Abstract
Double-strand DNA breaks are common events in eukaryotic cells, and there are two major pathways for repairing them: homologous recombination and nonhomologous DNA end joining (NHEJ). The diverse causes of DSBs result in a diverse chemistry of DNA ends that must be repaired. Across NHEJ evolution, the enzymes of the NHEJ pathway exhibit a remarkable degree of structural tolerance in the range of DNA end substrate configurations upon which they can act. In vertebrate cells, the nuclease, polymerases and ligase of NHEJ are the most mechanistically flexible and multifunctional enzymes in each of their classes. Unlike repair pathways for more defined lesions, NHEJ repair enzymes act iteratively, act in any order, and can function independently of one another at each of the two DNA ends being joined. NHEJ is critical not only for the repair of pathologic DSBs as in chromosomal translocations, but also for the repair of physiologic DSBs created during V(D)J recombination and class switch recombination. Therefore, patients lacking normal NHEJ are not only sensitive to ionizing radiation, but also severely immunodeficient.
Keywords: Ku, DNA-PKcs, Artemis, XRCC4, DNA ligase IV
THE BIOLOGICAL CONTEXT OF NHEJ
Unlike most other DNA repair and DNA recombination pathways, nonhomologous DNA end joining (NHEJ) in prokaryotes and eukaryotes evolved along themes of mechanistic flexibility, enzyme multifunctionality, and iterative processing in order to achieve repair a diverse range of substrate DNA ends at double-strand breaks (DSBs) (1-3). Except for very limited protein homology for the Ku protein in prokaryotes and eukaryotes (2), the actual nuclease, polymerase, and ligase components of NHEJ appear to have arisen independently, but converged on these same mechanistic themes in order to handle the challenge of joining two freely diffusing ends of diverse DNA end overhang configuration with a wide range of base or sugar oxidative damage (3).
Homology-Directed Repair versus Nonhomlogous DNA End Joining
When double-strand breaks arise in any organism, prokaryotic or eukaryotic, there are two major categories of DNA repair that can restore the duplex structure (Fig. 1). If the organism is diploid (even if the diploidy is only transient, as in replicating bacteria or replicating haploid yeast), then homology-directed repair can be used. The most common form of homology-directed repair is called homologous recombination (abbreviated HR), which has the longest sequence homology requirements between the donor and acceptor DNA. Other forms of homology-directed repair include single-strand annealing (abbreviated SSA) and breakage-induced replication, and these require shorter sequence homology relative to HR (4, 5).
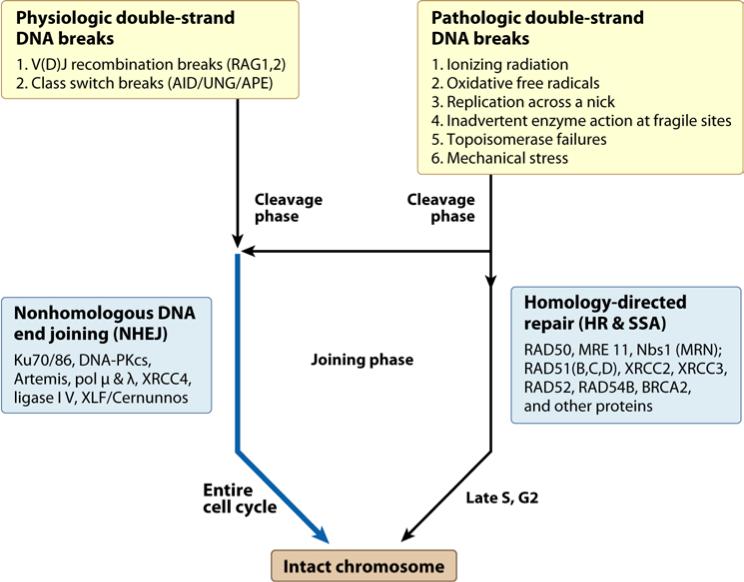
Figure 1. Causes and Repair of Double-Strand DNA Breaks.
Physiologic and pathologic causes of double-strand breaks in mammalian somatic cells are listed at the top. During S and G2 of the cell cycle, homology-directed repair is common because the two sister chromatids are in close proximity, providing a nearby homology donor. Homology-directed repair includes homologous recombination (HR) and single-strand annealing (SSA). At any time in the cell cycle, double-strand breaks can be repaired by nonhomologous DNA end joining (NHEJ). Proteins involved in the repair pathways are listed.
In nondividing haploid organisms or in diploid organisms that are not in S phase, a homology donor is not nearby. Hence, early in evolution, another form of double-strand break repair had an opportunity to provide survival advantage, and nonhomologous DNA end joining (NHEJ) includes a set of DNA enzymes that have the mechanistic flexibility to provide such an advantage (Table 1)(6).
Table 1.
Corresponding Enzymes in Prokaryotic and Eukaryotic NHEJ Components
| Functional component | Prokaryotes | Eukaryotes | |
|---|---|---|---|
| S. cerevisiae | Multicellular eukaryotes | ||
| Toolbelt protein | Ku (30-40kDa) | Ku 70/80 | Ku 70/80 |
| Polymerase | POL domain of LigD | Pol4 | Polmu and lambda |
| Nuclease | ? | Rad50:Mre11:Xrs2 (FEN-1) | Artemis:DNA-PKcs |
| Kinase/Phosphatase | PE domain of LigD | Tpp1 and others | PNK and others |
| Ligase | LIG domain of LigD | Nej1:Lif1:Dnl4 | XLF:XRCC4:DNA ligase IV |
How the cell determines whether HR or NHEJ will be used to repair a break is still an active area of investigation. The HR versus NHEJ determination may be somewhat operational (7). If a homologue is not present near a DSB during S/G2, then HR cannot proceed, and NHEJ is the only option. During S phase, the sister chromatid is physically very close, thereby provding a homology donor for HR. Outside of S/G2, NHEJ is indeed the markedly preferred option. The precise molecular events, beyond issues of proximity and possible competition between Ku and RAD51 or 52, are yet to be deciphered (7-9). Recent data from S. cerevisiae suggests that DNA ligase IV complex may be key in suppressing the DNA end resection needed to initiate HR (10).
Causes and Frequencies of Double-Strand Breaks
There are an estimated ten double-strand breaks (DSBs) per day per cell, based on metaphase chromosome and chromatid breaks in early passage primary human or mouse fibroblasts (11-13). Estimates of DSB frequency in nondividing cells are difficult to make because methods for assessing DSBs outside of metaphase are subject to even more caveats of interpretation.
In mitotic cells of multicellular eukaryotes, DSBs are all pathologic (accidental) except the specialized subset of physiologic DSBs in early lymphocytes of the vertebrate immune system (Fig. 1). Major pathologic causes of double-strand breaks in wild type cells include replication across a nick, giving rise to chromatid breaks during S phase. Such DSBs are ideally repaired by HR using the nearby sister chromatid.
All of the remaining pathologic forms of DSB are repaired by NHEJ because they usually occur when there is no nearby homology donor and/or because they occur outside of S phase. These causes include reactive oxygen species from oxidative metabolism, ionizing radiation, and inadvertent action of nuclear enzymes (14).
Reactive oxygen species (ROS) are a second major cause of DSBs (Fig. 1). During the course of normal oxidative respiration, mitochondria convert about ~0.1 to 1% of the oxygen to superoxide (O-2) (15). Superoxide dismutase in the mitochondrion (SOD2) or cytosol (SOD1) can convert this to hydroxyl free radicals, which may react with DNA to cause single-strand breaks. Two closely spaced lesions of this type on anti-parallel strands can cause a DSB. About 1022 free radicals or ROS species are produced in the human body each hour, and this represents about 109 ROS per cell per hour. A subset of the longer-lived ROS may enter the nucleus via the nuclear pores.
A third cause of DSBs is natural ionizing radiation of the environment. These include gamma rays and X-rays. At sea level, ~300 million ionizing radiation particles per hour pass through each person. As these traverse the body, they create free radicals along their path, primarily from water. When the particle comes close to a DNA duplex, clusters of free radicals damage DNA, generating double- and single-stranded breaks at a ratio of about 25 to 1 (16). About half of the ionizing radiation that strikes each of us comes from outside the earth. The other half of the radiation that strikes us comes from the decay of radioactive elements, primarily metals, within the earth.
A fourth cause of DSBs is inadvertent action by nuclear enzymes on DNA. These include failures of type II topoisomerases, which transiently break both strands of the duplex. If the topoisomerase fails to rejoin the strands, then a DSB results (17). Inadvertent action by nuclear enzymes of lymphoid cells, such as the RAG complex (composed of RAG1 and 2) and activation-induced deaminase (AID) are responsible for physiologic breaks for antigen receptor gene rearrangement; however, they sometimes accidentally cleave the DNA at off-target sites outside the antigen receptor gene loci (18). In humans, these account for about half of all of the chromosomal translocations that result in lymphoma.
Finally, physical or mechanical stress on the DNA duplex is a relevant cause of DSBs. In prokaryotes, this arises in the context of desiccation, which is quite important in nature (19). In eukaryotes, telomere failures can result in chromosomal fusions that have two centromeres, and this results in physical stress by the mitotic spindle (breakage/fusion/bridge cycles) with DSBs (20).
In addition to the above for mitotic cells, meiotic cells have an additional source of DSBs, which is physiologic and is caused by an enzyme called Spo11, a topoisomerase II-like enzyme (21). Spo11 creates DSBs to generate cross-overs between homologues during meiotic prophase I. These events are resolved by HR. Therefore, NHEJ is not relevant to Spo11 breaks. Interestingly, it is not clear that NHEJ occurs in vertebrate meiotic cells, because one group reports the lack of Ku70 in spermatogonia (22). Human spermatogonia remain in meiotic prophase I for about 3 weeks, and human eggs remain in meiotic prophase I for 12 to 50 years; hence, these cells can rely on HR during these long periods. Given the error-prone nature of NHEJ (see below), reliance on HR may be one way to minimize alterations to the germ line at frequencies that might be deleterious to a population.
MECHANISM OF NHEJ
The Substrates for NHEJ: Structural Diversity of DSB DNA Ends
Perhaps the most intriguing aspect of NHEJ is the diversity of substrates that it can accept and convert to joined products. This demands a remarkable level of mechanistic flexibility at the level of protein-substrate interaction and is unparalleled in most other biochemical processes. Though we have substantial amounts of information on the DNA end configurations at DSBs, there are limitations to the information because of the diverse manner in which ionizing radiation and ROS interact with DNA. Therefore, we know the most about the diversity of physiologic DSBs, specifically V(D)J recombination, because we know where the relevant enzymes initiate the cutting of the two DNA strands. In V(D)J recombination, we can examine many NHEJ outcomes from the same starting substrates. We can also vary the sequence of the two DNA ends being joined. All of these various overhangs are joined in vivo at about the same efficiency, regardless of sequence. Within this range of overhang variation then, NHEJ can accept a wide variety of overhang length, DNA end sequence, and DNA end chemistry.
Overview of the Proteins and Mechanism of Vertebrate NHEJ
Like most DNA repair processes, NHEJ requires a nuclease to resect damaged DNA, polymerases to fill-in new DNA, and a ligase to restore integrity to the DNA strands (Fig. 2). Functional correspondence between the NHEJ proteins of prokaryotes, yeast (along with plants and invertebrates), and vertebrates can be inferred (Table 1). Prokaryotic NHEJ has been reviewed recently (23), and comparisons have between prokaryotes and eukaryotes have been made (3). NHEJ in yeast, which appears to be similar for plants and invertebrates, has been thoroughly reviewed as well (24, 25). Hence, the discussion here will focus on NHEJ in vertebrates, with appropriate comparisons to prokaryotic and yeast NHEJ.
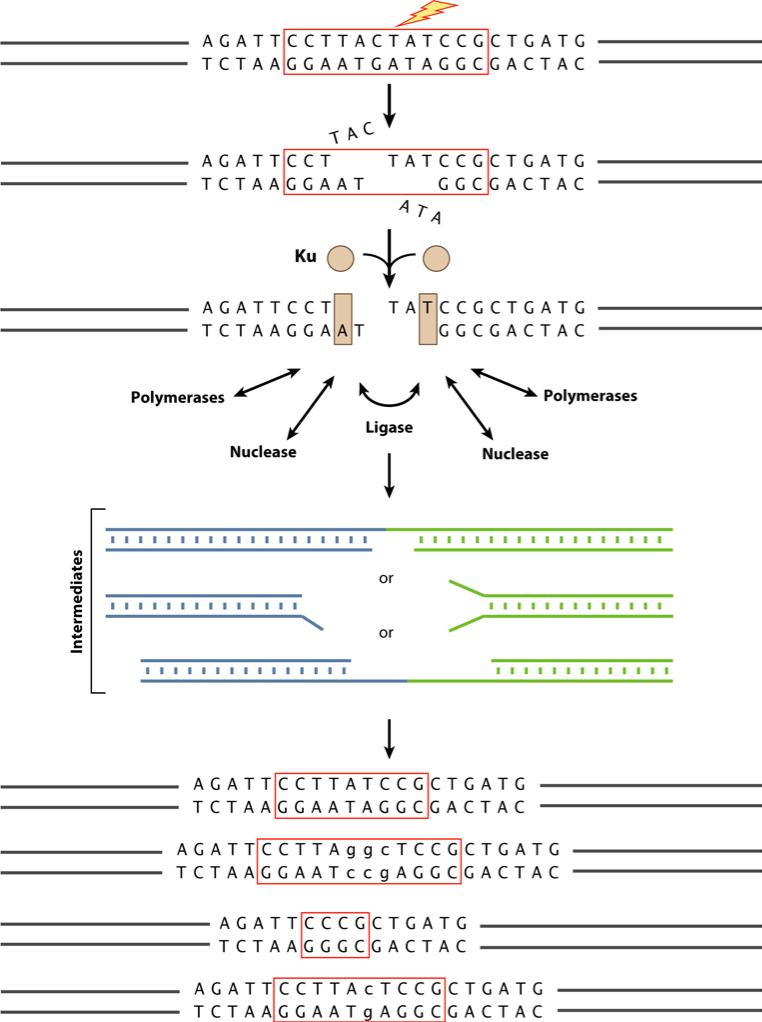
Figure 2. General Steps of Nonhomologous DNA End Joining.
Ku binding to the DNA ends at a DSB improves binding by nuclease, polymerase and ligase components. Flexibility in the loading of these enzymatic components, the option to load repeatedly (iteratively), and independent processing of the two DNA end all permit mechanistic flexibility for the NHEJ process. This mechanistic flexibility is essential to permit NHEJ to handle a very diverse array of DSB end configurations and to join them. In addition to the overall mechanistic flexibility, each component exhibits enzymatic flexibility and multifunctionality, as discussed in the text.
When a double-strand break arises in vertebrates, it is thought that Ku is the first protein to bind, based on its abundance (estimated at ~400,000 molecules per cell) and its strong equilibrium dissociation constant (~10-9 M) for duplex DNA ends of any configuration (Fig. 3 and 4a)(26-29). Ku is a toroidal protein, based on its crystal structure (30). Ku bound to a DNA end can be considered as a Ku:DNA complex, which serves as a node at which the nuclease, polymerases and ligase of NHEJ can dock (31). One can think of Ku as a toolbelt protein, similar to PCNA in DNA replication, where many proteins can dock. At a DSB, there are two DNA ends. Hence, it is presumed that there is a Ku:DNA complex at each of the two DNA ends being joined, thereby permitting each DNA end to be modified in preparation for joining.
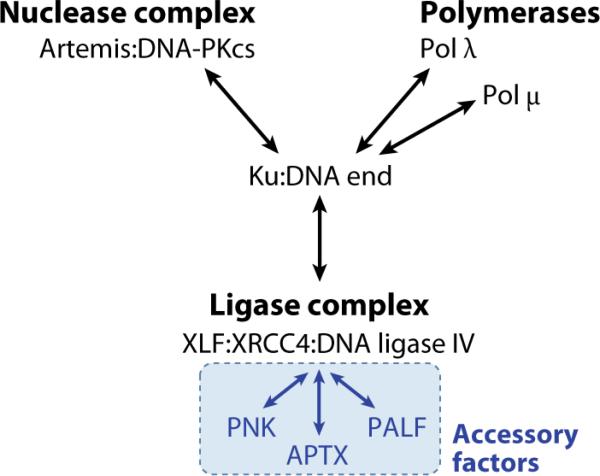
Figure 3. Interactions Between NHEJ Proteins.
Physical interactions between NHEJ components are summarized. In addition, interactions between XRCC4 and DNA-PKcs are discussed in the text, as are functional interactions.
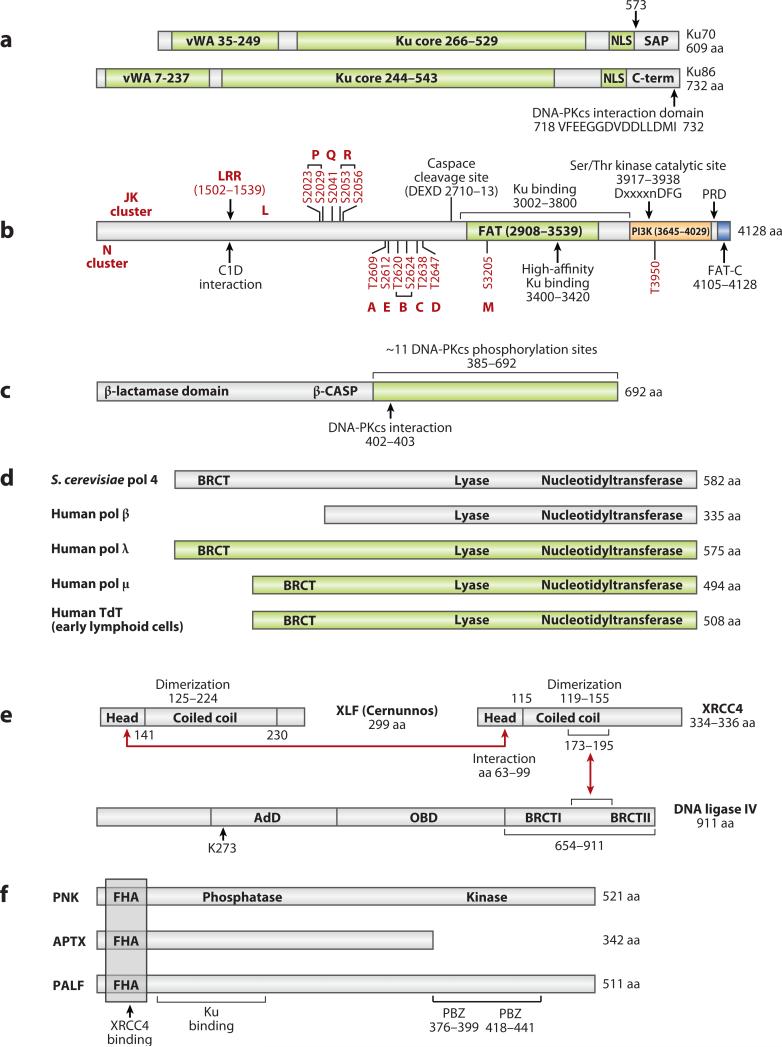
Figure 4. Diagrams of Domains within NHEJ Proteins.
a. Ku is a heterodimer of Ku70 and 86. vWA designates von Willebrand domains. SAP designates a SAF-A/B, Acinus, and PIAS domain and may be involved in DNA binding.
b. DNA-PKcs autophosphorylation sites are shown in red (90, 95, 96). The function of each phosphorylation site (A-E, L, M, P-R) and cluster (N and JK) are still under study. Adjacent phosphorylation sites that are linked by a bracket have not been functionally dissected from one another. LRR designates the leucine-rich region. The FAT-C domain is a FAT domain at the C-terminus. PI3K designates the PI3 kinase domain. PRD designates the PI3K regulatory domain.
c. Artemis is phosphorylated by DNA-PKcs at 11 sites within the C-terminal portion (green) (162, 163). Amino acids 156 to 385 share conserved sequence with those metallo-β-lactamases that act on nucleic acids (164). This region has been called the β-CASP domain (metallo-β-lactamase-associated CPSF Artemis SNM1 PSO2) (165).
d. POL × polymerase family. Pol mu and pol lambda are involved in NHEJ in mammalian somatic cells generally. TdT is only expressed in early lymphoid cells where it participates in NHEJ primarily in the context of V(D)J recombination.
e. The NHEJ ligase complex consists of XLF (Cernunnos), XRCC4, DNA ligase IV. The red arrows indicate the regions of physical interaction (118, 119). OBD in ligase IV is the oligo-binding domain, and AdB is the adenylation domain.
f. Polynucleotide kinase (PNK), Aprataxin (APTX), and PALF (APLF) are ancillary components that bind to XRCC4 of the ligase complex. The PBZ domain appears to be important for PARP-1 binding, for poly-ADPribose binding, and for nuclease activity. FHA designates the forkhead-associated domain.
Each Ku
DNA end complex can recruit the nuclease, polymerase and ligase activities in any order (31, 32). This flexibility is the basis for the diverse array of outcomes that can arise from identical starting ends. The processing of the two DNA ends may transiently terminate when there is some small extent of annealing between the two DNA ends. The processing may permanently terminate when one or both strands of the left and right duplexes are ligated.
Ku likely changes conformation when bound to a DNA end versus when Ku is in free solution. The basis for this inference is that Ku does not form stable complexes with DNA-PKcs in the absence of DNA ends (33), and the same appears to apply for its interactions with polymerases mu and lambda and with XRCC4:DNA ligase IV (34, 35). The crystal structure for Ku lacks the C-terminal 19 kDa (167 aa) of Ku86 (an important region of interaction with DNAPKcs and other proteins), and this may be a region for conformational change upon Ku binding to DNA (36).
The Artemis
DNA-PKcs complex has a diverse array of nuclease activities, including 5' endonuclease activity, 3' endonuclease activity, and hairpin opening activity, in addition to an apparent 5' exonuclease activity of Artemis alone (Fig. 4b and 4c and Suppl. Fig. 1)(37). The Artemis:DNA-PKcs complex is able to endonucleolytically cut a variety of types of damaged DNA overhangs (38, 39). Hence, there is no obvious need for additional nucleases, though the 3' exonuclease of PALF (APLF) and others are possibilities (see Future Questions section).
Polymerases mu and lambda are both able to bind to the Ku:DNA complexes by way of their BRCT domains located in the N-terminal portion of each polymerase (Fig. 4d)(32). Additional polymerases appear able to contribute when neither of these two polymerases is present (40, 41). As discussed later, pol mu is particularly well-suited for functioning in NHEJ because it is capable of template-independent synthesis, in addition to template-dependent synthesis. Pol lambda also has more flexibility than replicative polymerases.
A complex of XLF:XRCC4
DNA ligase IV is the most flexible ligase known, with the ability to ligate across gaps and ligate incompatible DNA ends (Fig. 4e)(42, 43). It can also ligate one strand when the other has a complex configuration (e.g., bearing flaps), and it can ligate single-stranded DNA, though with limited and substantial sequence preferences.
Therefore, the nuclease, polymerases and ligase of NHEJ all have much greater mechanistic flexibility than their counterparts in other repair pathways. This flexibility permits these structure-specific proteins to act on a wider range of starting DNA end structures. One consequence of such flexibility in vertebrates may be the substantial diversity of junctional outcomes observed, even from identical starting ends, as discussed in the next section.
Variation in NHEJ Products Even from Identical Starting Substrates
If we arbitrarily designate the two DNA ends as left and right, then the Ku bound at the left end could conceivably recruit the nuclease, and the Ku at the right DNA end might recruit the polymerase, or vice versa. It is likely that there are multiple rounds of action by the nuclease, polymerases, and ligase at the left and right ends of the DSB until the ‘top’ or ‘bottom’ strand is ligated. Therefore, the joining of the two ends is likely to be an iterative process with multiple possible routes all leading to a joining event, but with wide variation in the precise junctional sequence of the products.
Unlike pathologic causes of DSBs which generally cannot generate predictable pairs of starting DNA ends, V(D)J recombination always generates two hairpinned coding ends (see section on V(D)J recombination for a fuller discussion, including signal ends). Though these two coding ends can be opened in a few different ways, the predominant hairpin opening position is 2 nucleotides 3' of the hairpin tip in vivo (44) and in vitro (37). Hence, the two coding ends are often 3' overhangs of length 4 nucleotides each. Assuming that NHEJ within vertebrate B cells is representative of NHEJ in other tissue cell types (these junctions can be analyzed in cells that do not express terminal transferase), several inferences can be drawn from these relatively defined starting DNA ends.
First, the amount of nucleolytic resection (loss) from each DNA end varies, usually over a range of 0 to 14 bp; but there are less frequent examples with resection up to ~25 bp (45). The rare instances where there is loss greater than 25 bp may represent cases where the DNA end is released prematurely from whatever factors retain the two DNA ends in some proximity (see below). In vertebrate NHEJ, a complex of Artemis:DNA-PKcs is capable of endonucleolytically resecting a wide range of DNA end configurations. In yeast, plants and invertebrates, the MRX complex appears to be critical for some of the DNA end resection (24). The evolutionary inception of Artemis and DNA-PKcs coincides with the inception of V(D)J recombination (the vertebrate/invertebrate transition). The MRX nuclease system and the DNA-PKcs system both rely on the same conserved C-terminal tail for protein-protein interaction, also suggesting that the Artemis:DNA-PKcs complex may have evolved to replace the MRX complex for vertebrate NHEJ (46).
Second, nucleotide addition can occur at the DNA junction, even when terminal transferase is not present. In mammals, pol mu can add in a template-independent manner under physiologic conditions (7, 8). Mammalian pol lambda does not appear to add in a template-independent manner except when Mg2+ is replaced with Mn2+. (47, 48). The precise biochemical properties of the Pol X polymerases in other eukaryotes are not as clear. Interestingly, in bacteria, the polymerase activity intrinsic to the LigD protein is capable of adding one nucleotide or ribonucleotide in an entirely template-independent manner (23), perhaps reflecting convergent evolution.
Pol mu and pol lambda both seem to have much greater flexibility than most polymerases during template-dependent synthesis also (47, 48). The template-independent addition by pol mu would sometimes be expected to fold back on itself (42), and the resulting stem-loop structure might function as a primer/template substrate (see step 1 in Suppl. Fig. 2)(48). This may account for the observed inverted repeats at many NHEJ junctions from chromosomal translocations in humans (49, 50). Both pol mu and pol lambda can slip back on their template strand (51-53), and this may permit generation of direct repeats, accounting for such events seen in vivo (54-56). Direct repeats are also often seen at NHEJ junctions from human chromosomal translocations (49, 50). The direct and inverted repeats seen at these NHEJ junctions have been termed T-nucleotides, where the T stands for templated (49, 50).
Therefore, even from a relatively homogeneous set of starting DNA ends as substrates, there is substantial variation in the nucleotide resection from each end and variation in the amount of template-independent addition to the two DNA ends. These two sources of variation are the basis for the heterogeneity at the joining site.
Mechanistic Flexibility, Iterative Processing, and Independent Enzymatic Functions as Conserved Themes in NHEJ
In the context of considering diverse substrates and diverse joining products, it is worth noting an additional facet of that flexibility: the proteins involved and the order of their action in NHEJ can vary at either of the two DNA ends. Each DNA end, especially when bound by Ku, is best considered as a node at which any of the NHEJ proteins can dock. If one of the polymerases arrives first at the ‘left’ end, then this might be the first step at that end. However, if the nuclease binds first at that end, then resection will occur first (32).
In addition to the theme of mechanistic flexibility, there is the theme of iterative processing of the junction (32, 38). For any given joining event, there might, for example, be only three steps involving: one each involving a nuclease, a polymerase, and then a ligase. However, in another joining scheme, there might be 10 steps with multiple appearances by each enzyme activity. Hence, each of the enzymatic components might be involved not at all, once, or many times.
Related, but in addition to the theme of iterative processing is the independent function of the nuclease, polymerases, and ligase from one another and even from Ku. Each of the enzymatic activities has a substantial range and level of activity without one another, and even without Ku, when examined in purified biochemical systems. For example, Ku is entirely unnecessary in ligation of DNA ends by the ligase IV complex when those ends share 4 bp of terminal microhomology, but Ku is stimulatory for shorter microhomology lengths (42). Polymerases mu and lambda are able to carry out fill-in synthesis, and polymerase mu does not always require Ku or XRCC4:DNA ligase IV in order to have TdT-like activity at a DNA end (42). The Artemis:DNA-PKcs complex does not require Ku or any other component to carry out its endonucleolytic functions (37). Therefore, independent function of each enzymatic activity, along with iterative processing and mechanistic flexibility are all noteworthy features of vertebrate NHEJ. S. cerevisiae NHEJ manifests mechanistic flexibility, but within a narrower range of junctional outcomes than mammalian NHEJ (24, 57).
Enzymatic Revision of a Partially Completed Junction
In the context of iterative processing, the Artemis:DNA-PKcs nuclease complex is able to nick within the single-stranded portion of a gapped structure and within a bubble structure (38). Joinings where only one strand is ligated would often have a gapped configuration. Nicking of such a gap could permit nucleotides that were originally part of the arbitrarily designated left DNA end to become separated from that left end and become associated with the right DNA end. Then further nucleotide addition at the left end could separate these nucleotides from the left end. One can find potential examples of this at in vivo junctions. In other scenarios, with the flexibility of the ligase, there may be more nucleotides on the top strand than the bottom strand (42). The activated Artemis:DNA-PKcs complex can nick mismatched or bubble structures on either strand, thereby permitting additional rounds of junctional revision (38).
Terminal Microhomology Between the Initial Two DNA Ends Can Simplify the Protein Requirements of NHEJ
Based on both S. cerevisiae and mammalian in vivo NHEJ studies, the variation of the resulting junction is usually less when there is terminal microhomology at the ends (24, 57-60). This may reflect the involvement of fewer NHEJ proteins, and genetic studies support the view that not all of the NHEJ components are essential when the two DNA ends share terminal microhomology (60-66).
As mentioned, when the two DNA ends happen to share four nucleotide overhangs that are perfectly complementary, then the only component needed in purified biochemical systems is the XRCC4:DNA ligase IV (42). No nucleolytic resection or polymerase action is needed. In vitro, using purified proteins, XRCC4:ligase IV is adequate to join such a junction, and even ligase IV alone may be sufficient, all without Ku. Moreover, even ligase I or III alone is sufficient for such joins, though at a lower efficiency (42). In vivo, generation of defined DNA end configurations at DSBs is not simple, but there are two approaches that have been used. In S. cerevisiae, short oligonucleotide duplexes can be ligated onto the DNA ends of a linear plasmid, and then these can be transfected into cells (57). One can then harvest the joined circular molecules for analysis. Based on this, it is clear that the joining dependence is simplified when there is terminal microhomology at the DNA ends. A second system for generating defined DNA ends is V(D)J recombination, as mentioned earlier. These ends are not as precisely defined as in the yeast system (because the precise DNA end configuration depends on how hairpin intermediates are opened), but there is the advantage that the ends are actually generated inside the nucleus. In V(D)J recombination, the coding ends (defined in Suppl. Fig. 3) are usually configured with a four nucleotide 3' overhang. If the DNA ends are chosen to be complementary, then the dependence of the V(D)J recombination on Ku can be very minimal (60). However, NHEJ repairs such ends so as to a align the microhomology in a disportionate fraction of the joins (58, 67).
Terminal Microhomology Can Bias the Diversity of Joining Outcomes But Microhomology is Not Essential for NHEJ
One of the strengths of NHEJ is that microhomology does not appear to be essential in mammalian cells (58, 67). The joining of incompatible DNA ends may be a key selective advantage that drove further evolutionary development of NHEJ in higher eukaryotes. The fact that some of this evolution was convergent rather than divergent further illustrates the strength of this selective advantage. Most natural DSBs generate incompatible ends with little or no microhomology within the first few nucleotides. (S. cerevisiae shows mechanistically interesting differences from mammalian NHEJ insofar as yeast are very poor at blunt end ligation and perhaps more reliant on at least one base pair of terminal microhomology (24, 25, 68).) For mammalian NHEJ joins, the most common amount of observed terminal microhomology is zero nucleotides (45, 69). The next most common is one nucleotide, and longer microhomologies are less common in proportion to their length. As mentioned above, when microhomology is present, then usage of that microhomology for a given pair of DNA ends can be dominant (57-60). Overhangs with substantial terminal microhomology are uncommon in nature and primarily are limited to regions containing repetitive DNA. In wild type cells or neoplastic cells arising in normal animals, the most common amount of terminal microhomology at NHEJ junctions is zero. Exceptions to this arise from two circumstances. First, if the experimental system being used specifically positions terminal microhomology at or near the DNA ends, then the high-microhomology outcome might be observed. Second, in animals lacking a complete wild type NHEJ system, the NHEJ process may be slower, show more resection, and may seek end alignments that are stabilized by more terminal microhomology (2 or 3 bp), as discussed in the next section.
Alternative NHEJ
Ligase 4-independent Joining
It has been elegantly demonstrated in murine and yeast genetic studies that end joining can occur in the absence of ligase IV (57, 69-71). Insofar as the only remaining ligase activity in the cells is due to ligase I in S. cerevisiae or ligase I or III in vertebrate cells, these joinings must be done by ligase I or III. Most of these joinings rely more heavily on the use of terminal microhomology than NHEJ in wild type cells. In wild type cells, plots of end joining frequency versus microhomology length show a peak at zero nucleotides of microhomology and decline for increasing lengths (69). But for joinings without ligase IV, the peak is at 2.5 bp and the frequency declines on both sides of this peak (69-71).
In biochemical systems using purified NHEJ proteins, it has been shown that human ligase I and III are able to join DNA ends that are not fully compatible (e.g., joining across gaps in the ligated strand), though it is still substantially less efficient than joining by the XRCC4:ligase IV complex (42, 43). Though relatively inefficient, this joining by ligase I or III is somewhat more efficient with 2 or more bp of terminal microhomology to stabilize the ends. Therefore, in the absence of the ligase IV complex, it may not be surprising that the peak microhomology usage changes from zero to between 2 and 3 bp.
The in vivo joining efficiency by mammalian ligase IV relative to ligase I and III is difficult to measure. Two measurements have been done in murine cells in which the joining occurred at DNA ends that have some increased opportunities for terminal microhomology, the class switch recombination sequences. In one study, cells lacking ligase IV are removed from mice and stimulated in culture to undergo class switch recombination (70). Measurements of switch recombination can be done as early as 60 hrs after stimulation. In this case, end joining without ligase IV is reduced only 2.5-fold. In another case, a murine cell line was used to make the genetic knock out and measurements could be done as early as 24 hrs, at which time the joining without ligase IV was reduced about 9-fold (69). In both cases, the joining is almost certainly done by ligase I or III. The latter study suggests that the joining by ligase I or III is substantially less efficient at early times. In both studies, given sufficient time, the joining by ligase I or III improves to about half that of the wild type cells (where nearly all joining is likely -- though not proven -- to be due to ligase IV). Also, in both studies, the joining in wild type cells was much less dependent on terminal microhomology than joining in the ligase IV knock out cells. One reasonable interpretation of these two studies is that ligase IV Is more efficient (and perhaps faster) at joining incompatible DNA ends in vivo, but that ligase I or III can join ends at a lower efficiency, especially when terminal microhomology can stabilize the DNA ends.
In S. cerevisiae, end joining can occur in the absence of ligase IV, but it is at least 10-fold less efficient (24). Moreover, when the joining does occur, it tends to use microhomology (usually >4 bp) that is longer and more internal to the two DNA termini than is seen for wild type yeast (57).
Ku-independent Joining
Ku-independent end joining also occurs in both S. cerevisiae and mammalian cells. In yeast, such events can even be as efficient as end joining in the corresponding wild type cells (24, 57). For ligase IV mutants in yeast, the joining relies on longer microhomology (usually >4 bp) that is more internal to the two DNA ends. Ku-independent end joining also can be seen in mammalian cells (60). Even in vertebrate V(D)J recombination, when the two DNA ends share 4 bp of terminal microhomology, the dependence on Ku for joining efficiency can be small (2.5-fold) (60). This indicates that terminal microhomology can substitute for the presence of Ku. We do not know with certainty what Ku-independent joining means mechanistically, but one possibility is that the ligase IV complex holds the DNA ends and Ku stabilizes the ligase IV complex. But when Ku is absent, terminal microhomology may provide some of this stability, consistent with observations in biochemical systems (42).
Zero Microhomology Joins in the Absence of Ligase IV
For some ends joined in the absence of ligase IV, no microhomology is obvious (66). This raises the question whether ligase I or III can join ends with no terminal microhomology. XRCC4:ligase IV can ligate blunts ends (72), and ligase III is also able to do this at low efficiencies (35). All three ligases can do so when macromolecular volume excluders are present (35). Nevertheless, blunt end ligation is much less efficient for all three ligases.
Another explanation of such events is that they involve template-independent synthesis by the Pol X polymerase (42). As discussed, in mammalian cells, pol mu and pol lambda participate in NHEJ (42, 73, 74), as does POL4 in S. cerevisiae (75). In Mn2+ buffers, both pol mu and pol lambda can add nucleotides template-independently, and in the more physiologic Mg2+ buffers, pol mu still shows robust template-independent addition. Such template-independent activity could permit additions to DNA ends that provide microhomology with another end; because that addition would be random, it would not have been scored as microhomology. One could consider such inapparent microhomology as polymerase-generated microhomology.
In addition to template-independent synthesis by pol mu (which pol mu exhibits alone or in the context of other NHEJ proteins), pol mu together with Ku and XRCC4:ligase IV can synthesize across a discontinuous template, and this would also generate microhomology. However, this mechanism requires that the DNA end providing the ‘template’ have a 3' overhang to permit the polymerase to extend into that end. Hence, only a subset of DNA ends could be handled in this manner.
The ratio of template-independent versus template-dependent synthesis by pol mu at NHEJ junctions in such cases is not entirely clear, but both mechanisms occur under physiologic conditions in biochemical systems, and there is clearly some evidence for the template-independent pol mu addition within mammalian cells (41).
Nomenclature
In all organisms in which there is NHEJ, there are examples of DNA end joining in the absence of the major NHEJ ligase of that organism (76); even in mycobacteria, LigC can function in NHEJ when LigD is absent (77).) Given that the ligase is regarded as the signature enzymatic requirement of NHEJ, these joining events have been proposed to be due to ‘alternative NHEJ’, or ‘backup NHEJ’. As mentioned, at most of these end joinings, there is substantial terminal microhomology. Hence, in S. cerevisiae, this joining has also been called microhomology-mediated end joining (MMEJ); however, it is essentially an alternative NHEJ.
For eukaryotic end joining, one reasonable nomenclature is that NHEJ be the general term, and that exceptions simply be noted for their exception (e.g., ligase 4-independent NHEJ, Ku-independent NHEJ, or DNA-PKcs-independent NHEJ (i.e., X-independent NHEJ, where X is the omitted protein). Until a specific pathway is delineated, this is a practical solution. It is quite conceivable (even likely) that ligase 4-independent NHEJ is merely NHEJ in which ligase I or III completes the ligation at a somewhat lower efficiency than the ligase IV complex.
Evolutionary Comparisons of the NHEJ Mechanism Across Prokaryotes and Eukaryotes
Initially, NHEJ was thought to be restricted to eukaryotes because the best-studied prokaryote, E. coli, cannot recircularize linear plasmids. However, when bioinformatists discovered a distantly diverged Ku-like gene in prokaryotic genomes, the existence of a similar NHEJ pathway in bacteria became clear (2, 78, 79). The bacterial Ku homologue appears to form a homodimer with a structure similar to the ring-shaped eukaryotic Ku heterodimer (80). The gene for an ATP-dependent ligase named LigD was typically found to be adjacent to the Ku gene on the bacterial chromosome (2, 78). This linkage between Ku and an ATP-dependent ligase prompted further extensive studies and later defined a bacterial NHEJ pathway. In most bacterial species, unlike the eukaryotic NHEJ ligase IV, LigD is a multidomain protein that contains three components within a single polypeptide: a polymerase (POL) domain, a phosphoesterase (PE) domain, and a ligase (LIG) domain (23).
Why do not all bacteria have an NHEJ pathway? Bacterial NHEJ is nonessential under conditions of rapid proliferation because homologous recombination is active and a duplicate genome is present to provide homology donors (23, 81). Those bacteria that have the NHEJ pathway spend much of their life cycle in stationary phase at which point HR is not available for DSB repair for lack of homology donors. In addition, desiccation and dry heat are two naturally occurring physical processes that produce substantial numbers of DSBs in bacteria. Therefore, bacterial Ku and LigD are present in species that often form endospores, because NHEJ is important for repair of DSB arising during long periods of sporulation.
INDIVIDUAL PROTEINS OF VERTEBRATE NHEJ
Each of the individual NHEJ proteins carries an interesting detailed functional and structural literature, and more detailed reviews of each individual component are cited.
Ku
Ku was named based on protein gel mobilities (actually 70 kDa and 83 kDa) of a autoantigenic protein from a scleroderma patient with the initials K.U. Ku86 is also equivalently called Ku80. The toroidal shape of Ku is consistent with studies showing that purified Ku can bind at DNA ends but also slide internally at higher Ku concentrations (82). Ku can only load and unload at DNA ends. When linear molecules bearing Ku are circularized, the Ku proteins are trapped on the circular DNA. A minimal footprint size for Ku is ~14 bp at a DNA end (83). The key aspects of Ku in NHEJ have been discussed earlier, and the reader is referred to detailed reviews about Ku for additional information (84).
DNA-PKcs
DNA-PKcs has a molecular weight of 469 kDa and is 4128 aa. It is the largest protein kinase in biology, and the only one that is specifically activated by binding to duplex DNA ends of a wide variety of end configurations (33, 85-87). DNA-PKcs alone has an equilibrium dissociation constant of 3 × 10-9 M for blunt DNA ends, and this tightens to 3×10-11 M when Ku is also present at the end (88). Once bound, DNA-PKcs acquires serine/threonine kinase activity (89). But its initial phosphorylation target seems to be itself, with more than 15 autophosphorylation sites and probably an equal number yet to be defined (90). In addition to the relationship with Artemis discussed earlier, DNA-PKcs interacts with XRCC4 and phosphorylates (91) a very long list of proteins in vitro (26). In vivo evidence for functional effects of those additional protein phosphorylation targets is limited.
The best current structural information concerning DNA-PKcs alone is at 7A resolution by cryo-EM (92). At this resolution α-helices are resolved, but this cryo-EM structure only contains a fraction of the total number of α-helical densities expected and therefore could not definitively reveal which portions of the structure are related to the primary amino acid sequence of DNA-PKcs. The ‘crown’ in that structure is thought to contain the FAT domain and possibly parts of the kinase domain (Fig. 4b)(92). The ‘base’ in that structure is the same as ‘proximal claws 1 and 2’ in a cryo-EM structure of Ku:DNA-PKcs:DNA by another group and was shown to contain HEAT like repeats at 7A resolution (93). In the Ku:DNA-PKcs:DNA and DNA-PKcs:DNA structures, the path of the duplex DNA is not entirely certain, and it is not clear which side of Ku is bound to DNA-PKcs (93, 94). Positioning of the C-terminal portion of Ku when bound to DNA-PKcs is also not determined, which is important because this interaction activates DNA-PKcs and is defined at the primary sequence level (46). Continued work using cryo-EM and other structural methods will undoubtedly be of great value.
It is not clear whether DNA-PKcs remains bound to the DNA ends throughout all processing steps of NHEJ (31, 95). Phosphorylation at the ABCDE cluster appears to increase the ability of other proteins, such as ligases, to gain access the DNA ends, suggesting that DNA-PKcs may dissociate more readily after autophosphorylation at these sites (90, 95, 96).
DNA-PKcs interaction with other proteins is also important. As mentioned, DNA-PKcs is critical for the endonucleolytic activities of Artemis (31, 37, 39, 97, 98). Activated DNA-PKcs stimulates the ligase activity of XRCC4:DNA ligase IV (90, 95, 96). Interestingly, presence of XRCC4:DNA ligase IV stimulates the autophosphorylation activity of DNA-PKcs (96). Hence, DNA-PKcs may be critical for the nucleolytic step, but also stimulatory for the ligation step.
Artemis
The Artemis:DNA-PKcs complex has 5' endonuclease activity with a preference to nick a 5' overhang so as to leave a blunt duplex end (37). The Artemis:DNA-PKcs complex also has 3' endonuclease activity with a preference to nick a 3' overhang so as to leave a 4nt 3' overhang. In addition, the Artemis:DNA-PKcs complex has the ability to nick perfect DNA hairpins at a position that is 2 nts past the tip. These three seemingly diverse endonucleolytic activities at single- to double-strand DNA transitions are similar to one another if one infers the following model for binding of the Artemis:DNA-PKcs complex to DNA (Suppl. Fig. 1)(37). The complex appears to localize to a 4 nucleotide stretch of single-stranded DNA adjacent to a single-/double-strand transition, and then nick on the 3' side of that 4 nucleotide region. This would explain why 5' overhangs are preferably removed to generate a blunt DNA end, but 3' overhangs are nicked so as to preferably leave a 4 nucleotide 3' overhang. Moreover, it explains why a hairpin is nicked not at the tip, but 2' nucleotides 3' of the tip (37). In perfect DNA hairpins, the last two base pairs do not form well, which means the tip is actually similar in many ways to a 4 nucleotide single-stranded loop. Artemis nicks the hairpin on the 3' side of that loop. The opened hairpin then becomes a 3' overhang of 4 nucleotides.
In V(D)J recombination, null mutants of DNA-PKcs and of Artemis are very similar (63, 99-101). Both result in failure to open the DNA hairpins, but signal ends are joined. Biochemically, when a purified complex of Artemis:DNA-PKcs binds to an individual DNA hairpin molecule, that hairpin can activate the kinase activity of that DNA-PKcs protein (in cis) to phosphorylate itself and the bound Artemis within the C-terminal portion (96, 102). With respect to hairpin opening, and its other endonucleolytic activities, Artemis:DNA-PKcs functions as if it were a heterodimer in which mutation of either subunit results in failure of DNA end processing. A recent DNA-PKcs point mutation in a patient supports that view (103), as does a murine knock-in model that recreates a truncation mutant of Artemis that removes the C-terminal portion where the sites of DNA-PKcs phosphorylation are located.
Some DNA ends containing oxidative damage to the bases or sugars require nuclease action to remove the damaged nucleotides. In these cases, involvement of Artemis:DNA-PKcs appears critical (39, 98).
POL × Polymerases
Pol mu
Polymerase mu has several remarkable activities under physiologic buffer conditions. First, it can carry out template-dependent synthesis with dNTP and rNTP, and it has substantial template-independent synthesis capability, like TdT (104). No other higher eukaryotic polymerase has this range of activities. The ability to add rNTP may be important for NHEJ during G1, when dNTP levels are low, but rNTP levels are high (74). Incorporation of U into the junction might then mark the junction for possible revision using uracil glycosylases at a later point in time. (The highly homologous POL4 of S. cerevisiae also efficiently incorporates rNTPs (105)). Interestingly, the bacterial polymerase for NHEJ (part of LigD) has the ability to incorporate ribonucleotides as well (23).] Second, like many error-prone polymerases, polymerase mu can slip on the template strand (48, 51, 52). Third, and as mentioned earlier, pol mu, when together with Ku and XRCC4:DNA ligase IV, can polymerize across a discontinuous template strand, essentially crossing from one DNA end to another (106, 107). Fourth, and also mentioned earlier, pol mu has template-independent activity, which pol mu exhibits whether alone or together with Ku and XRCC4:DNA ligase IV (42).
Both the template-independent and the discontinous template polymerase activities are likely to be of great importance in the joining of two incompatible DNA ends. For example, in the case of two blunt DNA ends, the TdT-like activity of pol mu allows pol mu to add random nucleotides to each end. As soon as the resulting short 3' overhangs share even one nucleotide of complementarity (polymerase-generated microhomology), then ligation is much more efficient (42). (This type of microhomology would not have been present in the two original DNA end sequences, and in that sense, one could refer to it as polymerase-generated microhomology.) In contrast, in the mechanism where pol mu (with Ku and XRCC4:ligase IV present) crosses from one DNA end to the other (template-dependent synthesis across a discontinuous template strand), the duplex end receiving the new synthesis must be a 3' overhang to permit extension of the incoming polymerase (107). Hence, there are two mechanisms by which polymerase mu can create microhomology during the joining process (and these ‘reaction intermediates’ would not be scored as microhomology events based merely on the final DNA sequence of the junctional product).
Structural studies of polymerase mu, TdT and pol lambda are defining the basis for the intriguing differences between these three highly-related DNA polymerases (104). A region called loop 1 (and other positions, such as H329) is important for substituting for the template strand as TdT (always) and pol mu (sometimes) polymermize in their template-indepdendent mode (108, 109). Importantly, the crystal structures are on single-strand break DNA, and hence, we do not know how these enzymes configure on DSBs.
Polymerase Lambda
Mouse in vivo systems, crude extract NHEJ studies, and purified NHEJ systems support a role for pol lambda in NHEJ (41, 73, 110). Pol lambda functions primarily in a standard template-dependent manner in Mg2+ buffers, but has template-independent activity in Mn2+ (48, 104). The lyase domain in pol lambda is functional, whereas the one in pol mu does not appear to be functional. This permits pol lambda to function after action by a glycosylase to remove a damaged base. Like pol mu, pol lambda also slips on the template strand.
Terminal Deoxynucleotidyl Transferase
TdT or terminal transferase is only expressed in pro-B/pre-B and pro-T/pre-T stages of lymphoid differentiation (111). Like the other two Pol × polymerases of NHEJ, TdT has an N-terminal BRCT domain. (Polymerase beta is the only Pol × polymerase that is not involved in NHEJ, and it lacks any BRCT domain.) TdT only adds in a template-independent manner, consistent with a different loop 1 from pol mu and pol lambda (104). TdT has a preference to stack the incoming dNTP onto the base at the 3'OH, accounting for its tendency to add runs of purines or runs of pyrimidines (45). TdT also has a lower Km for dGTP, and this also biases its template-independent synthesis in vitro and in vivo (111, 112).
XLF, XRCC4 and DNA Ligase IV
DNA ligase IV, XRCC4:DNA ligase IV, and XLF:XRCC4:DNA ligase IV
DNA ligase IV (also called ligase 4 or DNL4) is mechanistically flexible. In the absence of XRCC4, DNA ligase IV appears to still be capable of ligating not only nicks, but even compatible (4 nt overhang) ends of duplex DNA (72). With XRCC4, ligase IV is able to ligate ends that share 2 bp of microhomology and have 1 nt gaps, but addition of Ku improves this 10-fold (42). When Ku is present, XRCC4:DNA ligase IV is able to ligate even incompatible DNA ends at low efficiency (42). When XLF if also added, then XLF:XRCC4:DNA ligase IV, in the presence of Ku, can ligate incompatible DNA ends much more efficiently (43, 113).
Even one nucleotide of terminal microhomology markedly increases the efficiency of ligation by Ku plus XRCC4:DNA ligase IV (43, 113). But some junctions formed within cells have no apparent microhomology (45). These could be cases where Ku plus XLF:XRCC4:DNA ligase IV ligate incompatible DNA ends or blunt ends. As mentioned earlier, pol mu may add either template-independently or across a discontinuous template strand from the left to the right DNA end, and either of these mechanisms would not be scored as use of microhomology upon inspection of the sequence of the joined product junction. As mentioned, one could call this polymerase-generated microhomology, and it is basically a reaction intermediate.
DNA ligase IV is predominately pre-adenylated as it is purified from crude extracts. The reader is referred to specialty reviews for more details (114).
XRCC4 and XLF (Cernunnos)
XRCC4 can tetramerize by itself, but it is unclear if this serves a function (115). The crystal structure demonstrates a globular head domain and a coiled coil C-terminus when it forms a dimer (116, 117).
The crystal structure of XLF (Cernunnos) suggests a structure similar to XRCC4, with a globular head domain and a coiled-coil C-terminus where multimerization is driven (118, 119). When XLF is missing in humans, patients are IR sensitive and lack V(D)J recombination (120, 121). In mice, the IR defect is the same as in humans, but the V(D)J recombination defect is less severe in pre-B cells and yet is severe in mouse embryonic fibroblasts (when given exogenous RAGs) from the same mice (122). Considering the biochemical role of XLF in the joining of incompatible DNA ends, it has been suggested that TdT in the pre-B cells can provide ‘occult’ or polymerase-generated microhomology, making joining less reliant on XLF, and this seems like a reasonable explanation of the data thus far (122).
Complexes of XLF, XRCC4, DNA ligase IV and Interaction with Other NHEJ Components
The interactions between XLF, XRCC4 and DNA ligase IV have been studied genetically and biochemically (120, 121, 123, 124). Gel filtration studies of XLF, XRCC4 and DNA ligase IV are most consistent with a stoichiometry of 2 XLF, 2 XRCC4 and 1 ligase IV (120). Complexes of XRCC4 and ligase IV are most consistent with a stoichiometry of 2 XRCC4 and 1 DNA ligase IV (115, 117). Further functional and structural work on the ligase complex will be of great value.
For both S. cerevisiae and in mammalian purified proteins, Ku is able to improve the binding of XRCC4:DNA ligase IV at DNA ends. This interaction requires both Ku70 and 86 and the first BRCT domain within the C-terminal portion of ligase IV (aa 644 to 748) (91). The presence of DNA-PKcs enhances this complex formation, perhaps through interactions with XRCC4 (125-127). XRCC4:DNA ligase IV is able to stimulate DNA-PKcs kinase activity (96). The ligase complex also stimulates the pol mu and lambda activities in the context of Ku (96). All of these findings suggest that the NHEJ components, while capable of acting independently, also evolved to function in a manner that is synergistic when in close proximity.
Polynucleotide Kinase (PNK), Aprataxin (APTX), and PNK-APTX-Like Factor (PALF or APLF)
PNK, APTX and PALF (also called APLF) all interact with XRCC4 (Fig. 2 and 4f). PNK and XRCC4 form a complex via the PNK FHA domain, but only after the CK2 kinase phosphorylates XRCC4 (128). This same interaction occurs between APLF and XRCC4, and APTX and XRCC4.
Polynucleotide Kinase
For pathologic breaks caused by ionizing radiation or free radicals, PNK plays an important role in several ways that illustrate a corollary theme of NHEJ: enzymatic multifunctionality (129-131). Mammalian PNK is both a kinase and a phosphatase. PNK has a kinase domain for adding a phosphate to a 5'OH. PNK has a phosphatase domain that is important for removing 3' phosphate groups, as can remain after some oxidative damage or partial processing (or after NIELS 1 or 2 remove an abasic sugar, leaving a 3' phosphate group). Interestingly, the short 3' overhang that the Artemis:DNA-PKcs complex leaves after cleaving a long 3' overhang represents an ideal substrate for PNK to add a 5' phosphate at a recessed 5'OH.
Removal of 3' Phosphoglycolate Groups
Oxidative damage often causes breaks that leave a 3'-phosphoglycolate group, and these can be removed in either of two major ways. First, Artemis:DNA-PKcs can remove such groups using its 3' endonucleolytic activity (39, 98). Second, 3'-phosphoglycolates can be converted to 3'-phosphate by tyrosyl DNA phosphodiesterase 1 (Tdp1), whose major role is the removal of tyrosyl-phosphate linkages that arise when topoisomerase fails to religate transient DNA single-strand break reaction intermediates. Then PNK can remove the 3'-phosphate group.
Aprataxin (APTX)
Aprataxin is important in deadenylation of aborted ligation products in which an AMP group is left at the 5' end of a nick or DSB due to a failed ligation reaction (132, 133).
PALF or APLF
PALF and APLF are the same protein (511 aa, 57 kDa). The PALF designation stands for PNK and APTX-like FHA protein (134). APLF stands for aprataxin- and PNK-like factor (135, 136). Previously, it was also called C2orf13. APLF is an endonuclease and a 3' exonuclease (134). This is interesting, given that Artemis lacks a 3' exonuclease.
PHYSIOLOGIC DNA RECOMBINATION SYSTEMS AND THE ROLE OF NHEJ
V(D)J Recombination
V(D)J recombination is one of the two physiologic systems for creating intentional DSBs in somatic cells, specifically in early B or T cells for purpose of generating antigen receptor genes. RAG1 and RAG2 (both expressed in early B and T cells only) form a complex that can bind sequence-specifically at recombination signal sequences (RSS) that consist of a heptamer and nonamer consensus sequence, separated by either 12- or 23-bp of nonconserved spacer sequence (Suppl. Fig. 3 or sidebar figure). (HMGB1 or 2 is thought to be part of this RAG complex, based on in vitro studies (137).) A given recombination reaction requires two such RSS sites, one 12-RSS and one 23-RSS (the 12/23 rule). The RAG complex initially nicks directly adjacent to each RSS and then uses that nick as a nucleophile to attack the anti-parallel strand at each of the non-RSS ends (138). The two non-RSS ends are called coding ends because these regions join to encode a new antigen receptor exon. The nucleophilic attack generates a DNA hairpin at each of the two coding ends. The NHEJ proteins take over at this point, beginning with the opening of the two hairpins by Artemis:DNA-PKcs, followed by NHEJ joining (37). Like vertebrate NHEJ generally, most coding ends do not share significant terminal microhomology (45, 139). The NHEJ junctions formed in V(D)J recombination have proven to be useful for understanding NHEJ more generally.
The DSBs at the two RSS ends are called signal ends, and these are blunt and 5' phosphorylated (140, 141). In cells that express terminal transferase, nucleotide addition can occur at these ends (142). But these ends only rarely suffer nucleolytic resection, presumably due to tight binding by the RAG complex (138). Joining of the two signal ends together to form a signal joint is also reliant on Ku and the ligase IV complex, but not dependent on Artemis or DNA-PKcs (65, 138). (The fact that DNA-PKcs is required for coding joint formation (for Artemis:DNA-PKcs opening of hairpins), but not for signal joint formation was a point of importance in the original description of scid mice (143). Scid mice have a mutant DNA-PKcs gene (144). Artemis null mice behave similarly in this respect (63).)
Class Switch Recombination
Class switch recombination (CSR) occurs only in B cells, after they have already completed V(D)J recombination. It is the second of the two physiologic forms of DSB formation in somatic cells (64). CSR is essential for mammalian B cells to change their immunoglobulin heavy chain gene from producing Igμ for IgM to Igγ, Igα or Igε for making IgG, IgA, or IgE, respectively (Suppl. Fig. 4). The process requires a B-cell specific cytidine deaminase called activation-induced deaminase (AID) which only acts to convert C to U within regions of single-stranded DNA (ssDNA). In mammalian CSR, the single-strandedness appears to be largely due to formation of kilobase-length R-loops that form at specialized CSR switch sequences due to the extremely (40-50%) G-rich RNA transcript that is generated at these specialized recombination zones (145, 146). This permits AID action on the nontemplate DNA strand. RNase H can resect portions of the RNA strand that pairs with the template strand, thereby exposing regions of ssDNA for AID action on that strand as well. Once AID introduces C to U changes in the switch region, then uracil glycosylase converts these to abasic sites, and APE1 can, in principal, nick at these abasic sites. Participation of other enzymes, such as Exo1, may assist in converting the nicks on the top and bottom strands into large overlapping gaps, resulting in DSBs. NHEJ is largely responsible for joining these DSBs, but, as mentioned, elegant work has demonstrated the role of either ligase I or III, when ligase IV is missing (70).
CHROMOSOMAL TRANSLOCATIONS, GENOME REARRANGEMENTS AND NHEJ
Chromosomal translocations and genome rearrangements can occur in somatic cells, most notably in cancer. In addition, such genome rearrangements can occur in germ cells, giving rise to heritable genome rearrangements. Though the breakage mechanisms vary, the joining mechanism is usually via NHEJ.
Neoplastic Chromosomal Rearrangements
The vast majority of genome rearrangement-related DSBs (translocations and deletions) in neoplastic cells are joined by NHEJ, though there is ample opportunity for participation of alternative ligases, if ligase IV is missing, as in experimental systems or extremely rare patients (66, 70). The breakage mechanisms in neoplastic cells include the following: random or near-random breakage mechanisms (due to ROS, ionizing radiation, or topoisomerase failures) in any cell type and V(D)J-type or CSR-type breaks in lymphoid cells (147). The lymphoid-specific breakage mechanisms can combine antigen-receptor loci with off-target loci at sequences that are similar to the RSS or CSR sequences (18, 148). In some lymphoid neoplasms, two off-target loci are recombined, and the breakage at each site of the two sites can occur by any of the above mechanisms. Sequential action by AID followed by the RAG complex at CpG sites appears likely in some of the most common breakage events (called CpG-type events) in human lymphoma (147). In both CSR-type and CpG-type breaks, AID requires single-stranded DNA to initiate C to U or meC to T changes, respectively. Departures from B-form DNA are relevant to such sites (149, 150).
Constitutional Chromosomal Rearrangements
The breakage mechanisms in germ cells are presumably due to random causes primarily (ROS, ionizing radiation, or topoisomerase failures). Deviations from B-DNA are known to be relevant at long inverted repeats where the most common constitutional translocations occur. The most common constitutional chromosomal rearrangement is the t(11;22) in the Emanuel Syndrome (151). In this case, inverted repeats result in cruciform formation, creating a DNA structure that is vulnerable to DNA enzymes that can act on various portions of the cruciform. Once broken, the DNA ends are likely joined by NHEJ, based on observed junctional sequence features.
During evolution, some of the chromosomal rearrangements that arise during speciation are almost certain to share themes with those discussed here, including breakage at sites of DNA structural variation and joining by NHEJ. Replication-based mechanisms are also likely to be very important for major genomic rearrangements (152, 153).
CHROMATIN AND NHEJ
It is not yet clear how much disassembly of histone octamers must occur at a DSB for NHEJ proteins to function. In contrast to homologous recombination (HR), where kilobases of DNA are involved and γ–H2AX alterations are important, NHEJ probably requires less than 30 bp of DNA on either side of a break.
If randomly distributed, 80% of DSBs would occur on DNA that is wrapped around histone octamers and 20% would occur internucleosomally. For those breaks within a nucleosome, one study showed that Ku can bind, implying that the duplex DNA can separate from the surface of the nucleosome sufficiently to permit Ku to bind (154).
Several studies propose that γ–H2AX is important for NHEJ (16, 155). Much of the evidence is based on immunolocalization studies where the damage site may contain a mixture of HR and NHEJ events within the 2000 angstrom confocal microscope section thickness. Differences in access within the euchromatic versus the heterochromatic regions are likely, but even early genetic insights concerning this are limited to yeast (156).
H2AX is only present, on average, in one of every ten nucleosomes because H2A is the predominant species in histone octamers (16). Therefore, most DSBs would occur about 5 nucleosomes away (about 1 kb) from the nearest octamer containing an H2AX that is eligible for conversion to γ–H2AX via phosphorylation by ATM or DNA-PKcs at serine 139 of H2AX. Given this substantial distance from the site of the enzymatic repair, it is not clear that such γ–H2AX phosphorylation events are critical for NHEJ.
When DNA-PKcs does phosphorylate H2AX, this increases vulnerability of H2AX to the histone exchange factor called FACT (which consists of a heterodimer of Spt16 and SSRP1). Phosphorylated H2AX (γ-H2AX) is more easily exchanged out of the octamer, thereby leaving only a tetramer of (H3)2(H4)2 at the site, and this is more sterically flexible, thereby perhaps permitting DNA repair factors to carry out their work (157).
PARP-1 is able to downregulate the activity of FACT by ADP-ribosylation of the Spt16 subunit of FACT. This may be able to shift the equilibrium of γ-H2AX and H2AX in the nucleosomes. That is, PARP-1 activation at a site of damage might shift the equilibrium toward retention of γ-H2AX in the region, perhaps thereby aiding in recruitment or retention of repair proteins (157).
Hence, FACT may initially act proximally at the most immediate nucleosome (or closest one) to exchange γ-H2AX out and leave an (H3)2(H4)2 tetramer at the site of damage for purposes of flexibility of the DNA. FACT may act more regionally (distally) to favor the retention of γ-H2AX for purposes of integrating the repair process with repair protein recruitment, protein retention, and cell cycle aspects (157).
CONCLUDING COMMENTS
Mechanistic flexibility by multifunctional enzymes and iterative processing of each DNA end are themes that apply to all NHEJ across billions of years of prokaryotic and eukaryotic evolution. Because much of this evolution was convergent, it illustrates that these themes are important for solving this particular biological problem: joining of heterogeneous DNA ends at a double-strand breaks.
SUMMARY POINTS
NHEJ evolved to repair DSBs that needed to be joined directly. In haploid stationary phase organisms, there is no homology donor, and HR is not an option. Evolutionarily, assuming that many organisms were haploid, NHEJ likely represents a very early evolutionary DNA repair strategy. Many prokaryotes have NHEJ. The lack of NHEJ in E. coli made it a suitable organism for molecular cloning, but gave the initial mis-impression that prokaryotes lack NHEJ.
In eukaryotes, most DSBs outside of S/G2 of the cell cycle are joined by NHEJ. This is based on time courses of DSBs remaining after IR in wild type versus NHEJ mutant mammalian cells in culture. Within S/G2 phases, homologous recombination is very active because the two sister chromatids are directly adjacent.
Key components of vertebrate NHEJ are Ku; DNA-PKcs; Artemis; Pol × polymerases (pol mu and lambda); and the ligase complex, consisting of XLF, XRCC4, DNA ligase IV. Polynucleotide kinase (PNK) is important in for a subset of NHEJ events.
- Predominantly convergent evolution of NHEJ in prokaryotes and eukaryotes yielded mechanisms that reflect key themes for NHEJ and the repair of double-strand breaks. These themes are:
- mechanistic flexibility in handling diverse DNA end configurations by the nuclease, polymease, and ligase activities. This flexibility manifests as multifunctionality of the enzymes. For example, Artemis:DNA-PKcs complex has 5' endo- and 3' endonuclease activities, as well hairpin opening activities. In multicellular eukaryotes, the NHEJ nucleases, polymerase, and ligases are the most mechanistically flexible enzymes in their respective categories.
- iterative processing of each DNA end. Each DNA end as well as incompletely ligated junctions can undergo multiple rounds of revision by the nuclease, polymerases and ligases. To some extent, each DNA end can undergo processing independently, but the ligase can ligate efficiently when there are one or more base pairs of potential annealing. Hence, the ligase complex is frequently ‘testing’ for compatibility between the two DNA ends, while the nuclease complex and polymerases are carrying out sequential rounds of modification.
When components of NHEJ are missing (e.g., genetically mutant yeast, mice or extremely rare human patients), the flexible nature of NHEJ permits substitutions by other enzymes. For example, when ligase IV is missing in S. cerevisiae or mice, ligase I (yeast) or ligases I or III in higher eukaryotes can provide for ligation. However, biochemical studies and some cellular studies indicate that the efficiency of these joinings is lower, and the magnitude to which they are lower depends on the amount of terminal microhomology shared between the two DNA ends being joined. The joins by ligase I and III in biochemical and in vivo studies indicate that these ligases are much more reliant on terminal microhomolgy than is ligase IV, which can ligate even incompatible DNA ends. Rather than designate such substitutions as separate pathways (e.g., alternative NHEJ, backup NHEJ, microhomology-mediated NHEJ), one can include them as part of NHEJ but designate them as such (e.g., ligase IV-independent or Ku-independent NHEJ).
Terminal microhomology of 1 to a few nucleotides that are shared between the two DNA ends improves the efficiency of joining by NHEJ and can often, but not always, bias the outcome of the joining process toward using that microhomology. However, NHEJ does not require any microhomology. In S. cerevisiae and in mammalian systems, some stretches of terminal microhomology are used and others are ignored, and the reasons for this may relate to how the nucleases and polymerases process the ends or how the ligase IV complex prefers to pair the ends at such sites of microhomology.
Many (even most in vertebrate cells) NHEJ junctions join with no apparent microhomology. Biochemical studies indicate that joining can occur with absolutely no microhomology via Ku plus XLF:XRCC4:DNA ligase IV in a fully incompatible joining. For in vivo joins, one cannot rule out occult (invisible) microhomology use. This can occur when pol mu adds in a template-independent manner and creates chance microhomology at one end, which happens to match with the other duplex DNA end. This is not so difficult, given that even one or two nucleotides of microhomology are very stimulatory for ligation, and given that the XRCC4:DNA ligase IV complex can ligate across gaps. A second mode of pol mu may allow it to cross a discontinuous template, when Ku and XRCC4:DNA ligase IV are present, and this would also create microhomology that was not present in the original two DNA ends (i.e., occult microhomology).
FUTURE QUESTIONS
Are the two DNA ends held in proximity during NHEJ or is there synapsis?
In biochemical systems, XRCC4:DNA ligase IV does not appear to require a dedicated protein to help it bring two DNA ends together. This is especially clear when there are 4 bp of terminal microhomology, in which case, addition of Ku does not markedly stimulate joining. However, at 2 bp or less of terminal microhomology, Ku does improve XRCC4:DNA ligase IV ligation. This could occur because Ku is known to stabilize XRCC4:DNA ligase IV at DNA ends, rather than for any intrinsic ability of Ku to bring DNA ends together (which appears to be minimal for Ku alone). Some data suggest that DNA-PKcs might help bring DNA ends together, but this is seen only at 30 mM monovalent salt or less and was not observed at higher salt concentrations. Nevertheless, this is an active area of research and is subject to further definition.
This issue is relevant to whether the two DNA ends generated at a single DSB (proximal) are joined more readily than two DNA ends that arise far apart (as in a chromosomal translocation where two DSBs far apart are involved). The issue of whether close DNA ends are joined more efficiently than ends that are far apart is a point of active study.
In what ways do the DNA damage response proteins mechanistically or functionally connect with the NHEJ enzymes?
NHEJ at a single DSB may be so rapid and physically confined that the damage response pathways (DDR) involving ATM, the RAD50:MRE11:NBS1 complex, γ-H2AX, and 53BP1 are not activated, but this is quite unclear and subject to speculation. Experimentally or with environmental extremes, a cell may be challenged with many DSBs, in which case, activation of the DDR pathways are increasingly likely. As these activate, the impact on the enzymology of NHEJ is not entirely clear. Obviously, competition between HR and NHEJ components may be one aspect that arises. With experimental systems in which each cell has hundreds or thousands of DSBs, titration out of NHEJ components becomes a concern, and at that point it is not clear how much such extreme experimental systems inform us about the typical situation for which NHEJ evolved, which is presumably one or a few DSBs per cell. This all assumes that cells with hundreds of DSBs are optimally allowed to die rather than recover, and death would seem best in a multicellular organism irradiated at high levels. This is an active area of research and subject to continuing study.
Other Candidate Participants in NHEJ
Are there additional NHEJ enzymatic components missing? Like most repair pathways, vertebrate NHEJ already has nuclease, polymerase, and ligase activities. Are their additional enzymes missing? Each new candidate must demonstrate some genetic role such as reduced IR resistance when missing. In addition, biochemical studies with purified proteins are important.
The Werner's 3' exonuclease/helicase enzyme has been proposed as one candidate, but the IR-sensitivity data fail to show a large effect (158). WRN does interact with Ku and PARP1, but it has been proposed that this may reflect a role in replication fork repair rather than NHEJ, and this seems reasonable (159). Nevertheless, further work on this and any other new NHEJ candidates will be interesting.
Metnase has been proposed as a possible NHEJ nuclease and helicase, but it also has decatenating activity (160, 161). Metnase is present in humans but not in apes, mice, or apparently any other vertebrates, and there is no yeast homologue. Moreover, there is no genetic knockout to demonstrate a role in NHEJ. The NHEJ biochemical studies have not been done with fully purified protein. Further, work may define what nuclear process to which Metnase contributes in human cells, but likely not a key role in NHEJ, given its absence from organisms other than humans.
LIST OF RELATED RESOURCES
Translocation and Mutation databases
One major online translocation database is GRABD: http://archive.uwcm.ac.uk/uwcm/mg/grabd/.
The Atlas of Genetics and Cytogenetics in Oncology and Haematology is an on-line journal and database. Some chromosomal translocation data is available on the web site: http://atlasgeneticsoncology.org//
A chromosomal translocation database related to the citation Cell 135: 1130-42 is at: http://lieber.usc.edu/data/2008_cell_135_1130/.
A small subset of p53 mutations may be due to DSB followed by NHEJ. All p53 mutations can be found at the IARC p53 mutation database: http://www-p53.iarc.fr.
Supplementary Material
Acknowledgements
I thank T. E. Wilson, D. Williams, W. An, N. Adachi, and K. Schwarz for comments. I thank Jiafeng Gu and Xiaoping Cui, as well as other current members of my lab for comments. I apologize to those not cited due to length restrictions.
ACRONYMS
- NHEJ
Nonhomologous DNA end joining
- HR
Homologous recombination
- HDR
Homology-directed recombination or repair
- MMEJ
Microhomology-mediated end joining
- IR
Ionizing radiation
- V(D)J
Variable (Diversity) Joining
- CSR
Class switch recombination
REFERENCES
- 1.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–4. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–74. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–5. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 5.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–50. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 6.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in S. cerevisiae. Mol. Cell. Biol. 1996;16:2164–73. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–9. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999;398:728–31. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 9.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. Embo J. 2006;25:1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–46. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 11.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nature Rev. Mol. Cell. Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 12.Lieber MR, Karanjawala ZE. Ageing, repetitive genomes and DNA damage. Nature Rev. Mol. Cell. Biol. 2004;5:69–75. doi: 10.1038/nrm1281. [DOI] [PubMed] [Google Scholar]
- 13.Martin GM, Smith AC, Ketterer DJ, Ogburn CE, Disteche CM. Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Israel J. Med. Sci. 1985;21:296–301. [PubMed] [Google Scholar]
- 14.Riballo E, Kuhne M, Rief N, Doherty A, Smith GCM, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–603. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington D.C.: 2006. 1118. [Google Scholar]
- 17.Adachi N, Suzuki H, Iiizumi S, Koyama H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 2003;278:35897–902. doi: 10.1074/jbc.M306500200. [DOI] [PubMed] [Google Scholar]
- 18.Mahowald GK, Baron JM, Sleckman BP. Collateral damage from antigen receptor gene diversification. Cell. 2008;135:1009–12. doi: 10.1016/j.cell.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Pitcher RS, Green AJ, Brzostek A, Korycka-Machala M, Dziadek J, Doherty AJ. NHEJ protects mycobacteria in stationary phase against the harmful effects of desiccation. DNA Repair (Amst) 2007;6:1271–6. doi: 10.1016/j.dnarep.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Murnane JP. Telomeres, chromosome instability, and cancer. DNA Repair (Amst) 2006;5:1082–92. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 21.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 22.Hamer G, Roepers-Gajadien HL, van Duyn-Goedhart A, Gademan IS, Kal HB, et al. Function of DNA-protein kinase catalytic subunit during the early meiotic prophase without Ku70 and Ku86. Biol Reprod. 2003;68:717–21. doi: 10.1095/biolreprod.102.008920. [DOI] [PubMed] [Google Scholar]
- 23.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–61. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 24.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Ann. Rev. Genet. 2005;39:431–51. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 25.Wilson TE. Encyclopedia of Chemical Biology. Wiley; 2008. Nonhomologous recombination. [Google Scholar]
- 26.Anderson CW, Carter TH. The DNA-Activated Protein Kinase-DNA-PK. In: Jessberger R, Lieber MR, editors. Molecular Analysis of DNA Rearrangements in the Immune System. Springer-Verlag; Heidelberg: 1996. pp. 91–112. [Google Scholar]
- 27.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986;261:10375–9. [PubMed] [Google Scholar]
- 28.Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku Protein to DNA. J. Biol. Chem. 1993;268:7594–601. [PubMed] [Google Scholar]
- 29.Falzon M, Fewell J, Kuff EL. EBP-80, a Transcription Factor Closely Resembling the Human Autoantigen Ku, Recognizes Single- to Double-Strand Transitions in DNA. J. Biol. Chem. 1993;268:10546–52. [PubMed] [Google Scholar]
- 30.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 2000;6:909–13. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 31.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, et al. A Biochemically Defined System for Mammalian Nonhomologous DNA End Joining. Molecular Cell. 2004;16:701–13. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy. EMBO J. 1997;16:5098–112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.NickMcElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku Recruits the XRCC4-Ligase IV Complex to DNA Ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IVXRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem. 2000;275:26196–205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 36.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 37.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis:DNA-PKcs complex in V(D)J recombination and in nonhomologous end joining. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs Endonuclease Can Cleave Gaps, Flaps, and Loops. DNA Repair. 2005;4:845–51. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Yannone SM, Khan IS, Zhou RZ, Zhou T, Valerie K, Povirk LF. Coordinate 5' and 3' endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:3354–65. doi: 10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining: evidence for a DNA polymerase beta (POL4)-dependent pathway. J. Biol. Chem. 1999;274:23599–609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 41.Bertocci B, DeSmet A, Weill J-C, Reynaud CA. Non-overlapping functions of polX family DNA polymerases, pol m, pol l, and TdT, during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 42.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. Embo J. 2007;26:1010–23. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–62. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlissel MS. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol. Cell. Biol. 1998;18:2029–37. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell. Biol. 1996;16:258–69. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 47.Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hubscher U. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 48.Ramadan K, Shevelev IV, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu, and terminal deoxynucleotidyl transferase. J. Mol. Biol. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 49.Jaeger U, Bocskor S, Le T, Mitterbauer G, Bolz I, et al. Follicular lymphomas BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–9. [PubMed] [Google Scholar]
- 50.Welzel N, T TL, Marculescu R, Mitterbauer G, Chott A, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–36. [PubMed] [Google Scholar]
- 51.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J. Biol. Chem. 2004;279:5360–8. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 52.Dominguez O, Ruiz JF, Lera TLd, Garcia-Diaz M, Gonzalez MA, et al. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–42. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bebenek B, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda: implications for function. J. Biol. Chem. 2003;278:34685–90. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 54.Roth D, Wilson J. Illegitimate Recombination in Mammalian Cells. In: Kucherlapapti R, Smith GR, editors. Genetic Recombination. American Society for Microbiology; Washington, D.C.: 1988. pp. 621–53. [Google Scholar]
- 55.Roth DB, Chang XB, Wilson JH. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989;9:3049–57. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 1986;6:4295–304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daley JM, Laan RLV, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005;280:29030–7. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 58.Gerstein RM, Lieber MR. Extent to which homology can constrain coding exon junctional diversity in V(D)J recombination. Nature. 1993;363:625–7. doi: 10.1038/363625a0. [DOI] [PubMed] [Google Scholar]
- 59.Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes and Development. 1993;7:1459–69. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 60.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Y, Chaudhurri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNAPKcs-null mutation reveals DNA-PK independent for Ku in V(D)J recombination. Immunity. 1998;9:367–76. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 62.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 63.Rooney S, Sekiguchi J, Zhu C, Cheng H-L, Manis J, et al. Leaky scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 2002;10:65–74. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 64.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 65.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–70. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 66.Wang JH, Alt FW, Gostissa M, Datta A, Murphy M, et al. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J Exp Med. 2008;205:3079–90. doi: 10.1084/jem.20082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boubnov NV, Wills ZP, Weaver DT. V(D)J Recombination Coding Junction Formation without DNA Homology: Processing of Coding End Termini. Mol. Cell. Biol. 1993;13:6957–68. doi: 10.1128/mcb.13.11.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV--deficient B cells. J Exp Med. 2008;205:2745–53. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–82. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 71.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204:1717–27. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–5. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 73.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, et al. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2003;279:805–11. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 74.NickMcElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–15. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tseng HM, Tomkinson AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 2002;277:45630–7. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- 76.Ferguson DO, Alt FW. DNA double-strand break repair and chromosomal translocations: lessons from animal models. Oncogene. 2001;20:5572–9. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- 77.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–27. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001;500:186–8. doi: 10.1016/s0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- 79.d'Adda di Fagagna F, Weller GR, Doherty AJ, Jackson SP. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 2003;4:47–52. doi: 10.1038/sj.embor.embor709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–9. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 81.Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous End-Joining in Bacteria: A Microbial Perspective. Annu Rev Microbiol. 2007;61:259–82. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- 82.deVries E, vanDriel W, Bergsma WG, Arnberg AC, vanderVliet PC. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 1989;208:65–78. doi: 10.1016/0022-2836(89)90088-0. [DOI] [PubMed] [Google Scholar]
- 83.Yoo S, Kimzey A, Dynan WS. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. Journal of Biological Chemistry. 1999;274:20034–9. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]
- 84.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell. Biol. 2004;5:367–78. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 85.Leuther KK, Hammarsten O, Kornberg RD, Chu G. Structure of the DNA-dependent protein kinase: implications for its regulation by DNA. EMBO J. 1999;18:1114–23. doi: 10.1093/emboj/18.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hammarsten O, Chu G. DNA-Dependent Protein Kinase: DNA Binding and Activation in the Absence of Ku. Proc. Natl. Acad. Sci. 1998;95:525–30. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammarsten O, DeFazio LG, Chu G. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J. Biol. Chem. 2000;275:1541–50. doi: 10.1074/jbc.275.3.1541. [DOI] [PubMed] [Google Scholar]
- 88.West RB, Yaneva M, Lieber MR. Productive and Nonproductive Complexes of Ku and DNA-PK at DNA Termini. Mol. Cell. Biol. 1998;18:5908–20. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hartley KO, Gell D, Smith GCM, Zhang H, Divecha N, et al. DNA-Dependent Protein Kinase Catalytic Subunit: A Relative of Phosphatidylinositol 3-kinase and the Ataxia Telangiectasia Gene Product. Cell. 1995;82:849–56. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 90.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 91.Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 2007;6:712–22. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 92.Williams DR, Lee K-J, Shi J, Chen DJ, Stewart PL. Cryoelectron microscopy structure of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) at subnanometer resolution reveals α-helices and insight into DNA binding. Structure. 2008;16:468–77. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–9. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 94.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol Chem. 1997;272:24763–6. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 95.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–50. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu H, Shimazaki N, Raval P, Gu J, Watanabe G, et al. A biochemically defined system for coding joint formation in human V(D)J recombination. Molecular Cell. 2008;31:485–97. doi: 10.1016/j.molcel.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–9. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3'-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem. 2007;282:3547–58. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 99.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, et al. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–65. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li L, Salido E, Zhou Y, Bhattacharyya S, Yannone SM, et al. Targeted disruption of the Artemis murine counterpart results in SCID and defective V(D)J recombination that is partially corrected with bone marrow transplantation. J Immunol. 2005;174:2420–8. doi: 10.4049/jimmunol.174.4.2420. [DOI] [PubMed] [Google Scholar]
- 101.Xiao Z, Dunn E, Singh K, Khan IS, Yannone SM, Cowan MJ. A non-leaky Artemis-deficient mouse that accurately models the human severe combined immune deficiency phenotype, including resistance to hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2009;15:1–11. doi: 10.1016/j.bbmt.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jovanovic M, Dynan WS. Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res. 2006;34:1112–20. doi: 10.1093/nar/gkj504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burg Mvd, Ijspeert H, Verkaik N, Turul T, Wiegant W, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J. Clin. Invest. 2009;119:91–8. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, et al. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–25. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem. 2005;280:20051–8. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 106.NickMcElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, et al. A gradient of template dependence defines distinct biological roles for family x polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–66. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 107.Davis BJ, Havener JM, Ramsden DA. End-bridging is required for pol mu to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–94. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, et al. Structural insight into the substrate specificity of DNA polymerase mu. Nature Struc. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 109.Juarez R, Ruiz JF, NickMcElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-insructed synthesis. Nucl. Acids Res. 2006;34:4572–82. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Povirk LF. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair (Amst) 2006;5:1199–212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 111.Chang LM, Bollum FJ. Molecular biology of terminal transferase. CRC Crit. Rev. Biochem. 1986;21:27–52. doi: 10.3109/10409238609113608. [DOI] [PubMed] [Google Scholar]
- 112.Kornberg A, Baker T. DNA Replication. W.H. Freeman; New York: 1992. [Google Scholar]
- 113.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–6. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–99. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 115.Modesti M, Junop MS, Ghirlando R, Rakt Mvd, Gellert M, et al. Tetramerization and DNA ligase IV interaction of the DNA double-strand break protein XRCC4 are mutually exclusive. J. Mol. Biol. 2003;334:215–28. doi: 10.1016/j.jmb.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 116.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the XRCC4 DNA repair protein and implications for end joining. EMBO J. 2000;19:5962–70. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, et al. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–9. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 118.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol Cell. 2007;28:1093–101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 119.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, et al. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. Embo J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 121.Buck D, Malivert L, deChasseval R, Barraud A, Fondaneche M-C, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 122.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, et al. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell. 2008;31:631–40. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deshpande RA, Wilson TE. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 2007;6:1507–16. doi: 10.1016/j.dnarep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, Villartay JPd. Cernunnos interacts with the XRCC4/DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor NEJ1. J. Biol. Chem. 2006;281:13857–60. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 125.Hsu HL, Yannone SM, Chen DJ. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair (Amst) 2002;1:225–35. doi: 10.1016/s1568-7864(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 126.Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J Biol Chem. 2004;279:37282–90. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 127.Leber R, Wise TW, Mizuta R, Meek K. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 1998;273:1794–801. doi: 10.1074/jbc.273.3.1794. [DOI] [PubMed] [Google Scholar]
- 128.Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–85. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bernstein NK, Karimi-Busheri F, Rasouli-Nia A, Mani R, Dianov G, et al. Polynucleotide kinase as a potential target for enhancing cytotoxicity by ionizing radiation and topoisomerase I inhibitors. Anticancer Agents Med Chem. 2008;8:358–67. doi: 10.2174/187152008784220311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–70. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 131.Williams RS, Bernstein N, Lee MS, Rakovszky ML, Cui D, et al. Structural basis for phosphorylation-dependent signaling in the DNA-damage response. Biochem Cell Biol. 2005;83:721–7. doi: 10.1139/o05-153. [DOI] [PubMed] [Google Scholar]
- 132.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–6. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 133.Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J Biol Chem. 2008;283:33994–4001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. Embo J. 2007;26:2094–103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol. 2007;27:3793–803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 137.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunolog. Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 138.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 139.Feeney AJ. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J. Immunol. 1992;149:222–9. [PubMed] [Google Scholar]
- 140.Schlissel M, Constantinescu A, Morrow T, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5'-phosphorylated, RAG-dependent, and cell cycle regulated. Genes & Dev. 1993;7:2520–32. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 141.Roth DB, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl. Acad. Sci. 1993;90:10788–92. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc. Natl. Acad. Sci. 1988;85:8588–92. doi: 10.1073/pnas.85.22.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bosma MJ, Carroll AM. The scid mouse mutant: Definition, characterization, and potential uses. Ann. Rev. Immun. 1991;9:323–50. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 144.Blunt T, Finnie NJ, Taccioli GE, Smith GCM, Demengeot J, et al. Defective DNA-Dependent Protein Kinase Activity is Linked to V(D)J Recombination and DNA Repair Defects Associated with the Murine scid Mutation. Cell. 1995;80:813–23. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 145.Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol. 2003;4:442–51. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 146.Shinkura R, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. The influence of transcriptional orientation on endogenous switch region function. Nature Immunol. 2003;4:435–41. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 147.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–42. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 Stimulates V(D)J RAG Complex for Both Nicking and Hairpinning in Trans in Addition to Tethering in Cis: Implications for Translocations. Mol. Cell. 2009;34:535–44. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tsai AG, Engelhart AE, Hatmal MM, Houston SI, Hud NV, et al. Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J Biol Chem. 2008;284:7157–64. doi: 10.1074/jbc.M806866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair. 2003;2:1163–74. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 151.Kurahashi H, Inagaki H, Ohye T, Kogo H, Kato T, Emanuel BS. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–45. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 153.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roberts SA, Ramsden DA. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J Biol Chem. 2007;282:10605–13. doi: 10.1074/jbc.M611125200. [DOI] [PubMed] [Google Scholar]
- 155.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 156.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–44. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Heo K, Kim H, Choi SH, Choi J, Kim K, et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 158.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, et al. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 2001;276:38242–8. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 159.Li B, Comai L. Displacement of DNA-PKcs from DNA ends by the Werner syndrome protein. Nucl. Acids Res. 2002;30:3653–61. doi: 10.1093/nar/gkf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, et al. The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair (Amst) 2008;7:1927–37. doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Williamson EA, Rasila KK, Corwin LK, Wray J, Beck BD, et al. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 2008;36:5822–31. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-PKcs phosphorylation sites of human artemis. J. Biol. Chem. 2005;280:33839–46. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 163.Pannicke U, Ma Y, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease Artemis. EMBO J. 2004;23:1987–97. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 165.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.