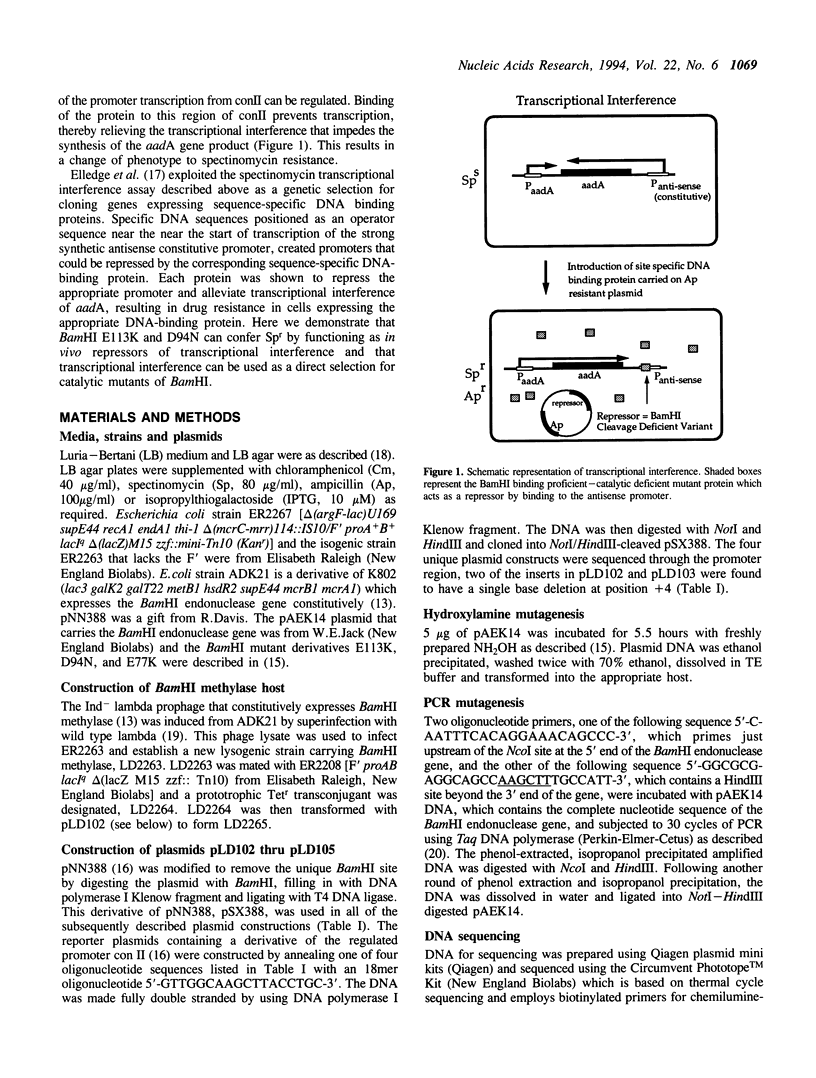
Abstract
Variants of BamHI endonuclease in which the glutamate 113 residue has been changed to lysine or the aspartate 94 to asparagine were shown to behave as repressor molecules in vivo. This was demonstrated by placing a BamHI recognition sequence, GGATCC, positioned as an operator sequence in an antisense promoter for the aadA gene (spectinomycin resistance). Repression of this promoter relieved the inhibition of expression of spectinomycin resistance. This system was then used to select new binding proficient/cleavage deficient BamHI variants. The BamHI endonuclease gene was mutagenized either by exposure to hydroxylamine or by PCR. The mutagenized DNA was reintroduced into E. coli carrying the aadA gene construct, and transformants that conferred spectinomycin resistance were selected. Twenty Spr transformants were sequenced. Thirteen of these were newly isolated variants of the previously identified D94 and E113 residues which are known to be involved in catalysis. The remaining seven variants were all located at residue 111 and the glutamate 111 residue was shown to be involved with catalysis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brooks J. E., Benner J. S., Heiter D. F., Silber K. R., Sznyter L. A., Jager-Quinton T., Moran L. S., Slatko B. E., Wilson G. G., Nwankwo D. O. Cloning the BamHI restriction modification system. Nucleic Acids Res. 1989 Feb 11;17(3):979–997. doi: 10.1093/nar/17.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Nathan P. D., Landry D., Sznyter L. A., Waite-Rees P., Ives C. L., Moran L. S., Slatko B. E., Benner J. S. Characterization of the cloned BamHI restriction modification system: its nucleotide sequence, properties of the methylase, and expression in heterologous hosts. Nucleic Acids Res. 1991 Feb 25;19(4):841–850. doi: 10.1093/nar/19.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 1989 Feb;3(2):185–197. doi: 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Sugiono P., Guarente L., Davis R. W. Genetic selection for genes encoding sequence-specific DNA-binding proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3689–3693. doi: 10.1073/pnas.86.10.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- JACOB F., SUSSMAN R., MONOD J. [On the nature of the repressor ensuring the immunity of lysogenic bacteria]. C R Hebd Seances Acad Sci. 1962 Jun 13;254:4214–4216. [PubMed] [Google Scholar]
- Jack W. E., Greenough L., Dorner L. F., Xu S. Y., Strzelecka T., Aggarwal A. K., Schildkraut I. Overexpression, purification and crystallization of BamHI endonuclease. Nucleic Acids Res. 1991 Apr 25;19(8):1825–1829. doi: 10.1093/nar/19.8.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Pavco P. A., Steege D. A. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990 Jun 15;265(17):9960–9969. [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Wolfes H., Pieper U., Geiger R., Urbanke C., Winkler F. K., Pingoud A. Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry. 1991 Jul 2;30(26):6416–6422. doi: 10.1021/bi00240a011. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. Y., Schildkraut I. Protecting recognition sequences on DNA by a cleavage-deficient restriction endonuclease. Biotechniques. 1993 Aug;15(2):310–315. [PubMed] [Google Scholar]
- Zebala J. A., Choi J., Barany F. Characterization of steady state, single-turnover, and binding kinetics of the TaqI restriction endonuclease. J Biol Chem. 1992 Apr 25;267(12):8097–8105. [PubMed] [Google Scholar]
- Zhou Y. H., Zhang X. P., Ebright R. H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991 Nov 11;19(21):6052–6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]