we assessed genome-wide transcriptomic alterations in the colonic mucosa of Tlr2−/− mice; 328 transcripts with increase and 277 with decrease were identified (for processed array data, see http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE21845). Therefore, ∼1.8% (605/33,074) of all murine transcripts was affected significantly by homozygous Tlr2 deletion. Tlr2 transcript levels were the lowest, by at least an order of magnitude, in the Tlr2−/− tissues within the decreased transcript group serving as internal control. Ontology analysis revealed that genes involved in immune responses were enriched (P=0.0038; Supplemental Table S1) in the induced Tlr2-deficiency group compared to the rest of the mouse genome. We confirmed the significant expression difference in Ifit2 (Fig. 3B) and Spon2 (Fig. 4C) within the immune response-associated group. A trend for increased Fas expression was also supported (Fig. 4A). Similar transcriptomic changes to IBD-associated genes with differential expression in human colonic mucosa were also validated for Anpep (ref. 40 and Fig. 1B) and Stat1 (refs. 40, 41 and Fig. 4D). In addition, decreased expression of Lgals2 was verified in the Tlr2−/− group (Fig. 4B). An analogous decrease in Lgals2 was detected in the colonic mucosa of mice with experimental acute colitis, and human recombinant GAL-2 has been shown to ameliorate the induced inflammation (42). Consequently, transcriptomic changes in the noninflamed colonic mucosa of Tlr2−/− animals appear to mimic those of mammalian colitis at a number of genes (Anpep, Lgals2, and Stat1), consistent with the notion of Tlr2 being an important modifier of mammalian gut inflammation.
Figure 3.
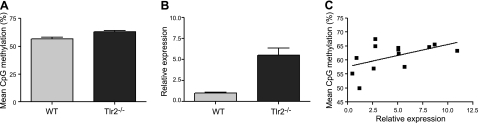
Tlr2-dependent DNA methylation changes at Ifit2 correlate with its expression. A) Average methylation of 2 CpG sites in the downstream vicinity (see Materials and Methods) of the IRF3 transcription factor binding site in the Ifit2-promoter region increased in Tlr2−/− colonic mucosa (n=10/group; P=0.0026). B) Ifit2 expression increased in Tlr2−/− colonic mucosa (WT, n=9; Tlr2−/−, n=10; P=0.0001). C) Methylation at Ifit2 positively correlates with expression (r=0.5; n=13; P=0.04).
Figure 4.
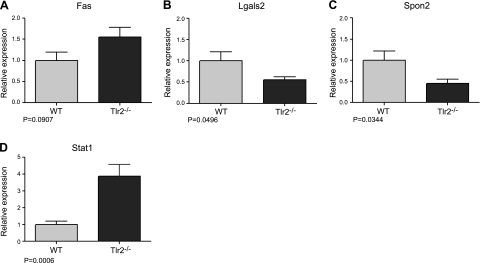
Validation of transcript level changes by real time RT-PCR. A) Fas expression increased in Tlr2−/− mucosa, with a trend approaching significance (n=10/group). B–D) Significant expression changes were confirmed in Tlr2−/− colonic mucosa for Lgals2 (WT, n=9; Tlr2−/−, n=10; B), Spon2 (n=10/group; C), and Stat1 (WT, n=10; Tlr2−/−, n=8; D).
Overlaps between epigenomic and transcriptomic changes
All genes (upstream and downstream) nearest to the SmaI/XmaI intervals with methylation change in the Tlr2−/− animals were identified, and their overlap with the genes that concurrently had transcriptomic alterations was examined. As assessed by the microarrays, of the 387 genomic intervals with a change in methylation, only 20 (5.2%) were associated with an expression change at a neighboring transcript (Table 2). The major reason for this restricted overlap is likely secondary to the inherent limitations of the microarray methods employed, regardless of their obvious advantages (7). Therefore, we first confirmed the inverse correlation between DNA methylation and expression at Anpep (Fig. 1C). Then we decided to investigate the promoter methylation of genes with a prominent expression change in Tlr2−/− colonic mucosa, specifically Stat1 (Fig. 4D) and Ifit2 (Fig. 3B). We could not identify significant DNA methylation alteration in association with Stat1 (data not shown), but the methylation of 2 CpG sites nearest to the IRF3 transcription factor binding site in the Ifit2 promoter (see Materials and Methods) correlated positively with its expression (Fig. 3C). This finding strongly indicates that significant epigenomic changes can associate with the identified transcriptomic changes of this study (even if not detected by the MSAM) and likely vice versa, underscoring the independent value of the Tlr2-deficiency-dependent compendiums. Notably, 67% (14/21) of the DNA methylation and gene expression changes correlated inversely (such as in the case of Anpep), thereby following the usual pattern of association between this epigenetic modification and transcriptional regulation (Table 2). In the meantime, increasingly more exceptions to this rule are recognized (32). Therefore, it is not surprising to find direct association between DNA methylation and gene expression changes, such as here for Ifit2. In addition, a number of the observed DNA methylation changes occurred quite distantly from the associated gene with altered expression in agreement with the recognition that epigenetic changes, such as histone modifications at enhancer elements, further than 200 kb from TSSs can correlate with cell type gene expression (43), and imprinting control elements can regulate allele specific expression over hundreds of kilobases (44).
Table 2.
Genes with overlap between SmaI/XmaI interval methylation and transcriptomic change identified with the microarrays
| Gene | SmaI/XmaI interval | TSS | TES | Methylation | Expression |
|---|---|---|---|---|---|
| Abcg5 | chr17:85094508-85094630 | −11,306 | −36,995 | ↑ | ↓ |
| Abcg8 | chr17:85094508-85094630 | 13,098 | −5,104 | ↑ | ↓ |
| Anpep | chr7:86986986-86987196 | 147 | −20,401 | ↑ | ↓ |
| Arl3 | chr19:46648639-46649271 | −380 | −43,357 | ↓ | ↓ |
| Cmah | chr13:24508684-24508983 | 1,842 | −60,321 | ↑ | ↓ |
| Cyp3a16 | chr5:146267013-146267525 | −36,677 | −70,092 | ↓ | ↓ |
| Cyp3a41a | chr5:146267013-146267525 | 78,330 | 52,263 | ↓ | ↓ |
| Dll1 | chr17:15512946-15513849 | −610 | −9,080 | ↓ | ↑ |
| Elavl3 | chr9:21855078-21856138 | 859 | −36,159 | ↓ | ↑ |
| Gad2 | chr2:22547946-22562379 | 77,316 | 5,765 | ↓ | ↑ |
| Hpcal4 | chr4:122879405-122881036 | 20,473 | 8,278 | ↓ | ↑ |
| Mid1 | chrX:166427168-166428555 | 110,308 | −898 | ↑ | ↑ |
| Mid1 | chrX:166439486-166440819 | 122,599 | 11,423 | ↑ | ↑ |
| Mnd1 | chr3:83705005-83706654 | 253,788 | 186,026 | ↓ | ↑ |
| Nsg1 | chr5:38497269-38497579 | 53,282 | 31,007 | ↑ | ↑ |
| Rab42 | chr4:131864258-131864382 | −5,049 | −6,212 | ↑ | ↓ |
| Rev1 | chr1:38684220-38685381 | −498,293 | −57,5170 | ↓ | ↑ |
| Sardh | chr2:27189061-27189187 | −86,792 | −145,222 | ↑ | ↓ |
| Slc32a1 | chr2:158439864-158442039 | 4,458 | −532 | ↓ | ↑ |
| St3gal4 | chr9:34832759-34832929 | 91,551 | 20,320 | ↑ | ↑ |
| Ufm1 | chr3:53525026-53526994 | 141,719 | 133,120 | ↑ | ↓ |
| Zfp281 | chr1:138849567-138850127 | −328,370 | −333,860 | ↓ | ↑ |
Intervals associated with two different genes are underscored. TSS, distance (bp) of SmaI/XmaI interval midpoint from transcription start site relative to the gene; TES, distance (bp) of SmaI/XmaI interval midpoint from transcription end site relative to the gene; 21 genes associated with 20 intervals.
Tlr2 deficiency associates with colonic mucosal microbiomic changes
We next turned to metagenomic analyses, since our goal was to identify potential associations between the epithelial epigenome and the mucosal microbiota with regard to Tlr2 deficiency. The relative abundance of several phyla was significantly altered between the Tlr2−/− and WT groups. Firmicutes comprised a lower proportion (averaging 73%) in Tlr2−/− mucosa than in WT (88%; P=0.012). Proteobacteria (6.4 vs. 0.1%; P=0.05), Bacteroidetes (17.1 vs. 10.88%; P=0.09) and Actinobacteria (1.52 vs. 0.52%; P=0.015) were all in higher proportion in the Tlr2−/− samples.
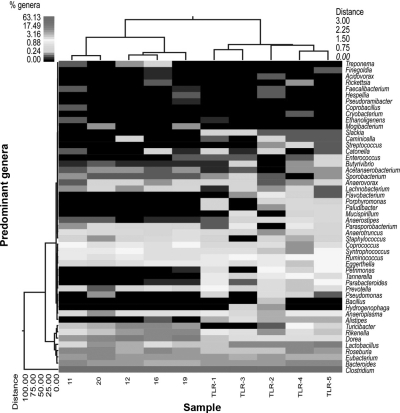
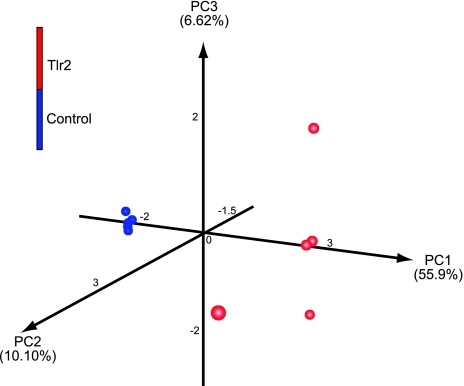
A significant separation was also present at the genus level between mutants and WT. Figure 5 provides a comprehensive overview of this data at the genus taxonomic level, encompassing all of the predominant and ubiquitous genera (n=50). The Tlr2−/− group showed a definite partitioning from the WT strains. The maximum relative Manhattan distance within the Tlr2−/− group was 0.88, and 1.50 in the WT group. In the meantime, the relative distance was 3.0 between the Tlr2−/− and WT groups, demonstrating a clear separation of the mutant animals with respect to colonic mucosal bacteriome. To determine further whether this was a true separation, we utilized UniFrac-based (45) PCA, which revealed clustering, with the primary 3 PCA vectors describing 55.99, 10.10, and 6.62% of the variation among the samples, respectively (Fig. 6). FDR-corrected t test analysis showed a significant difference between 13 genera in Tlr2−/− mucosa compared to WT (Supplemental Table S2). PCA analysis provided plotted examination of the data (Supplemental Fig. S2). Based on these analyses, we could ascertain that bacteria related to Dorea and Turicibacter were significantly more abundant in the WT, while Alistipes, Slackia, Anaerostipes, Parabacteroides, and Lactobacillus species were more common in the Tlr2−/− group. Clostridium was the dominant genus in all samples.
Figure 5.
Dual hierarchical clustering dendogram of the most predominant and ubiquitous 50 genera among mucosal samples. Heat map depicts the relative percentage of each genus for each sample; color scale for heat map at top left. Tlr2−/− animals were from 3 different litters (samples Tlr1 and Tlr2; Tlr3 and Tlr5; and Tlr4, respectively), while WT animals were from 2 different litters (samples 11 and 12; and 16, 19, and 20, respectively). Therefore, interindividual (within the same litter) variation exceeded that of interlitter variation within the two groups of animals.
Figure 6.
Principal component analysis of UniFrac distance metric. Three-dimensional visualization of the PCA analysis using the top 3 vectors clearly shows the significant clustering distance between the two groups. Red balls represent Tlr2−/− samples; blue balls represent WT samples.
The metagenomic investigations at the species level also revealed notable separation between the Tlr2−/− and WT animals. Of the 226 distinguished species, 22 (10%) were significantly affected at an FDR < 5% (Table 3). Approximately half (10/22=45.5%) of these species were predicted by RDP-based taxonomy to be within the genus Clostridium. Dual hierarchal clustering indeed showed a notable separation of the Tlr2 mutant and WT groups on the basis of clostridial species present in one or the other group (Supplemental Fig. S3 and Table 3).
Table 3.
Bacterial species with significant difference in abundance between Tlr2−/− (TLR-1–5) and WT (11–20) mice in colonic mucosal scrapings
| Species | WT |
Tlr |
P | FDR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 16 | 19 | 20 | 1 | 2 | 3 | 4 | 5 | |||
| Clostridium populeti | 4.52 | 3.18 | 3.72 | 3.17 | 2.41 | 0.32 | 0.33 | 0.19 | 0.13 | 0.21 | 9.31E-06 | 0.00 |
| Clostridium hiranonis | 0.01 | 0.02 | 0.03 | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 4.27E-05 | 0.00 |
| Clostridium xylanovorans | 2.55 | 1.92 | 2.13 | 1.35 | 1.32 | 0.19 | 0.10 | 0.08 | 0.33 | 0.09 | 5.28E-05 | 0.00 |
| Eubacterium xylanophilum | 0.75 | 0.64 | 0.51 | 0.52 | 0.39 | 0.21 | 0.13 | 0.16 | 0.09 | 0.09 | 0.0001 | 0.01 |
| Clostridium lituseburense | 1.84 | 2.15 | 4.16 | 4.14 | 2.42 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.000187 | 0.01 |
| Dorea formicigenerans | 6.34 | 10.97 | 10.18 | 15.16 | 7.65 | 2.05 | 0.46 | 1.49 | 1.57 | 1.35 | 0.000251 | 0.01 |
| Eubacterium contortum | 0.72 | 1.51 | 1.16 | 1.77 | 1.13 | 0.19 | 0.13 | 0.22 | 0.40 | 0.26 | 0.000281 | 0.01 |
| Eubacterium ventriosum | 1.01 | 1.49 | 2.04 | 1.37 | 0.99 | 0.11 | 0.38 | 0.16 | 0.33 | 0.51 | 0.000387 | 0.01 |
| Clostridium amygdalinum | 0.61 | 0.52 | 0.23 | 0.25 | 0.36 | 0.04 | 0.13 | 0.00 | 0.07 | 0.00 | 0.001047 | 0.03 |
| Clostridium sporosphaeroides | 0.92 | 0.68 | 0.76 | 0.50 | 0.71 | 0.19 | 0.56 | 0.11 | 0.22 | 0.02 | 0.00138 | 0.03 |
| Alistipes shahii | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.67 | 2.59 | 0.15 | 2.08 | 3.00 | 0.001657 | 0.03 |
| Slackia faecicanis | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.11 | 0.04 | 0.11 | 0.04 | 0.05 | 0.00178 | 0.03 |
| Turicibacter sanguinis | 1.93 | 9.13 | 7.05 | 4.81 | 3.26 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.001856 | 0.03 |
| Anaerotruncus colihominis | 0.65 | 0.52 | 0.26 | 0.52 | 0.41 | 0.27 | 0.21 | 0.12 | 0.20 | 0.19 | 0.001869 | 0.03 |
| Clostridium algidixylanolyticum | 0.13 | 0.07 | 0.20 | 0.28 | 0.24 | 0.61 | 0.36 | 0.45 | 0.84 | 0.44 | 0.002536 | 0.04 |
| Tannerella forsythensis | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.63 | 0.71 | 0.10 | 0.91 | 1.25 | 0.002716 | 0.04 |
| Clostridium leptum | 0.11 | 0.06 | 0.00 | 0.01 | 0.01 | 0.42 | 0.23 | 0.18 | 0.22 | 0.12 | 0.003522 | 0.05 |
| Alistipes putredinis | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.51 | 0.25 | 0.23 | 0.49 | 1.02 | 0.003948 | 0.05 |
| Clostridium disporicum | 0.02 | 0.24 | 0.34 | 0.54 | 0.32 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.004036 | 0.05 |
| Eggerthella lenta | 0.08 | 0.05 | 0.06 | 0.04 | 0.00 | 0.13 | 0.15 | 0.27 | 0.47 | 0.28 | 0.004254 | 0.05 |
| Lactobacillus animalis | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.18 | 2.40 | 7.93 | 3.61 | 9.31 | 0.004417 | 0.05 |
| Clostridium clostridioforme | 2.08 | 1.38 | 0.49 | 0.68 | 0.75 | 2.45 | 1.73 | 3.84 | 2.44 | 2.44 | 0.005128 | 0.05 |
Abundance values are percentages of total. Samples 11–20 are from WT mice; samples Tlr1–5 are from Tlr2−/− mice. Note that 10 of 20 species are from the genus Clostridium.
Bacterial diversity was analyzed by the rarefaction, Shannon index, and Chao1 methods at the species and genus level. Each of the estimates provided significantly lower indices in the WT group (P<0.008) at the 5% level (data not shown), indicating a marked separation of the Tlr2−/− animals from WT with respect to colonic mucosal bacterial genus diversity.
One limitation of our metagenomic investigation was that the studied WT and Tlr2−/− animals were not littermates. However, they were reared within the same housing room from birth, and animal relocation experiments provide evidence for major similarities in gastrointestinal tract microbiota of nonlittermate mice exposed to the environment of the same room at early ages (46). The similar microbiome results between nonlittermate Tlr2−/− mice (relative Manhattan distance 0.88), and WT animals (Manhattan distance 1.5) supports this observation and argues that the environmental influences within the room of our animal facility induced a rather uniform colonic mucosal bacteriome within genetically identical mice. Indeed, interindividual gut microbiota variation within the same litter exceeded that of interlitter variation in the Tlr2−/− and WT groups (Fig. 5) indicating that litter relationship is not a major driving force for mucosal microbial separation in our experimental groups. Based on these findings, we are confident that the observed highly significant metagenomic separation of Tlr2−/− colonic mucosa from WT is secondary to the genetic defect of the animals. Several important observations have been made recently with regard to the role of Tlr2 in colitis (15, 22), colon cancer (47), and metabolic syndrome (23) in nonlittermate Tlr2−/− mice, similarly to the approach utilized herein. Therefore, our results on the remarkable mucosal microbiome separation in the knockout animals provide a layer of added complexity to Tlr2 biology that should be taken into consideration when interpreting the findings of those studies.
DISCUSSION
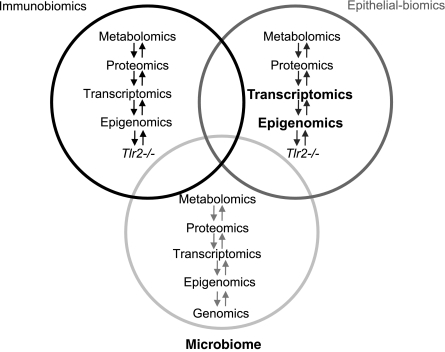
Common human diseases, such as IBDs, are vastly multifaceted with respect to molecular, cellular, and organic pathophysiology. The expanding number of susceptibility genes (48), along with significant monozygotic twin discordance rates (49) and variable microbial associations (50, 51), underscore the diversified nature of this disease group. Consequently, an elaborate, environmentally sensitive interplay between the intestinal microbiota, the mucosa, and the immune system has been recognized in relation to IBDs (5, 7). To highlight and examine this intriguing complexity, we addressed the colonic mucosal epigenomic, transcriptomic, and metagenomic consequences of Tlr2 loss, a molecule important in mammalian gut inflammation. The studies were performed in young adult mice to model the time period when the onset of IBDs peaks in humans. All these large “omic” groups were significantly influenced by the absence of Tlr2 (Fig. 7). Namely, whole genomic interrogation of DNA methylation changes showed that ∼1.4% of the colonocyte genome is modified epigenetically in Tlr2−/− mice. These changes were associated more with CpG islands, gene promoters, DNase I hypersensitive regions, and CTCF-binding sites than control regions. DNase-hypersensitive regions and CTCF-binding sites have been linked to colorectal cancer development (52, 53). TLR2 has also been implicated to play a role in tumorigenesis, including colonic malignancy (54, 55), showing the potential importance of our findings for colorectal oncology. Recent results in Tlr2−/− mice on augmented colitis-induced cancer development support these observations (47).
Figure 7.
“Omic” Venn diagram of large organic categories involved in IBD pathogenesis in the background of Tlr2 deficiency. This study confirmed significant colonic epithelium-associated epigenomic, transcriptomic, and microbiome changes in Tlr2−/− mice (bold and larger font) compared to WT.
In addition to epigenomic changes in response to Tlr2 deficiency, we observed that ∼1.8% of all murine transcript levels were altered in the Tlr2−/− colonic mucosa. Genes involved in immune regulation were enriched in this group of transcripts, supporting the role of Tlr2 as a significant immune response modifier in mammalian large bowel. The importance of Tlr2 for the expression changes is supported by observations where the inhibition of Toll-like receptor-associated MyD88 signaling increased IRF3 phosphorylation, and induced Ifit2 (56). We specifically investigated overlaps between epigenetic and transcriptional changes at the promoter of this gene. Methylation of Ifit2 increased in the vicinity of the IRF3-binding site, unraveling epigenetic mechanisms potentially involved in the regulation of this inflammatory pathway. Similarly, at Anpep, an inverse correlation between increased DNA methylation and decreased expression was determined. The decreased expression of this molecule has not only been observed in IBDs (40), but also in invasive colorectal tumor cells (57). In the meantime, undetectable tumor expression of Anpep associates with increased survival rates in patients with colorectal cancer (58). These studies repeatedly emphasize the relevance of our findings not only for intestinal inflammation, but also for oncology. Epigenetic patterns can potentially be modified, prevented or even reversed by therapeutic interventions (59). Therefore, the Tlr2 dependent epigenomic and transcriptomic compendiums presented here may serve as future pharmacologic targets.
We assessed metagenomic modifications in addition to epigenomic and transcriptomic changes secondary to the loss of Tlr2 in the colonic mucosa of mice (Fig. 7), since genetic alteration-induced microbiota changes (29) and dynamic interactions between intestinal bacterial composition and mucosal inflammation are increasingly recognized (51). A significant proportion of the colonic mucosa-associated microbiome was modified both in number and composition in Tlr2−/− mice. Namely, Proteobacteria and Actinobacteria were more abundant in Tlr2−/− samples similar to that observed in a subset of colonic samples from patients with IBD (50). In the meantime, Bacteroidetes were also more abundant in the Tlr2-knockout animals, while conflicting results have been obtained for this group with respect to human IBD (50, 60). Changes in mucosal microbiota in the mutant mice at the genus level (Supplemental Table S2) showed some resemblance of that observed previously in UC. Namely, Lactobacillus species were more abundant in Tlr2−/− murine mucosa, just as in patients with UC (61). Similarly, Prevotella (62) and Ruminococcus were more plentiful (63), although the latter results were obtained from feces and not mucosa of UC probands.
A major redistribution of clostridial species was observed in the Tlr2 mutants, along with an increase in mucosal Fas expression. Although Clostridium difficile was not detected during our investigations, this organism can induce Fas-mediated acute epithelial injury (64). The latter observation and our results of altered Fas expression in association with clostridial redistribution highlight the possible interactions between the bacterial microbiota and mucosal immune responses. Moreover, recent studies have revealed dynamic relationships between Helicobacter pylori and mucosal DNA methylation (9), emphasizing the potential for epithelial cells to respond epigenetically to the associated microbiota.
Tlr2 has been shown to be critical for the development of diet-induced metabolic syndrome utilizing the same mice strains from the same room of our facility (23), while this multiorganic derangement has been linked to altered gut microbiota in Tlr5-deficient animals (29). Therefore, our findings have implications for the etiology of metabolic syndrome by raising the possibility that the identified mucosal microbial composition variation in Tlr2−/− mice may contribute to their previously observed protection against obesity and associated metabolic changes on a Western type of diet (23).
The effect of homozygous Tlr2 deletion on chemically [dextran sulfate sodium (DSS)]-induced colitis has been previously studied in mice. Both decreased (25) and augmented (65) colitis severity was observed, depending on the background of the animals and their commensal flora (65). In C56BL/6 mice, however, histologically comparable inflammation was found on DSS challenge (66). In agreement with this later finding, we could not detect notable differences in weight loss and colonic length (accepted measures of colitis severity in DSS experiments) throughout and 9 d following 5 d of 3% DSS exposure in drinking water between the Tlr2−/− and WT animals (data not shown). Therefore, the epigenomic, transcriptomic, and microbiomic changes observed in this study appear to largely compensate for the loss of Tlr2 and highlight the remarkable capacity for an orchestrated reprogramming of the mucosal inflammatory network to overcome significant genetic challenges in the mammalian bowel. Future investigations are needed to determine how isolated epigenetic or microbial changes can affect other molecular components of the intestinal defense system. Such studies will likely require single bacterial species inoculation (monocolonization) of germ-free animals along with detailed molecular explorations.
This report includes the first study employing high-throughput technologies testing the epigenome, transcriptome. and metagenome concomitantly to interrogate the consequences of a single protein loss in mammals. Our results emphasize the sophisticated network of molecular and cellular interactions that bridge genotype to phenotype in higher organisms and have potential implications for a wide array of common human disorders involving immunological and metabolic disturbances.
Supplementary Material
Acknowledgments
The authors thank Stefi Lee for her outstanding technical support.
R.K. was supported by a young investigator joint award from the Crohn's and Colitis Foundation of America–Children's Digestive Health and Nutrition Foundation/North American Society of Pediatric Gastroenterology Hepatology and Nutrition, (CCFA 2426), and the Broad Medical Research Program, the Broad Foundation (IBD-0252). C.W.S. was supported by a U.S. Department of Agriculture/Agriculture Research Service Child Health Research Center, Current Research Information System Project grant (6250-51000-055). DNaseI mapping data were funded through a National Human Genome Research Initiative ENCODE grant (U54HG004592) to John A. Stamatoyannopoulos (University of Washington, Seattle, WA, USA). J.V. was supported by the National Institute of Diabetes, Digestive, and Kidney Disease (R01 DK065075, UH3 DK083990, and P30 DK56338) and the National Center for Complementary and Alternative Medicine (R01 AT004326). The authors declare no conflicts of interest.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Backhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005) Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 2. Maslowski K. M., Vieira A. T., Ng A., Kranich J., Sierro F., Yu D., Schilter H. C., Rolph M. S., Mackay F., Artis D., Xavier R. J., Teixeira M. M., Mackay C. R. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 4. Preidis G. A., Versalovic J. (2009) Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology 136, 2015–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sartor R. B. (2006) Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 390–407 [DOI] [PubMed] [Google Scholar]
- 6. Barnett M., Bermingham E., McNabb W., Bassett S., Armstrong K., Rounce J., Roy N. (2010) Investigating micronutrients and epigenetic mechanisms in relation to inflammatory bowel disease. Mutat. Res. 690, 71–80 [DOI] [PubMed] [Google Scholar]
- 7. Kellermayer R., Balasa A., Zhang W., Lee S., Mirza S., Chakravarty A., Szigeti R., Laritsky E., Tatevian N., Smith C. W., Shen L., Waterland R. A. (2010) Epigenetic maturation in colonic mucosa continues beyond infancy in mice. Hum. Mol. Genet. 19, 2168–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balasa A., Gathungu G., Kisfali P., Smith E. O., Cho J. H., Melegh B., Kellermayer R. (2010) Assessment of DNA methylation at the interferon regulatory factor 5 (IRF5) promoter region in inflammatory bowel diseases. Int. J. Colorectal. Dis. 25, 553–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sepulveda A. R., Yao Y., Yan W., Park D. I., Kim J. J., Gooding W., Abudayyeh S., Graham D. Y. (2010) CpG methylation and reduced expression of O6-methylguanine DNA methyltransferase is associated with Helicobacter pylori infection. Gastroenterology 138, 1836–1844 [DOI] [PubMed] [Google Scholar]
- 10. Vamadevan A. S., Fukata M., Arnold E. T., Thomas L. S., Hsu D., Abreu M. T. (2010) Regulation of Toll-like receptor 4-associated MD-2 in intestinal epithelial cells: a comprehensive analysis. Innate Immun. 16, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Z., Schluesener H. J. (2006) Mammalian toll-like receptors: from endogenous ligands to tissue regeneration. Cell. Mol. Life Sci. 63, 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lundin A., Bok C. M., Aronsson L., Bjorkholm B., Gustafsson J. A., Pott S., Arulampalam V., Hibberd M., Rafter J., Pettersson S. (2008) Gut flora, Toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine. Cell. Microbiol. 10, 1093–1103 [DOI] [PubMed] [Google Scholar]
- 13. Takeda K., Kaisho T., Akira S. (2003) Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 14. Mowat A. M. Does TLR2 regulate intestinal inflammation? Eur. J. Immunol. 40, 318–320 [DOI] [PubMed] [Google Scholar]
- 15. Ey B., Eyking A., Gerken G., Podolsky D. K., Cario E. (2009) TLR2 mediates gap junctional intercellular communication through connexin-43 in intestinal epithelial barrier injury. J. Biol. Chem. 284, 22332–22343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Messlik A., Schmechel S., Kisling S., Bereswill S., Heimesaat M. M., Fischer A., Gobel U., Haller D. (2009) Loss of Toll-like receptor 2 and 4 leads to differential induction of endoplasmic reticulum stress and proapoptotic responses in the intestinal epithelium under conditions of chronic inflammation. J. Proteome Res. 8, 4406–4417 [DOI] [PubMed] [Google Scholar]
- 17. Shan-Shan Z., Yu-Lan L. (2009) Therapeutic effects of mucosal tolerance on experimental colitis in rats. Eur. J. Gastroenterol. Hepatol. 21, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 18. McMahon L. A., House A. K., Catchpole B., Elson-Riggins J., Riddle A., Smith K., Werling D., Burgener I. A., Allenspach K. (2010) Expression of Toll-like receptor 2 in duodenal biopsies from dogs with inflammatory bowel disease is associated with severity of disease. Vet. Immunol. Immunopathol. 135, 158–163 [DOI] [PubMed] [Google Scholar]
- 19. Szebeni B., Veres G., Dezsofi A., Rusai K., Vannay A., Mraz M., Majorova E., Arato A. (2008) Increased expression of Toll-like receptor (TLR) 2 and TLR4 in the colonic mucosa of children with inflammatory bowel disease. Clin. Exp. Immunol. 151, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frolova L., Drastich P., Rossmann P., Klimesova K., Tlaskalova-Hogenova H. (2008) Expression of Toll-like receptor 2 (TLR2), TLR4, and CD14 in biopsy samples of patients with inflammatory bowel diseases: upregulated expression of TLR2 in terminal ileum of patients with ulcerative colitis. J. Histochem. Cytochem. 56, 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pierik M., Joossens S., Van Steen K., Van Schuerbeek N., Vlietinck R., Rutgeerts P., Vermeire S. (2006) Toll-like receptor-1, -2, and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm. Bowel. Dis. 12, 1–8 [DOI] [PubMed] [Google Scholar]
- 22. Podolsky D. K., Gerken G., Eyking A., Cario E. (2009) Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology 137, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Himes R. W., Smith C. W. (2010) Tlr2 is critical for diet-induced metabolic syndrome in a murine model. FASEB J. 24, 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehses J. A., Meier D. T., Wueest S., Rytka J., Boller S., Wielinga P. Y., Schraenen A., Lemaire K., Debray S., Van Lommel L., Pospisilik J. A., Tschopp O., Schultze S. M., Malipiero U., Esterbauer H., Ellingsgaard H., Rutti S., Schuit F. C., Lutz T. A., Boni-Schnetzler M., Konrad D., Donath M. Y. (2010) Toll-like receptor 2-deficient mice are protected from insulin resistance and beta cell dysfunction induced by a high-fat diet. Diabetologia 53, 1795–1806 [DOI] [PubMed] [Google Scholar]
- 25. Heimesaat M. M., Fischer A., Siegmund B., Kupz A., Niebergall J., Fuchs D., Jahn H. K., Freudenberg M., Loddenkemper C., Batra A., Lehr H. A., Liesenfeld O., Blaut M., Gobel U. B., Schumann R. R., Bereswill S. (2007) Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS ONE 2, e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loh G., Brodziak F., Blaut M. (2008) The Toll-like receptors TLR2 and TLR4 do not affect the intestinal microbiota composition in mice. Environ. Microbiol. 10, 709–715 [DOI] [PubMed] [Google Scholar]
- 27. Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. (2005) Diversity of the human intestinal microbial flora. Science 308, 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swidsinski A., Ladhoff A., Pernthaler A., Swidsinski S., Loening-Baucke V., Ortner M., Weber J., Hoffmann U., Schreiber S., Dietel M., Lochs H. (2002) Mucosal flora in inflammatory bowel disease. Gastroenterology 122, 44–54 [DOI] [PubMed] [Google Scholar]
- 29. Vijay-Kumar M., Aitken J. D., Carvalho F. A., Cullender T. C., Mwangi S., Srinivasan S., Sitaraman S. V., Knight R., Ley R. E., Gewirtz A. T. (2010) Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 31. Chesa P. G., Rettig W. J., Melamed M. R. (1986) Expression of cytokeratins in normal and neoplastic colonic epithelial cells. Implications for cellular differentiation and carcinogenesis. Am. J. Surg. Pathol. 10, 829–835 [DOI] [PubMed] [Google Scholar]
- 32. Waterland R. A., Kellermayer R., Rached M. T., Tatevian N., Gomes M. V., Zhang J., Zhang L., Chakravarty A., Zhu W., Laritsky E., Zhang W., Wang X., Shen L. (2009) Epigenomic profiling indicates a role for DNA methylation in early postnatal liver development. Hum. Mol. Genet. 18, 3026–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey M. T., Walton J. C., Dowd S. E., Weil Z. M., Nelson R. J. (2010) Photoperiod modulates gut bacteria composition in male Siberian hamsters (Phodopus sungorus). Brain Behav. Immun. 24, 577–584 [DOI] [PubMed] [Google Scholar]
- 34. Dowd S. E., Zaragoza J., Rodriguez J. R., Oliver M. J., Payton P. R. (2005) Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST). BMC Bioinformatics. 6, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., Kulam-Syed-Mohideen A. S., McGarrell D. M., Marsh T., Garrity G. M., Tiedje J. M. (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J., Sahl J. W., Stres B., Thallinger G. G., Van Horn D. J., Weber C. F. (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolcott R. D., Gontcharova V., Sun Y., Zischakau A., Dowd S. E. (2009) Bacterial diversity in surgical site infections: not just aerobic cocci any more. J. Wound Care. 18, 317–323 [DOI] [PubMed] [Google Scholar]
- 38. Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001) Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 39. Al-Shahrour F., Minguez P., Tarraga J., Montaner D., Alloza E., Vaquerizas J. M., Conde L., Blaschke C., Vera J., Dopazo J. (2006) BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 34, W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carey R., Jurickova I., Ballard E., Bonkowski E., Han X., Xu H., Denson L. A. (2008) Activation of an IL-6:STAT3-dependent transcriptome in pediatric-onset inflammatory bowel disease. Inflamm. Bowel Dis. 14, 446–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu F., Dassopoulos T., Cope L., Maitra A., Brant S. R., Harris M. L., Bayless T. M., Parmigiani G., Chakravarti S. (2007) Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm. Bowel Dis. 13, 807–821 [DOI] [PubMed] [Google Scholar]
- 42. Paclik D., Berndt U., Guzy C., Dankof A., Danese S., Holzloehner P., Rosewicz S., Wiedenmann B., Wittig B. M., Dignass A. U., Sturm A. (2008) Galectin-2 induces apoptosis of lamina propria T lymphocytes and ameliorates acute and chronic experimental colitis in mice. J. Mol. Med. 86, 1395–1406 [DOI] [PubMed] [Google Scholar]
- 43. Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fitzpatrick G. V., Soloway P. D., Higgins M. J. (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32, 426–431 [DOI] [PubMed] [Google Scholar]
- 45. Lozupone C., Hamady M., Knight R. (2006) UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 7, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Friswell M. K., Gika H., Stratford I. J., Theodoridis G., Telfer B., Wilson I. D., McBain A. J. (2010) Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE 5, e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lowe E. L., Crother T. R., Rabizadeh S., Hu B., Wang H., Chen S., Shimada K., Wong M. H., Michelsen K. S., Arditi M. (2010) Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS ONE 5, e13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lees C. W., Satsangi J. (2009) Genetics of inflammatory bowel disease: implications for disease pathogenesis and natural history. Expert Rev. Gastroenterol. Hepatol. 3, 513–534 [DOI] [PubMed] [Google Scholar]
- 49. Halfvarson J., Bodin L., Tysk C., Lindberg E., Jarnerot G. (2003) Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology 124, 1767–1773 [DOI] [PubMed] [Google Scholar]
- 50. Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104, 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Packey C. D., Sartor R. B. (2009) Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr. Opin. Infect. Dis. 22, 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shiraga T., Winpenny J. P., Carter E. J., McCarthy V. A., Hollingsworth M. A., Harris A. (2005) MZF-1 and DbpA interact with DNase I hypersensitive sites that correlate with expression of the human MUC1 mucin gene. Exp. Cell Res. 308, 41–52 [DOI] [PubMed] [Google Scholar]
- 53. Nakagawa H., Chadwick R. B., Peltomaki P., Plass C., Nakamura Y., de La Chapelle A. (2001) Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A. 98, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang B., Zhao J., Shen S., Li H., He K. L., Shen G. X., Mayer L., Unkeless J., Li D., Yuan Y., Zhang G. M., Xiong H., Feng Z. H. (2007) Listeria monocytogenes promotes tumor growth via tumor cell toll-like receptor 2 signaling. Cancer Res. 67, 4346–4352 [DOI] [PubMed] [Google Scholar]
- 55. Yoshioka T., Morimoto Y., Iwagaki H., Itoh H., Saito S., Kobayashi N., Yagi T., Tanaka N. (2001) Bacterial lipopolysaccharide induces transforming growth factor beta and hepatocyte growth factor through toll-like receptor 2 in cultured human colon cancer cells. J. Int. Med. Res. 29, 409–420 [DOI] [PubMed] [Google Scholar]
- 56. Selvarajoo K., Takada Y., Gohda J., Helmy M., Akira S., Tomita M., Tsuchiya M., Inoue J., Matsuo K. (2008) Signaling flux redistribution at toll-like receptor pathway junctions. PLoS ONE 3, e3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wiese A. H., Auer J., Lassmann S., Nahrig J., Rosenberg R., Hofler H., Ruger R., Werner M. (2007) Identification of gene signatures for invasive colorectal tumor cells. Cancer Detect. Prev. 31, 282–295 [DOI] [PubMed] [Google Scholar]
- 58. Hashida H., Takabayashi A., Kanai M., Adachi M., Kondo K., Kohno N., Yamaoka Y., Miyake M. (2002) Aminopeptidase N is involved in cell motility and angiogenesis: its clinical significance in human colon cancer. Gastroenterology 122, 376–386 [DOI] [PubMed] [Google Scholar]
- 59. Smith A. E., Ford K. G. (2007) Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Res. 35, 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gophna U., Sommerfeld K., Gophna S., Doolittle W. F., Veldhuyzen van Zanten S. J. (2006) Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44, 4136–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fyderek K., Strus M., Kowalska-Duplaga K., Gosiewski T., Wedrychowicz A., Jedynak-Wasowicz U., Sladek M., Pieczarkowski S., Adamski P., Kochan P., Heczko P. B. (2009) Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J. Gastroenterol. 15, 5287–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kleessen B., Kroesen A. J., Buhr H. J., Blaut M. (2002) Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 37, 1034–1041 [DOI] [PubMed] [Google Scholar]
- 63. Andoh A., Sakata S., Koizumi Y., Mitsuyama K., Fujiyama Y., Benno Y. (2007) Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm. Bowel Dis. 13, 955–962 [DOI] [PubMed] [Google Scholar]
- 64. Kim H., Rhee S. H., Pothoulakis C., Lamont J. T. (2007) Inflammation and apoptosis in Clostridium difficile enteritis is mediated by PGE2 up-regulation of Fas ligand. Gastroenterology 133, 875–886 [DOI] [PubMed] [Google Scholar]
- 65. Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. (2004) Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 [DOI] [PubMed] [Google Scholar]
- 66. Cario E., Gerken G., Podolsky D. K. (2007) Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 132, 1359–1374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.